Metabolomic Biomarkers for Detection, Prognosis and Identifying Recurrence in Endometrial Cancer
Abstract
1. Introduction
2. Metabolomics and EC Biomarker Discovery
Metabolomic Platforms for EC Biomarker Research
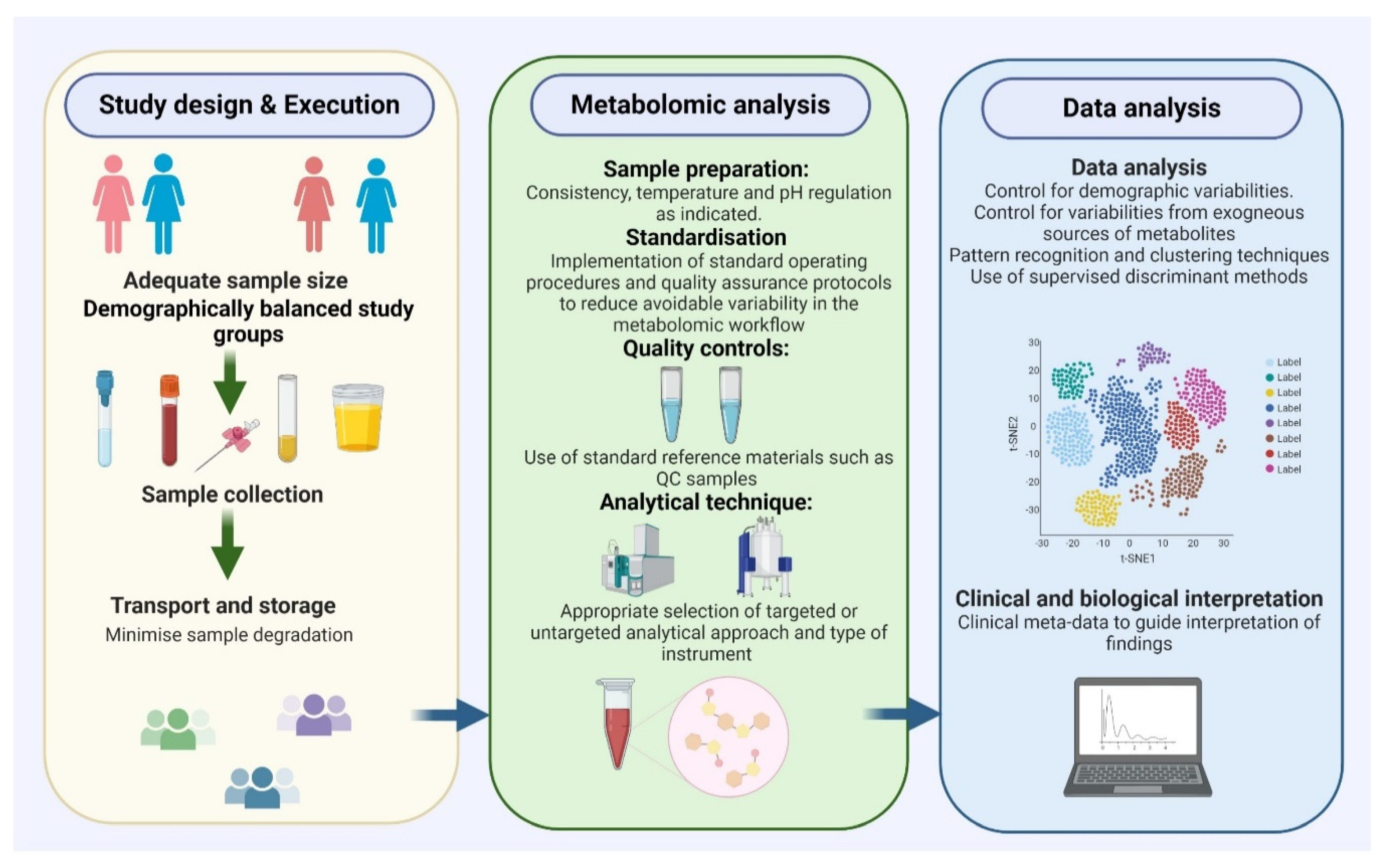
3. Challenges and Important Considerations in Metabolomic Biomarker Research
3.1. Patient Selection
3.2. Consistent and Effective Sample Preparation
3.3. Appropriate Analytical and Statistical Methods
4. Endometrial Cancer Metabolomic Biomarkers in Biofluids
4.1. Blood-Based Metabolite Biomarkers
4.1.1. Blood-Based Diagnostic Metabolomic EC Biomarkers
4.1.2. Blood-Based Predictive and Prognostic Metabolomic EC Biomarkers
| Metabolite | Group/Sub-Class | Potential Clinical Utility | Biochemical Function and Summary of Evidence |
|---|---|---|---|
| 2-oleoylglycerol [71] | Conjugated lipids | Prognosis Monitor disease recurrence | Produced by lipolysis. Have signalling functions. Activate G-protein-coupled GPR119. |
| 3-hydroxybutyrate [77,93] | Fatty acid metabolite | Diagnosis/early detection | Marker of mitochondrial fatty acid beta-oxidation. Synthesised in the liver from acetyl-CoA. Source of energy during low glucose levels. |
| Acylcarnitines [71,72] | Conjugated lipids (Fatty acyls) | Diagnosis Prognosis | Fatty acid transport through the mitochondrial membrane via the carnitine shuttle. Long-chain fatty acids important for tissues and enriched in hypoxic tissues. Role in beta-oxidation. |
| Asparagine [87] | Non-essential amino acid | Diagnosis | Amino donor in urea, pyrimidine and purine synthesis. Supports protein synthesis during glutamine starvation. Also found in CVF fluids. |
| Bile acids [71] | Steroid acids | Diagnosis Prognosis (Recurrence after surgery) | Increase myometrial sensitivity to hormones, have pro-inflammatory properties and modulate cholesterol homeostasis. Act with steroids to promote EC growth, involved in signalling. |
| Bradykinin [71] | Polypeptide | Diagnosis Prognosis (Elevated in Type 1 EC) | Promotes inflammation, a vasodilator. Causes the release of prostacyclin and nitric oxide. Activates phospholipase D. Triggers kinin-activated pathways. |
| Ceramides [71,113] | Lipids | Diagnosis Prognosis (Linked to Type 2 EC recurrence) | Composed of sphingosine and a fatty acid. Involved in cell signalling, differentiation, proliferation and programmed cell death. |
| Cholines/acylcholines [71] | Conjugated lipids | Diagnosis Prognosis (Elevated in Type 2 EC) | Choline is necessary for the production of acetylcholine, a neurotransmitter and S-adenosyl methionine, a methyl donor in homocysteine synthesis. Acylcholines enhance penetration of estradiol in tissues. Also found in tissues and CVF fluids [79]. |
| Cystathionine [71] | Modified amino acid | Diagnosis | Intermediate in the synthesis of cysteine. Product of homocysteine. |
| Estrogen metabolites [99,100,101] | Hormone | Diagnosis Prognosis | Modulates growth of the endometrium by inducing proliferation. |
| Glycine [71] | Amino acid | Prognosis (Elevated in Type 2 EC) | Proteinogenic amino acid. Integral to the formation of alpha-helices in secondary protein structure. Inhibitory neurotransmitter. |
| Heme [71] | Iron-containing porphyrin | Diagnosis Prognosis (Elevated in Type 2 EC) | A viable source of electrons during electron transfer. Modifications in Heme synthesis related pathways such as tetra-hydrofolate serine glycine pathway implicated in EC. |
| Hexadecadienyl carnitine/phosphatidylcholine with diacyl residue C38:1 [98] | Carnitine/choline | Prognosis (LVSI) | Carnitine-phosphatidylcholine ratio shown to be associated with presence/absence of LVSI. |
| Hexadecanoylcarnitine/phosphatidylcholine with acyl-alkyl residue C40:1 [98] | Carnitine/choline | Diagnosis/early detection | Carnitine-phosphatidylcholine ratio with potential for EC detection. |
| Homocysteine [93] | Amino acid | Diagnosis/detection Prognosis | Homologue of cysteine, a product of methionine. Sensitivity of DNA. High levels correlate with increased risk of malignant epithelial tumours. |
| Hydroxypropionyl carnitine [98] | Carnitine | Prognosis (Survival) | Fatty acid transport through the mitochondrial membrane via the carnitine shuttle. Long-chain fatty acids important fuels for tissues. |
| Hydroxysphingomyelins C14:1/hydroxysphingomyelins C24:1 [98] | Sphingomyelins | Prognosis (Myometrial invasion) | Sphingomyelin is involved in signal transduction. Degradation leads to the production of ceramide/ is involved in the apoptotic signalling pathway. |
| Indoleacetic acid [92] | Indoles | Diagnosis/early detection | Involved in cell proliferation/division, migration, invasion and autophagy. |
| Isoleucine [87] | Essential amino acid | Diagnosis | Alpha-amino acid useful in the biosynthesis of proteins. Associated with insulin resistance. Both glucogenic and ketogenic. Also found in CVF fluids [79]. |
| Isovalerate [71] | Fatty acid | Diagnosis /early detection | Salt of isovaleric acid. Also known as 3-methyl butanoate. |
| Lactic acid [93] | Organic acid (Alpha-hydroxy acid) | Diagnosis Prognosis | Synthetic intermediate in metabolic pathways. Produced by pyruvate when the rate of demand for energy is high. Warburg effect. Low pH suppresses T function, promotes angiogenesis. Increases interleukin-8. |
| Linoleic acid [71,72,93] | Essential fatty acid | Diagnosis (Lower levels in EC) Prognosis | Unclear role in tumorigenesis. Promotes growth of mammary tumours in rodent models. |
| Lyso-platelet-activating factor [92] | Phospholipid | Diagnosis/early detection | Induced lipid mediator. Potent phospholipid activator and mediator of inflammation, platelet aggregation and leukocyte functions. Linked to skin cancer. |
| Methionine sulfoxide [111] | Essential amino acid | Prognosis (survival) | Methionine is a precursor for succinyl-CoA, homocysteine, cysteine, creatine and carnitine. Met-SO is an oxidised form of methionine. |
| Monoacylglycerols [71] | Glyceride | Diagnosis Prognosis | Glycerols linked to fatty acid. Act primarily as surfactants. Favour estrogenic environment. |
| Myristic acid [71,93] | Free fatty acid | Diagnosis (Lower levels in EC) Prognosis | Saturated fatty acids are strongly related to cholesterol concentrations. Correlate with rising triglycerides in plasma. |
| Phenylalanine [71,87] | Essential amino acid | Diagnosis/early detection | Precursor for tyrosine, dopamine and norepinephrine. Inhibits proliferation without affecting apoptosis or autophagy. Also found in CVF fluids [79] |
| Phosphatidylcholine with diacyl C42:0/phosphatidylcholine with acyl-alkyl C44:5 [98] | Lipid-like (Choline derivatives) | Diagnosis/early detection | Specific choline derivative ratios shown to predict EC. |
| Phosphatidylcholine with diacyl residue sum C34:4/phosphatidylcholine with acyl-alkyl C38:3 [98] | Lipid-like (Choline derivatives) | Prognosis (LVSI) | Specific choline derivative ratios are associated with presence/ absence of LVSI. |
| Phosphatidylcholine with diacyl residueC40:2/Phosphatidylcholine with diacyl residue C42:6 [98] | Choline derivatives | Prognosis | Specific choline derivative ratios are associated with myometrial invasion. |
| Phosphocholine [92] | Phospholipid | Diagnosis/early detection Prognosis | Plays a role in biosynthesis of cell membranes. Surrogate marker for cell proliferation, inhibition of invasion and migration. Protects against TNF-induced apoptosis. Also found in CVF fluids [79]. |
| Progesterone [93] | Hormone | Diagnosis | Anti-estrogenic effect and associated with estrogen sensitivity of ECs. |
| Proline/tyrosine [98] | Amino acids | Diagnosis/early detection | Involved in the biosynthesis of proteins. |
| Sarcosine [71] | Biogenic amine | Prognosis (Elevated in Type 2 EC) | Intermediate in the metabolism of choline to glycine. |
| Spermine [71] | Biogenic amine | Diagnosis/early detection Prognosis | Likely originating from EC cells. Involved in cellular metabolism. |
| Sphingolipids [71] | Sphingolipids | Diagnosis Prognosis | Fatty acid derivatives of sphingosine which occur in cell membranes, especially of the brain and nervous tissues. Also found in EC tissues [113]. |
| Stearic acid [72,93] | Fatty acid | Diagnosis/early detection | Saturated fatty acid with surfactant properties. In vitro inhibition of cancer cell growth. Downregulated in EC. |
| Sulfated androgens [71] | Sulfated androgens | Diagnosis Prognosis | Sulfated androgens implicated in Type 1 EC. Role in sexual development of males. |
| Tetradecadienoylcarnitine [77] | Carnitine | Diagnosis/early detection | Energy metabolism and fatty acid transport. |
| Threonine [93] | Amino acid | Diagnosis/early detection | Amino acid involved in protein biosynthesis. |
| Valine [93] | Amino acid | Diagnosis | An amino acid used in the biosynthesis of proteins. |
4.2. Tissue-Based Metabolomic Biomarkers
| Metabolite | Group/Sub-Class | Potential Clinical Utility | Biochemical Function and Summary of Evidence |
|---|---|---|---|
| 13Z- Docosenamide [80] | Primary fatty amide | Diagnosis Prognosis | An amide of docosenoic acid. Unclear mechanism relating to EC development and progression. |
| 1-palmitoyl-2-linoleoyl-glycero-3phosphocholine [80] | Diacylglycerol and phospholipid | Diagnosis Prognosis | Component of biological membranes. Involved in membrane-mediated cell signalling. |
| 5,8,11-eicosatrienoic acid [80] | Straight chain fatty acid | Diagnosis Prognosis | Belong to eicosanoids, synthesised from oxidised polyunsaturated fatty acids, mediate cell–cell communication and inflammatory immune response. |
| Arachidonic acid [80] | Polyunsaturated fatty acid | Diagnosis Prognosis | Present in phospholipids of membranes, plays roles in the synthesis of prostaglandins and leukotrienes. |
| Capric acid [118] | Saturated fatty acid | Diagnosis | Downregulated in EC. Role in cell signaling, energy storage, membrane stability. In vitro inhibition of cancer proliferation. |
| Cholines/acylcholines [78,80] | Conjugated lipids | Diagnosis Prognosis (Elevated in Type 2 EC) | Acylcholines enhance penetration of estradiol in tissues. Seen in blood [71] and CVF [79]. |
| Glutamate/arginine/Tryptophan [80] | Amino acids | Diagnosis Prognosis | Bio-active amino acids. Metabolic fuels. Also reported in plasma [87]. |
| Glycerophosphocholines [78,80] | Natural choline | Diagnosis Prognosis | Biosynthetic precursors of acetylcholine. Up to 70% increase in EC tissues. |
| Hypoxanthine [80] | Purine metabolite | Prognosis (myometrial invasion) | Purine derivative, a constituent of nucleic acids present in the anticodon of tRNA. |
| Inosine [78,80] | Purine metabolite | Diagnosis Prognosis | Nucleoside found in tRNAs and essential for translation of the genetic code in wobble base pairs. Imbalance in isoleucine–alanine ratio. |
| Monoacylglycerol [118] | Acylglycerol | Diagnosis | Monoacylglycerol 24:0 significantly downregulated in EC tissues. Modulates cellular processes including proliferation and apoptosis. |
| Oleamide [80] | Fatty acid amide | Diagnosis Prognosis (Increased in grade 3 EC) | Mechanism of action is unclear. Modulator of neurotransmitter and voltage-gated ion channel activity. |
| Palmitic amide [80] | Amide | Diagnosis Prognosis | Primary fatty acid amide. |
| Phosphatidic acid [80,115] | Phospholipids | Diagnosis Prognosis | Anionic phospholipids important in cell signalling and activation of lipid-gated ion channels. |
| Phosphatidylethanolamines [80] | Phospholipids | Diagnosis Prognosis | Phospholipids found in biological membranes. Involved in membrane fusion and cytokinesis/cell division. Regulate membrane curvature. |
| Phosphatidylglycerol [80] | Phospholipids | Diagnosis Prognosis | Glycerophospholipid and pulmonary surfactant. Activates lipid-gated ion channels. |
| Phosphatidylinositols [80] | Phospholipids | Diagnosis Prognosis | Acidic phospholipids involved in lipid signalling, cell signalling and membrane trafficking. |
| Phosphatidylserine [80] | Phospholipids | Diagnosis Prognosis | Role in cell signalling, especially in brain cells. |
| Picolinic acid [80] | Pyridine derivative | Diagnosis Prognosis | Catabolite of tryptophan through the kynurenine pathway. Unclear function. Possible immunological and anti-proliferative/ anti-tumoral effects. |
| Sphingolipids [113] | Sphingolipid | Diagnosis Prognosis | Fatty acid derivatives of sphingosine. Also reported in blood [71]. |
| Stearamide [80] | Endocannabinoid | Diagnosis Prognosis | Endocannabinoids regulate cell proliferation, differentiation and cell survival. |
| Taurine [80] | Amino sulfonic acid | Prognosis (Type 1 EC) | Amino sulfonic acids, naturally occurring, found in muscles, brain, eyes and heart. Decreased in high-grade EC. |
| UDP-N-acetyl-d–galactosamine [80] | Hexosamine | Diagnosis Prognosis | Linked to the metabolism of glucose, fatty acids, and amino acids. |
| Vaccenic acid [80] | Fatty acid | Diagnosis Prognosis | Trans fatty acid which in mammals is converted into rumenic acid, where it shows anti-carcinogenic properties. |
| Xanthine [80] | Purine metabolite | Prognosis (Myometrial invasion) | Product of purine degradation, created from guanine by the actions of guanine deaminase. |
4.3. Urine Based Metabolomics Biomarkers
4.4. EC Detection in Minimally Invasive Genital Samples
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the Global Cancer Incidence and Mortality in 2018: GLOBOCAN Sources and Methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Crosbie, E.; Morrison, J. The Emerging Epidemic of Endometrial Cancer: Time to Take Action. Cochrane Database Syst. Rev. 2014, 12, ED000095. [Google Scholar]
- CRUK. Uterine Cancer Incidence Statistics. 2020. Available online: www.Cancerresearchuk.Org (accessed on 1 June 2020).
- Sundar, S.; Balega, J.; Crosbie, E.; Drake, A.; Edmondson, R.; Fotopoulou, C.; Gallos, I.; Ganesan, R.; Gupta, J.; Johnson, N. BGCS Uterine Cancer Guidelines: Recommendations for Practice. Eur. J. Obstet. Gynecol. Reprod. Biol. 2017, 213, 71–97. [Google Scholar] [CrossRef] [PubMed]
- WHO. Obesity and Overweight. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 18 June 2020).
- Hales, C.M.; Carroll, M.D.; Fryar, C.D.; Ogden, C.L. Prevalence of Obesity and Severe Obesity among Adults: United States, 2017–2018. NCHS Data Brief No 360, February 2020. Available online: https://www.cdc.gov/nchs/products/databriefs/db360.htm (accessed on 18 June 2020).
- Gynaecologists. Royal College of Obstetricians and Gynaecologists (RCOG) Scientific Impact Paper No. 32 on Endometrial Cancer in Obese Women. London, UK, 2012. Available online: https://www.rcog.org.uk/globalassets/documents/guidelines/scientific-impact-papers/sip_32.pdf (accessed on 18 June 2020).
- Colombo, N.; Creutzberg, C.; Amant, F.; Bosse, T.; González-Martín, A.; Ledermann, J.; Marth, C.; Nout, R.; Querleu, D.; Mirza, M.R. ESMO-ESGO-ESTRO Consensus Conference on Endometrial Cancer: Diagnosis, Treatment and Follow-Up. Int. J. Gynecol. Cancer 2016, 26, 2–30. [Google Scholar] [CrossRef] [PubMed]
- Vandecaveye, V. Advances in Endometrial Cancer Diagnosis. In Management of Endometrial Cancer; Springer: Berlin/Heidelberg, Germany, 2020; pp. 49–58. [Google Scholar]
- Casadio, P.; Magnarelli, G.; Alletto, A.; Guasina, F.; Morra, C.; Talamo, M.R.; La Rosa, M.; Su, H.; Frisoni, J.; Seracchioli, R. Endometrial Cancer. In Atlas of Hysteroscopy; Springer: Berlin/Heidelberg, Germany, 2020; pp. 125–152. [Google Scholar]
- Clark, T.J.; Voit, D.; Gupta, J.K.; Hyde, C.; Song, F.; Khan, K.S. Accuracy of Hysteroscopy in the Diagnosis of Endometrial Cancer and Hyperplasia: A Systematic Quantitative Review. JAMA 2002, 288, 1610–1621. [Google Scholar] [CrossRef] [PubMed]
- Njoku, K.; Chiasserini, D.; Whetton, A.D.; Crosbie, E.J. Proteomic Biomarkers for the Detection of Endometrial Cancer. Cancers 2019, 11, 1572. [Google Scholar] [CrossRef]
- Rižner, T.L. Discovery of Biomarkers for Endometrial Cancer: Current Status and Prospects. Expert Rev. Mol. Diagn. 2016, 16, 1315–1336. [Google Scholar] [CrossRef]
- Badrick, E.; Cresswell, K.; Ellis, P.; Crosbie, P.; Hall, P.S.; O’Flynn, H.; Martin, R.; Leighton, J.; Brown, L.; Makin, D. Top Ten Research Priorities for Detecting Cancer Early. Lancet Public Health 2019, 4, e551. [Google Scholar] [CrossRef]
- Bokhman, J.V. Two Pathogenetic Types of Endometrial Carcinoma. Gynecol. Oncol. 1983, 15, 10–17. [Google Scholar] [CrossRef]
- Ceppi, L.; Dizon, D.S.; Birrer, M.J. Endometrial Cancer Genetic Classification and Its Clinical Application. In Management of Endometrial Cancer; Springer: Berlin/Heidelberg, Germany, 2020; pp. 23–47. [Google Scholar]
- Levine, D.A.; Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Endometrial Carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Yang, W.; Lum, A.; Senz, J.; Boyd, N.; Pike, J.; Anglesio, M.; Kwon, J.S. Confirmation of ProMisE: A Simple, Genomics-based Clinical Classifier for Endometrial Cancer. Cancer 2017, 123, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Stelloo, E.; Bosse, T.; Nout, R.A.; MacKay, H.J.; Church, D.N.; Nijman, H.W.; Leary, A.; Edmondson, R.J.; Powell, M.E.; Crosbie, E.J. Refining Prognosis and Identifying Targetable Pathways for High-Risk Endometrial Cancer; a TransPORTEC Initiative. Mod. Pathol. 2015, 28, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Yen, T.-T.; Wang, T.-L.; Fader, A.N.; Shih, I.-M.; Gaillard, S. Molecular Classification and Emerging Targeted Therapy in Endometrial Cancer. Int. J. Gynecol. Pathol. 2020, 39, 26–35. [Google Scholar] [CrossRef]
- Vermij, L.; Smit, V.; Nout, R.; Bosse, T. Incorporation of Molecular Characteristics into Endometrial Cancer Management. Histopathology 2020, 76, 52–63. [Google Scholar] [CrossRef]
- Njoku, K.; Abiola, J.; Russell, J.; Crosbie, E.J. Endometrial Cancer Prevention in High-Risk Women. Best Pract. Res. Clin. Obstet. Gynaecol. 2020, 65, 66–78. [Google Scholar] [CrossRef]
- Wan, Y.L.; Beverley-Stevenson, R.; Carlisle, D.; Clarke, S.; Edmondson, R.J.; Glover, S.; Holland, J.; Hughes, C.; Kitchener, H.C.; Kitson, S. Working Together to Shape the Endometrial Cancer Research Agenda: The Top Ten Unanswered Research Questions. Gynecol. Oncol. 2016, 143, 287–293. [Google Scholar] [CrossRef]
- Hardiman, G. An Introduction to Systems Analytics and Integration of Big Omics Data. Genes 2020, 11, 245. [Google Scholar] [CrossRef]
- Lockhart, D.J.; Winzeler, E.A. Genomics, Gene Expression and DNA Arrays. Nature 2000, 405, 827–836. [Google Scholar] [CrossRef]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-Omics Approaches to Disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef]
- Jacob, M.; Lopata, A.L.; Dasouki, M.; Abdel Rahman, A.M. Metabolomics toward Personalized Medicine. Mass Spectrom. Rev. 2019, 38, 221–238. [Google Scholar] [CrossRef]
- Wishart, D.S.; Mandal, R.; Stanislaus, A.; Ramirez-Gaona, M. Cancer Metabolomics and the Human Metabolome Database. Metabolites 2016, 6, 10. [Google Scholar] [CrossRef]
- Yang, X.; Wang, J. The Role of Metabolic Syndrome in Endometrial Cancer: A Review. Front. Oncol. 2019, 9, 744. [Google Scholar] [CrossRef] [PubMed]
- Altadill, T. Metabolomic Pathway Alterations in Endometrial Cancer. In Proceedings of the AACR Annual Meeting, Washington, DC, USA, 1–5 April 2017. [Google Scholar]
- Crosbie, E.J.; Zwahlen, M.; Kitchener, H.C.; Egger, M.; Renehan, A.G. Body Mass Index, Hormone Replacement Therapy, and Endometrial Cancer Risk: A Meta-Analysis. Cancer Epidemiol. Prev. Biomark. 2010, 19, 3119–3130. [Google Scholar] [CrossRef]
- Perry, R.J.; Shulman, G.I. Mechanistic Links between Obesity, Insulin, and Cancer. Trends Cancer 2020, 6, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Kitson, S.J.; Evans, D.G.; Crosbie, E.J. Identifying High-Risk Women for Endometrial Cancer Prevention Strategies: Proposal of an Endometrial Cancer Risk Prediction Model. Cancer Prev. Res. 2017, 10, 1–13. [Google Scholar] [CrossRef]
- Cai, Y.; Chang, Y.; Liu, Y. Multi-Omics Profiling Reveals Distinct Microenvironment Characterization of Endometrial Cancer. Biomed. Pharmacother. 2019, 118, 109244. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Yang, Y.; Zhang, Y.; Jiang, S.; Li, X.; Wan, J. Identification of Prognostic Immune-Related Genes in the Tumor Microenvironment of Endometrial Cancer. Aging 2020, 12, 3371. [Google Scholar] [CrossRef]
- Giannone, G.; Attademo, L.; Scotto, G.; Genta, S.; Ghisoni, E.; Tuninetti, V.; Aglietta, M.; Pignata, S.; Valabrega, G. Endometrial Cancer Stem Cells: Role, Characterization and Therapeutic Implications. Cancers 2019, 11, 1820. [Google Scholar] [CrossRef]
- Felix, A.; Weissfeld, J.; Edwards, R.; Linkov, F. Future Directions in the Field of Endometrial Cancer Research: The Need to Investigate the Tumor Microenvironment. Eur. J. Gynaecol. Oncol. 2010, 31, 139. [Google Scholar]
- Pinu, F.R.; Beale, D.J.; Paten, A.M.; Kouremenos, K.; Swarup, S.; Schirra, H.J.; Wishart, D. Systems Biology and Multi-Omics Integration: Viewpoints from the Metabolomics Research Community. Metabolites 2019, 9, 76. [Google Scholar] [CrossRef]
- Gomez-Casati, D.F.; Busi, M.V. Molecular Basis of Clinical Metabolomics. In Clinical Molecular Medicine; Elsevier: Amsterdam, The Netherlands; Academic Press: Cambridge, MA, USA, 2020; pp. 47–55. [Google Scholar]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N. HMDB 4.0: The Human Metabolome Database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Kohler, I.; Hankemeier, T.; van der Graaf, P.H.; Knibbe, C.A.J.; van Hasselt, J.G.C. Integrating Clinical Metabolomics-Based Biomarker Discovery and Clinical Pharmacology to Enable Precision Medicine. Eur. J. Pharm. Sci. 2017, 109, S15–S21. [Google Scholar] [CrossRef] [PubMed]
- Armitage, E.G.; Barbas, C. Metabolomics in Cancer Biomarker Discovery: Current Trends and Future Perspectives. J. Pharm. Biomed. Anal. 2014, 87, 1–11. [Google Scholar] [CrossRef]
- Basetti, M. Cancer Metabolism. Metabolites 2017, 7, 41. [Google Scholar] [CrossRef]
- Costas, L.; Frias-Gomez, J.; Guardiola, M.; Benavente, Y.; Pineda, M.; Pavón, M.Á.; Martínez, J.M.; Climent, M.; Barahona, M.; Canet, J. New Perspectives on Screening and Early Detection of Endometrial Cancer. Int. J. Cancer 2019, 145, 3194–3206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Wu, X.; Wang, X. Urine Metabolomics. Clin. Chim. Acta 2012, 414, 65–69. [Google Scholar] [CrossRef]
- Paraskevaidi, M.; Morais, C.L.M.; Lima, K.M.G.; Ashton, K.M.; Stringfellow, H.F.; Martin-Hirsch, P.L.; Martin, F.L. Potential of Mid-Infrared Spectroscopy as a Non-Invasive Diagnostic Test in Urine for Endometrial or Ovarian Cancer. Analyst 2018, 143, 3156–3163. [Google Scholar] [CrossRef]
- Bingol, K. Recent Advances in Targeted and Untargeted Metabolomics by NMR and MS/NMR Methods. High-Throughput 2018, 7, 9. [Google Scholar] [CrossRef]
- Tolstikov, V.; Moser, A.J.; Sarangarajan, R.; Narain, N.R.; Kiebish, M.A. Current Status of Metabolomic Biomarker Discovery: Impact of Study Design and Demographic Characteristics. Metabolites 2020, 10, 224. [Google Scholar] [CrossRef]
- Le, A.; Mak, J.; Cowan, T.M. Metabolic Profiling by Reversed-Phase/Ion-Exchange Mass Spectrometry. J. Chromatogr. B 2020, 1143, 122072. [Google Scholar] [CrossRef] [PubMed]
- Damiani, C.; Gaglio, D.; Sacco, E.; Alberghina, L.; Vanoni, M. Systems Metabolomics: From Metabolomic Snapshots to Design Principles. Curr. Opin. Biotechnol. 2020, 63, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Bracewell-Milnes, T.; Saso, S.; Abdalla, H.; Nikolau, D.; Norman-Taylor, J.; Johnson, M.; Holmes, E.; Thum, M.-Y. Metabolomics as a Tool to Identify Biomarkers to Predict and Improve Outcomes in Reproductive Medicine: A Systematic Review. Hum. Reprod. Update 2017, 23, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Banach, P.; Suchy, W.; Dereziński, P.; Matysiak, J.; Kokot, Z.J.; Nowak-Markwitz, E. Mass Spectrometry as a Tool for Biomarkers Searching in Gynecological Oncology. Biomed. Pharmacother. 2017, 92, 836–842. [Google Scholar] [CrossRef] [PubMed]
- Djukovic, D.; Raftery, D.; Gowda, N. Mass Spectrometry and NMR Spectroscopy Based Quantitative Metabolomics. In Proteomic and Metabolomic Approaches to Biomarker Discovery; Elsevier: Amsterdam, The Netherlands; Academic Press: Cambridge, MA, USA, 2020; pp. 289–311. [Google Scholar]
- Theodoridis, G.A.; Gika, H.G.; Plumb, R.; Wilson, I.D. Liquid Chromatographic Methods Combined with Mass Spectrometry in Metabolomics. In Proteomic and Metabolomic Approaches to Biomarker Discovery; Elsevier: Amsterdam, The Netherlands; Academic Press: Cambridge, MA, USA, 2020; pp. 149–169. [Google Scholar]
- López-Gonzálvez, Á.; Godzien, J.; García, A.; Barbas, C. Capillary Electrophoresis Mass Spectrometry as a Tool for Untargeted Metabolomics. In High-Throughput Metabolomics; Springer: Berlin/Heidelberg, Germany, 2019; pp. 55–77. [Google Scholar]
- Wishart, D.S. NMR Metabolomics: A Look Ahead. J. Magn. Reson. 2019, 306, 155–161. [Google Scholar] [CrossRef]
- Cameron, J.M.; Bruno, C.; Parachalil, D.R.; Baker, M.J.; Bonnier, F.; Butler, H.J.; Byrne, H.J. Vibrational Spectroscopic Analysis and Quantification of Proteins in Human Blood Plasma and Serum. In Vibrational Spectroscopy in Protein Research; Elsevier: Amsterdam, The Netherlands, 2020; pp. 269–314. [Google Scholar]
- Perez, E.A. Biomarkers and Precision Medicine in Oncology Practice and Clinical Trials. In Advancing the Science of Cancer in Latinos; Springer: Berlin/Heidelberg, Germany, 2020; pp. 113–123. [Google Scholar]
- Chang, J.Y.H.; Ladame, S. Diagnostic, Prognostic, and Predictive Biomarkers for Cancer. In Bioengineering Innovative Solutions for Cancer; Elsevier: Amsterdam, The Netherlands, 2020; pp. 3–21. [Google Scholar]
- Page, M.J. Confounding and Other Concerns in Meta-Epidemiological Studies of Bias. J. Clin. Epidemiol. 2020, 123, 133–134. [Google Scholar] [CrossRef]
- McKeigue, P. Sample Size Requirements for Learning to Classify with High-Dimensional Biomarker Panels. Stat. Methods Med. Res. 2019, 28, 904–910. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Lin, W.; Broadhurst, D.; Begley, P.; Brown, M.; Zelená, E.; Vaughan, A.A.; Halsall, A.; Harding, N.; Knowles, J.; et al. Molecular phenotyping of a UK population: Defining the human serum metabolome. Metabolomics 2015, 11, 9–26. [Google Scholar] [CrossRef] [PubMed]
- González-Domínguez, R.; González-Domínguez, Á.; Sayago, A.; Fernández-Recamales, Á. Recommendations and Best Practices for Standardizing the Pre-Analytical Processing of Blood and Urine Samples in Metabolomics. Metabolites 2020, 10, 229. [Google Scholar] [CrossRef]
- Lucas-Torres, C.; Bernard, T.; Huber, G.; Berthault, P.; Nishiyama, Y.; Kandiyal, P.S.; Elena-Herrmann, B.; Molin, L.; Solari, F.; Bouzier-Sore, A.-K. General Guidelines for Sample Preparation Strategies in HR-ΜMAS NMR-Based Metabolomics of Microscopic Specimens. Metabolites 2020, 10, 54. [Google Scholar] [CrossRef]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of Bioactive Metabolites Using Activity Metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef]
- Dinges, S.S.; Hohm, A.; Vandergrift, L.A.; Nowak, J.; Habbel, P.; Kaltashov, I.A.; Cheng, L.L. Cancer Metabolomic Markers in Urine: Evidence, Techniques and Recommendations. Nat. Rev. Urol. 2019, 16, 339–362. [Google Scholar] [CrossRef] [PubMed]
- Jolliffe, I.T.; Cadima, J. Principal Component Analysis: A Review and Recent Developments. Philos. Trans. R. Soc. Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Lee, L.C.; Liong, C.-Y.; Jemain, A.A. Partial Least Squares-Discriminant Analysis (PLS-DA) for Classification of High-Dimensional (HD) Data: A Review of Contemporary Practice Strategies and Knowledge Gaps. Analyst 2018, 143, 3526–3539. [Google Scholar] [CrossRef] [PubMed]
- Ueda, Y.; Enomoto, T.; Kimura, T.; Miyatake, T.; Yoshino, K.; Fujita, M.; Kimura, T. Serum Biomarkers for Early Detection of Gynecologic Cancers. Cancers 2010, 2, 1312–1327. [Google Scholar] [CrossRef] [PubMed]
- Audet-Delage, Y.; Villeneuve, L.; Grégoire, J.; Plante, M.; Guillemette, C. Identification of Metabolomic Biomarkers for Endometrial Cancer and Its Recurrence after Surgery in Postmenopausal Women. Front. Endocrinol. 2018, 9, 87. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, M.M.; Falk, R.T.; Stevens, R.D.; Gunter, M.J.; Bain, J.R.; Pfeiffer, R.M.; Potischman, N.; Lissowska, J.; Peplonska, B.; Brinton, L.A. Analysis of Serum Metabolic Profiles in Women with Endometrial Cancer and Controls in a Population-Based Case-Control Study. J. Clin. Endocrinol. Metab. 2012, 97, 3216–3223. [Google Scholar] [CrossRef] [PubMed]
- Strimbu, K.; Tavel, J.A. What Are Biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463. [Google Scholar] [CrossRef] [PubMed]
- Engerud, H.R. Molecular Markers to Predict Prognosis and Guide Therapy in Endometrial Cancer. Ph.D. Thesis, University of Bergen, Bergen, Norway, 2020. [Google Scholar]
- Gentry-Maharaj, A.; Karpinskyj, C. Current and Future Approaches to Screening for Endometrial Cancer. Best Pract. Res. Clin. Obstet. Gynaecol. 2020, 65, 79–97. [Google Scholar] [CrossRef]
- Raffone, A.; Troisi, J.; Boccia, D.; Travaglino, A.; Capuano, G.; Insabato, L.; Mollo, A.; Guida, M.; Zullo, F. Metabolomics in Endometrial Cancer Diagnosis: A Systematic Review. Acta Obstet. Gynecol. Scand. 2020. [Google Scholar] [CrossRef]
- Bahado-Singh, R.O.; Lugade, A.; Field, J.; Al-Wahab, Z.; Han, B.; Mandal, R.; Bjorndahl, T.C.; Turkoglu, O.; Graham, S.F.; Wishart, D. Metabolomic Prediction of Endometrial Cancer. Metabolomics 2018, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Trousil, S.; Lee, P.; Pinato, D.J.; Ellis, J.K.; Dina, R.; Aboagye, E.O.; Keun, H.C.; Sharma, R. Alterations of Choline Phospholipid Metabolism in Endometrial Cancer Are Caused by Choline Kinase Alpha Overexpression and a Hyperactivated Deacylation Pathway. Cancer Res. 2014, 74, 6867–6877. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.-C.; Chen, K.; Chiu, C.-Y.; Lu, K.-Y.; Lu, H.-Y.; Chiang, M.-H.; Tsai, C.-K.; Lo, C.-J.; Cheng, M.-L.; Chang, T.-C. Metabolomic Biomarkers in Cervicovaginal Fluid for Detecting Endometrial Cancer through Nuclear Magnetic Resonance Spectroscopy. Metabolomics 2019, 15, 146. [Google Scholar] [CrossRef] [PubMed]
- Altadill, T.; Dowdy, T.M.; Gill, K.; Reques, A.; Menon, S.S.; Moiola, C.P.; Lopez-Gil, C.; Coll, E.; Matias-Guiu, X.; Cabrera, S. Metabolomic and Lipidomic Profiling Identifies the Role of the RNA Editing Pathway in Endometrial Carcinogenesis. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Loftsson, T.; Somogyi, G.; Bodor, N. Effect of Choline Esters and Oleic Acid on the Penetration of Acyclovir, Estradiol, Hydrocortisone, Nitroglycerin, Retinoic Acid and Trifluorothymidine across Hairless Mouse Skin in Vitro. Acta Pharm. Nord. 1989, 1, 279–286. [Google Scholar]
- Chughtai, K.; Jiang, L.; Greenwood, T.R.; Glunde, K.; Heeren, R.M.A. Mass Spectrometry Images Acylcarnitines, Phosphatidylcholines, and Sphingomyelin in MDA-MB-231 Breast Tumor Models. J. Lipid Res. 2013, 54, 333–344. [Google Scholar] [CrossRef]
- Qin, H.; Ruan, Z. The Role of Monoacylglycerol Lipase (MAGL) in the Cancer Progress. Cell Biochem. Biophys. 2014, 70, 33–36. [Google Scholar] [CrossRef]
- Paraskevaidi, M.; Morais, C.L.M.; Ashton, K.M.; Stringfellow, H.F.; McVey, R.J.; Ryan, N.A.J.; O’Flynn, H.; Sivalingam, V.N.; Kitson, S.J.; MacKintosh, M.L. Detecting Endometrial Cancer by Blood Spectroscopy: A Diagnostic Cross-Sectional Study. Cancers 2020, 12, 1256. [Google Scholar] [CrossRef]
- O’Connell, T.M. The Complex Role of Branched Chain Amino Acids in Diabetes and Cancer. Metabolites 2013, 3, 931–945. [Google Scholar] [CrossRef]
- Lieu, E.L.; Nguyen, T.; Rhyne, S.; Kim, J. Amino Acids in Cancer. Exp. Mol. Med. 2020, 1–16. [Google Scholar] [CrossRef]
- Ihata, Y.; Miyagi, E.; Numazaki, R.; Muramatsu, T.; Imaizumi, A.; Yamamoto, H.; Yamakado, M.; Okamoto, N.; Hirahara, F. Amino Acid Profile Index for Early Detection of Endometrial Cancer: Verification as a Novel Diagnostic Marker. Int. J. Clin. Oncol. 2014, 19, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, N. Use of “AminoIndex Technology” for Cancer Screening. Ningen Dock 2012, 26, 911–922. [Google Scholar]
- Mikami, H.; Kimura, O.; Yamamoto, H.; Kikuchi, S.; Nakamura, Y.; Ando, T.; Yamakado, M. A Multicentre Clinical Validation of AminoIndex Cancer Screening (AICS). Sci. Rep. 2019, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Miyagi, E.; Numazaki, R.; Nakanishi, T.; Kataoka, F.; Saruki, N.; Ihata, Y. Diagnostic Performance and Clinical Utility of Novel Gynecologic Cancer Screening Method Based on “AminoIndex Technology”. Ningen Dock 2012, 26, 749–755. [Google Scholar]
- Suzuki, Y.; Tokinaga-Uchiyama, A.; Mizushima, T.; Maruyama, Y.; Mogami, T.; Shikata, N.; Ikeda, A.; Yamamoto, H.; Miyagi, E. Normalization of Abnormal Plasma Amino Acid Profile-Based Indexes in Patients with Gynecological Malignant Tumors after Curative Treatment. BMC Cancer 2018, 18, 973. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Wang, Q.; Su, Y.; Xuan, X.; Liu, Y.; Chen, W.; Qian, Y.; Lash, G.E. Identification and Functional Analyses of Differentially Expressed Metabolites in Early Stage Endometrial Carcinoma. Cancer Sci. 2018, 109, 1032–1043. [Google Scholar] [CrossRef]
- Troisi, J.; Sarno, L.; Landolfi, A.; Scala, G.; Martinelli, P.; Venturella, R.; Di Cello, A.; Zullo, F.; Guida, M. Metabolomic Signature of Endometrial Cancer. J. Proteome Res. 2018, 17, 804–812. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of Tumors in the Body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef]
- Narayanan, S.; Santhoshkumar, A.; Ray, S.; Harihar, S. Reprogramming of Cancer Cell Metabolism: Warburg and Reverse Warburg Hypothesis. In Cancer Cell Metabolism: A Potential Target for Cancer Therapy; Springer: Berlin/Heidelberg, Germany, 2020; pp. 15–26. [Google Scholar]
- Polet, F.; Feron, O. Endothelial Cell Metabolism and Tumour Angiogenesis: Glucose and Glutamine as Essential Fuels and Lactate as the Driving Force. J. Intern. Med. 2013, 273, 156–165. [Google Scholar] [CrossRef]
- Shen, W.; Gao, C.; Cueto, R.; Liu, L.; Fu, H.; Shao, Y.; Yang, W.Y.; Fang, P.; Choi, E.T.; Wu, Q. Homocysteine-Methionine Cycle Is a Metabolic Sensor System Controlling Methylation-Regulated Pathological Signaling. Redox Biol. 2020, 28, 101322. [Google Scholar] [CrossRef]
- Knific, T.; Vouk, K.; Smrkolj, Š.; Prehn, C.; Adamski, J.; Rižner, T.L. Models Including Plasma Levels of Sphingomyelins and Phosphatidylcholines as Diagnostic and Prognostic Biomarkers of Endometrial Cancer. J. Steroid Biochem. Mol. Biol. 2018, 178, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Brinton, L.A.; Trabert, B.; Anderson, G.L.; Falk, R.T.; Felix, A.S.; Fuhrman, B.J.; Gass, M.L.; Kuller, L.H.; Pfeiffer, R.M.; Rohan, T.E. Serum Estrogens and Estrogen Metabolites and Endometrial Cancer Risk among Postmenopausal Women. Cancer Epidemiol. Prev. Biomark. 2016, 25, 1081–1089. [Google Scholar] [CrossRef] [PubMed]
- Audet-Walsh, E.; Lepine, J.; Gregoire, J.; Plante, M.; Caron, P.; Teˆtu, B.; Ayotte, P.; Brisson, J.; Villeneuve, L.; Belanger, A. Profiling of Endogenous Estrogens, Their Precursors, and Metabolites in Endometrial Cancer Patients: Association with Risk and Relationship to Clinical Characteristics. J. Clin. Endocrinol. Metab. 2011, 96, E330–E339. [Google Scholar] [CrossRef] [PubMed]
- Küçük, O.; Churley, M.; Goodman, M.T.; Franke, A.; Custer, L.; Wilkens, L.R.; St Pyrek, J. Increased Plasma Level of Cholesterol-5 Beta, 6 Beta-Epoxide in Endometrial Cancer Patients. Cancer Epidemiol. Prev. Biomark. 1994, 3, 571–574. [Google Scholar]
- Zeleniuch-Jacquotte, A.; Shore, R.E.; Afanasyeva, Y.; Lukanova, A.; Sieri, S.; Koenig, K.L.; Idahl, A.; Krogh, V.; Liu, M.; Ohlson, N. Postmenopausal Circulating Levels of 2-and 16α-Hydroxyestrone and Risk of Endometrial Cancer. Br. J. Cancer 2011, 105, 1458–1464. [Google Scholar] [CrossRef] [PubMed]
- Potischman, N.; Hoover, R.N.; Brinton, L.A.; Siiteri, P.; Dorgan, J.F.; Swanson, C.A.; Berman, M.L.; Mortel, R.; Twiggs, L.B.; Barrett, R.J. Case—Control Study of Endogenous Steroid Hormones and Endometrial Cancer. JNCI J. Natl. Cancer Inst. 1996, 88, 1127–1135. [Google Scholar] [CrossRef]
- Colombo, I.; Lheureux, S.; Oza, A.M. Summary of Management Guidelines for Endometrial Cancer. In Management of Endometrial Cancer; Springer: Berlin/Heidelberg, Germany, 2020; pp. 133–149. [Google Scholar]
- Hofman, Z.L.M. Bradykinin Driven Inflammation. Ph.D. Thesis, Utrecht University, Utrecht, The Netherlands, 2020. [Google Scholar]
- Fiorito, V.; Chiabrando, D.; Petrillo, S.; Bertino, F.; Tolosano, E. The Multifaceted Role of Heme in Cancer. Front. Oncol. 2019, 9, 1540. [Google Scholar] [CrossRef] [PubMed]
- Saddoughi, S.A.; Song, P.; Ogretmen, B. Roles of Bioactive Sphingolipids in Cancer Biology and Therapeutics. In Lipids in Health and Disease; Springer: Berlin/Heidelberg, Germany, 2008; pp. 413–440. [Google Scholar]
- Cheng, M.; Bhujwalla, Z.M.; Glunde, K. Targeting Phospholipid Metabolism in Cancer. Front. Oncol. 2016, 6, 266. [Google Scholar] [CrossRef]
- Lakhani, N.J.; Sarkar, M.A.; Venitz, J.; Figg, W.D. 2-Methoxyestradiol, a Promising Anticancer Agent. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2003, 23, 165–172. [Google Scholar] [CrossRef]
- Li, L.; Heldin, N.-E.; Grawé, J.; Ulmsten, U.; Fu, X. Induction of Apoptosis or Necrosis in Human Endometrial Carcinoma Cells by 2-Methoxyestradiol. Anticancer Res. 2004, 24, 3983–3990. [Google Scholar]
- Strand, E.; Tangen, I.L.; Fasmer, K.E.; Jacob, H.; Halle, M.K.; Hoivik, E.A.; Delvoux, B.; Trovik, J.; Haldorsen, I.S.; Romano, A. Blood Metabolites Associate with Prognosis in Endometrial Cancer. Metabolites 2019, 9, 302. [Google Scholar] [CrossRef] [PubMed]
- Martínez, Y.; Li, X.; Liu, G.; Bin, P.; Yan, W.; Más, D.; Valdivié, M.; Hu, C.-A.A.; Ren, W.; Yin, Y. The Role of Methionine on Metabolism, Oxidative Stress, and Diseases. Amino Acids 2017, 49, 2091–2098. [Google Scholar] [CrossRef] [PubMed]
- Knapp, P.; Baranowski, M.; Knapp, M.; Zabielski, P.; Błachnio-Zabielska, A.U.; Górski, J. Altered Sphingolipid Metabolism in Human Endometrial Cancer. Prostaglandins Other Lipid Mediat. 2010, 92, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Jové, M.; Gatius, S.; Yeramian, A.; Portero-Otin, M.; Eritja, N.; Santacana, M.; Colas, E.; Ruiz, M.; Pamplona, R.; Matias-Guiu, X. Metabotyping Human Endometrioid Endometrial Adenocarcinoma Reveals an Implication of Endocannabinoid Metabolism. Oncotarget 2016, 7, 52364. [Google Scholar] [CrossRef] [PubMed]
- Eritja, N.; Jové, M.; Fasmer, K.E.; Gatius, S.; Portero-Otin, M.; Trovik, J.; Krakstad, C.; Sol, J.; Pamplona, R.; Haldorsen, I.S. Tumour-Microenvironmental Blood Flow Determines a Metabolomic Signature Identifying Lysophospholipids and Resolvin D as Biomarkers in Endometrial Cancer Patients. Oncotarget 2017, 8, 109018. [Google Scholar] [CrossRef] [PubMed]
- Rolin, J.; Maghazachi, A.A. Effects of Lysophospholipids on Tumor Microenvironment. Cancer Microenviron. 2011, 4, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Connor, K.M.; SanGiovanni, J.P.; Lofqvist, C.; Aderman, C.M.; Chen, J.; Higuchi, A.; Hong, S.; Pravda, E.A.; Majchrzak, S.; Carper, D. Increased Dietary Intake of ω-3-Polyunsaturated Fatty Acids Reduces Pathological Retinal Angiogenesis. Nat. Med. 2007, 13, 868–873. [Google Scholar] [CrossRef] [PubMed]
- Shafiee, M.N.; Ortori, C.A.; Barrett, D.A.; Mongan, N.P.; Abu, J.; Atiomo, W. Lipidomic Biomarkers in Polycystic Ovary Syndrome and Endometrial Cancer. Int. J. Mol. Sci. 2020, 21, 4753. [Google Scholar] [CrossRef]
- Njoku, K.; Chaiserini, D.; Jones, E.; Barr, C.; O’Flynn, H.; Whetton, A.; Crosbie, E. Urinary Biomarkers and Their Potential for the Non-Invasive detection of Endometrial Cancer. Front. Oncol. 2020. [Google Scholar]
- Shao, X.; Wang, K.; Liu, X.; Gu, C.; Zhang, P.; Xie, J.; Liu, W.; Sun, L.; Chen, T.; Li, Y. Screening and Verifying Endometrial Carcinoma Diagnostic Biomarkers Based on a Urine Metabolomic Profiling Study Using UPLC-Q-TOF/MS. Clin. Chim. Acta 2016, 463, 200–206. [Google Scholar] [CrossRef]
- Zhao, H.; Jiang, Y.; Liu, Y.; Yun, C.; Li, L. Endogenous Estrogen Metabolites as Biomarkers for Endometrial Cancer via a Novel Method of Liquid Chromatography-Mass Spectrometry with Hollow Fiber Liquid-Phase Microextraction. Horm. Metab. Res. 2015, 47, 158–164. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Roux, A.; Thévenot, E.A.; Seguin, F.; Olivier, M.-F.; Junot, C. Impact of Collection Conditions on the Metabolite Content of Human Urine Samples as Analyzed by Liquid Chromatography Coupled to Mass Spectrometry and Nuclear Magnetic Resonance Spectroscopy. Metabolomics 2015, 11, 1095–1105. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zegels, G.; Van Raemdonck, G.A.A.; Tjalma, W.A.A.; Van Ostade, X.W.M. Use of Cervicovaginal Fluid for the Identification of Biomarkers for Pathologies of the Female Genital Tract. Proteome Sci. 2010, 8, 63. [Google Scholar] [CrossRef] [PubMed]
- Njoku, K.; Crosbie, E.J. Does the Vaginal Microbiome Drive Cervical Carcinogenesis? BJOG An Int. J. Obstet. Gynaecol. 2020, 127, 181. [Google Scholar] [CrossRef] [PubMed]
- Ytre-Hauge, S.; Husby, J.A.; Magnussen, I.J.; Werner, H.M.J.; Salvesen, Ø.O.; Bjørge, L.; Trovik, J.; Stefansson, I.M.; Salvesen, H.B.; Haldorsen, I.S. Preoperative Tumor Size at MRI Predicts Deep Myometrial Invasion, Lymph Node Metastases, and Patient Outcome in Endometrial Carcinomas. Int. J. Gynecol. Cancer 2015, 25, 459–466. [Google Scholar] [CrossRef] [PubMed]

| Metabolite | Group/Sub-Class | Potential Clinical Utility | Biochemical Function and Summary of Evidence |
|---|---|---|---|
| Acetylcysteine [120] | Amino acid metabolite | Diagnosis | Precursor of the anti-oxidant glutathione. Able to reduce free radicals. Found to be downregulated in EC. |
| Estrogens [121] | Hormones | Diagnosis | Female sex hormones, endometrial proliferation. 4-hydroxyestrone found to be elevated in EC. 2-methoxyestrone and 2-methoxyestradiol were downregulated in EC. |
| Isobutyrylglycine [120] | Acyl glycine | Diagnosis | Minor metabolite of fatty acids and known urinary metabolite. A conjugate acid of N-isobutyrylglycinate. Found to be upregulated in EC. |
| N-acetylserine [120] | Amino acid | Diagnosis | Acetylation of the serine amino acid N-terminal. Found to be upregulated in EC. |
| Porphobilinogen [120] | Amine | Diagnosis | Pyrrole intermediate in the synthesis of porphyrin. Found to be downregulated in EC. |
| Urocanic acid [120] | Deamination product | Diagnosis | Breakdown product of histidine. Found to be upregulated in EC. |
| Metabolite | Group/Sub-Class | Potential Clinical Utility | Biochemical Function and Summary of Evidence |
|---|---|---|---|
| Fumarate [79] | Organic acid (Dicarboxylate) | Diagnosis/early detection | Intermediate in the citric acid cycle. Converted to malate. Citric cycle releases stored energy through the oxidation of acetyl-CoA. |
| Malate [79] | Dicarboxylic acid | Diagnosis/early detection | Intermediate in the citric acid cycle |
| Isoleucine [79] | Essential amino acid | Diagnosis | Alpha-amino acid useful in the biosynthesis of proteins. Associated with insulin resistance. Both glucogenic and ketogenic. Reported in serum [87]. |
| Asparagine [79] | Non-essential amino acid | Diagnosis/early detection | Amino donor in urea, pyrimidine and purine synthesis. Supports protein synthesis during glutamine starvation. Reported in serum [87]. |
| Aspartate [79] | Non-essential amino acid | Diagnosis | Involved in protein synthesis and neurotransmission. |
| Cholines/acylcholines [79] | Conjugated lipids | Diagnosis Prognosis (elevated in Type 2 EC) | Necessary for homocysteine synthesis. Acylcholines enhance penetration of estradiol in tissues. Reported in tissue/serum [78,80]. |
| Phenylalanine [79] | Essential amino acid | Diagnosis Early detection | Precursor for tyrosine, dopamine and norepinephrine. Inhibits proliferation without affecting apoptosis or autophagy. Also reported in plasma [71,92]. |
| Phosphocholine [79,92] | Phospholipid | Diagnosis Prognosis (high-grade EC) | Plays a role in biosynthesis of cell membranes. Surrogate marker for cell proliferation, inhibition of invasion and migration. Protects against TNF-induced apoptosis. Elevated in CVF of EC patients. Also seen in plasma [92]. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Njoku, K.; Sutton, C.J.J.; Whetton, A.D.; Crosbie, E.J. Metabolomic Biomarkers for Detection, Prognosis and Identifying Recurrence in Endometrial Cancer. Metabolites 2020, 10, 314. https://doi.org/10.3390/metabo10080314
Njoku K, Sutton CJJ, Whetton AD, Crosbie EJ. Metabolomic Biomarkers for Detection, Prognosis and Identifying Recurrence in Endometrial Cancer. Metabolites. 2020; 10(8):314. https://doi.org/10.3390/metabo10080314
Chicago/Turabian StyleNjoku, Kelechi, Caroline J.J Sutton, Anthony D. Whetton, and Emma J. Crosbie. 2020. "Metabolomic Biomarkers for Detection, Prognosis and Identifying Recurrence in Endometrial Cancer" Metabolites 10, no. 8: 314. https://doi.org/10.3390/metabo10080314
APA StyleNjoku, K., Sutton, C. J. J., Whetton, A. D., & Crosbie, E. J. (2020). Metabolomic Biomarkers for Detection, Prognosis and Identifying Recurrence in Endometrial Cancer. Metabolites, 10(8), 314. https://doi.org/10.3390/metabo10080314





