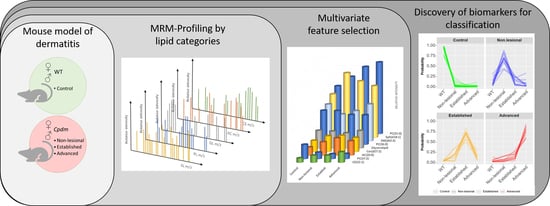
Lipidomic Profiling of the Epidermis in a Mouse Model of Dermatitis Reveals Sexual Dimorphism and Changes in Lipid Composition before the Onset of Clinical Disease
Abstract
:1. Introduction
2. Results
2.1. Association of Sex and Genotype to the Lipid Composition of the Mice Skin
2.1.1. Selection of Predictive Lipid Categories for Sex and Genotype
2.1.2. Individual Lipid Ions Feature Selection for Genotype
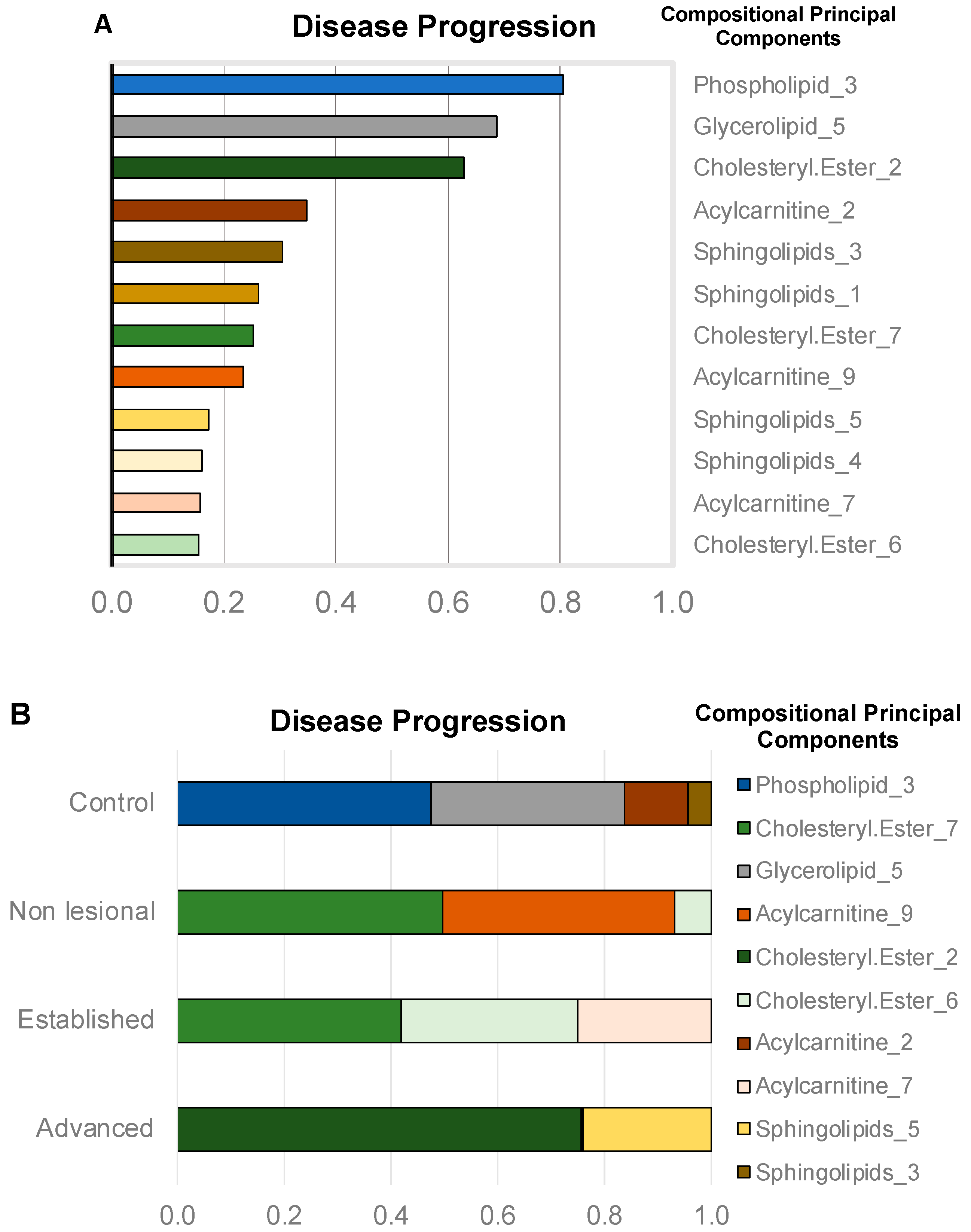
2.2. Selection of Features Associated with Disease Progression
2.2.1. Compositional Principal Component Analysis and Data Visualization
2.2.2. Compressed-Feature Selection for Disease Progression
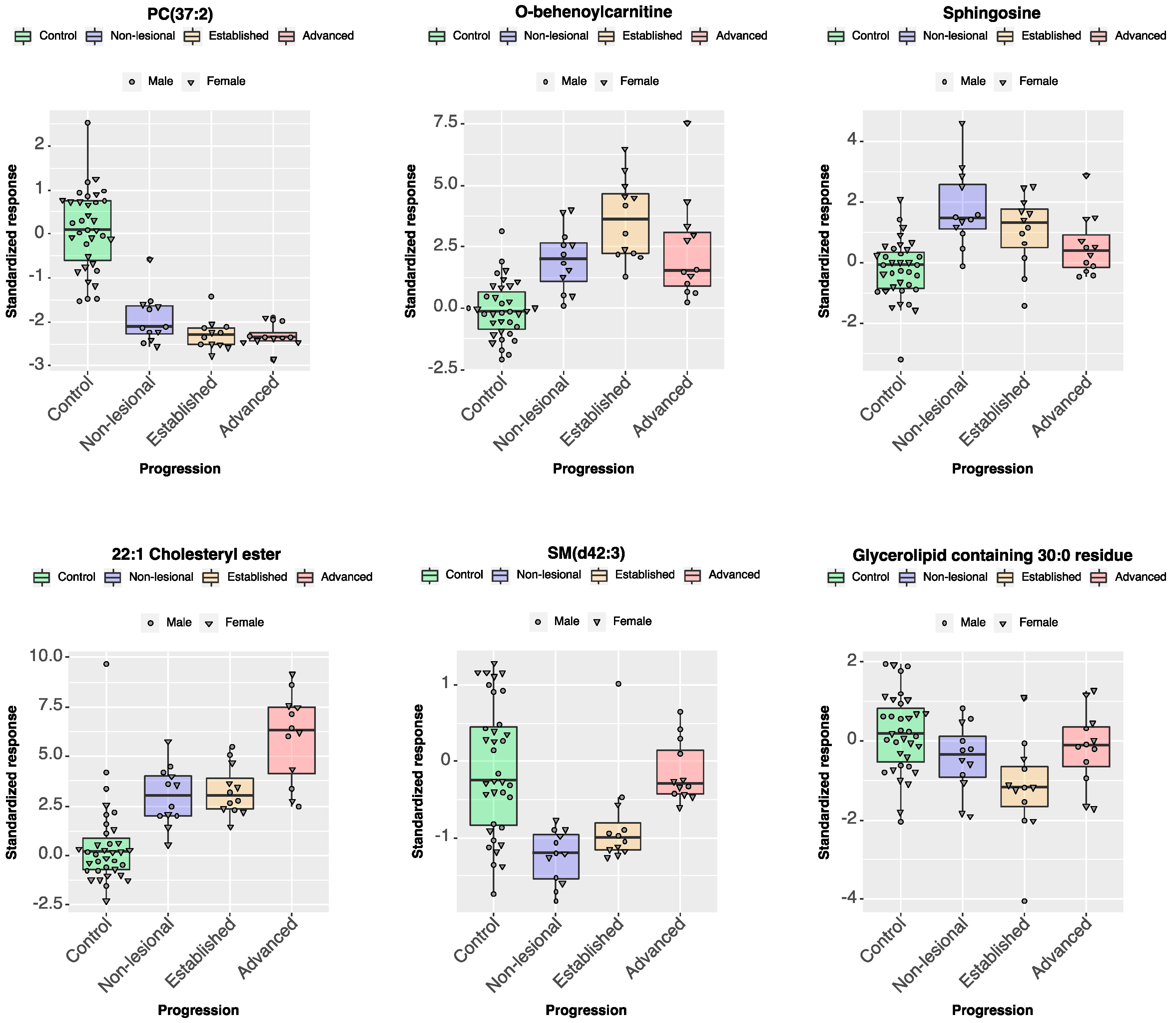
2.2.3. Individual Lipid Ions Feature Selection for Disease Progression
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Epidermis Isolation and Lipid Extraction
4.3. MRM-Profiling Method Development and Sample Screening
4.4. Data Analysis
4.4.1. Selection of Predictive Lipid Categories
4.4.2. Feature Selection of Predictive Individual Lipid Ions
4.4.3. Predictive Elastic Net Regression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Thijs, J.L.; Strickland, I.; Bruijnzeel-Koomen, C.A.; Nierkens, S.; Giovannone, B.; Csomor, E.; Sellman, B.R.; Mustelin, T.; Sleeman, M.A.; De Bruin-Weller, M.; et al. Moving toward endotypes in atopic dermatitis: Identification of patient clusters based on serum biomarker analysis. J. Allergy Clin. Immunol. 2017, 140, 730–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunner, P.M.; Guttman-Yassky, E. Racial differences in atopic dermatitis. Ann. Allergy Asthma Immunol. 2019, 122, 449–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ardern-Jones, M.; Bieber, T. Biomarkers in atopic dermatitis: It is time to stratify. Br. J. Dermatol. 2014, 171, 207–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elias, P.M.; Gruber, R.; Crumrine, D.; Menon, G.; Williams, M.L.; Wakefield, J.S.; Holleran, W.M.; Uchida, Y. Formation and functions of the corneocyte lipid envelope (CLE). Biochim. Biophys. Acta (BBA) Bioenerg. 2013, 1841, 314–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elias, P.M. Epidermal Lipids, Barrier Function, and Desquamation. J. Investig. Dermatol. 1983, 80, S44–S49. [Google Scholar] [CrossRef]
- Malajian, D.; Guttman-Yassky, E. New pathogenic and therapeutic paradigms in atopic dermatitis. Cytokine 2015, 73, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Czarnowicki, T.; He, H.; Krueger, J.G.; Guttman-Yassky, E. Atopic dermatitis endotypes and implications for targeted therapeutics. J. Allergy Clin. Immunol. 2019, 143, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Brunner, P.M.; Guttman-Yassky, E.; Leung, D.Y. The immunology of atopic dermatitis and its reversibility with broad-spectrum and targeted therapies. J. Allergy Clin. Immunol. 2017, 139, S65–S76. [Google Scholar] [CrossRef] [Green Version]
- Kaufman, B.P.; Guttman-Yassky, E.; Alexis, A.F. Atopic dermatitis in diverse racial and ethnic groups-Variations in epidemiology, genetics, clinical presentation and treatment. Exp. Dermatol. 2018, 27, 340–357. [Google Scholar] [CrossRef] [Green Version]
- Ong, P.Y.; Leung, D.Y.M. The Infectious Aspects of Atopic Dermatitis. Immunol. Allergy Clin. N. Am. 2010, 30, 309–321. [Google Scholar] [CrossRef] [Green Version]
- O’Regan, G.M.; Sandilands, A.; McLean, W.I.; Irvine, A.D. Filaggrin in atopic dermatitis. J. Allergy Clin. Immunol. 2008, 122, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, J.; Langan, S.; Williams, H.C. What are the best outcome measurements for atopic eczema? A systematic review. J. Allergy Clin. Immunol. 2007, 120, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Chopra, R.; Vakharia, P.P.; Sacotte, R.; Patel, N.; Immaneni, S.; White, T.; Kantor, R.; Hsu, D.Y.; Silverberg, J.I. Severity strata for Eczema Area and Severity Index (EASI), modified EASI, Scoring Atopic Dermatitis (SCORAD), objective SCORAD, Atopic Dermatitis Severity Index and body surface area in adolescents and adults with atopic dermatitis. Br. J. Dermatol. 2017, 177, 1316–1321. [Google Scholar] [CrossRef] [PubMed]
- Silverberg, J.I.; Gelfand, J.M.; Margolis, D.J.; Fonacier, L.; Boguniewicz, M.; Schwartz, L.B.; Simpson, E.; Grayson, M.H.; Ong, P.Y.; Fuxench, Z.C.C.; et al. Severity strata for POEM, PO-SCORAD, and DLQI in US adults with atopic dermatitis. Ann. Allergy, Asthma Immunol. 2018, 121, 464.e3–468.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Futamura, M.; Leshem, Y.A.; Thomas, K.S.; Nankervis, H.; Williams, H.C.; Simpson, E.; Information, P.E.K.F.C. A systematic review of Investigator Global Assessment (IGA) in atopic dermatitis (AD) trials: Many options, no standards. J. Am. Acad. Dermatol. 2016, 74, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Elias, P.M.; Hatano, Y.; Williams, M.L. Basis for the barrier abnormality in atopic dermatitis: Outside-inside-outside pathogenic mechanisms. J. Allergy Clin. Immunol. 2008, 121, 1337–1343. [Google Scholar] [CrossRef] [Green Version]
- Van Smeden, J.; Janssens, M.; Gooris, G.; Bouwstra, J. The important role of stratum corneum lipids for the cutaneous barrier function. Biochim. Biophys. Acta (BBA) Mol. Cell Boil. Lipids 2014, 1841, 295–313. [Google Scholar] [CrossRef]
- Janssens, M.; Van Smeden, J.; Gooris, G.S.; Bras, W.; Portale, G.; Caspers, P.J.; Vreeken, R.J.; Hankemeier, T.; Kezic, S.; Wolterbeek, R.; et al. Increase in short-chain ceramides correlates with an altered lipid organization and decreased barrier function in atopic eczema patients[S]. J. Lipid Res. 2012, 53, 2755–2766. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, A.; Serizawa, S.; Ito, M.; Sato, Y. Stratum corneum lipid abnormalities in atopic dermatitis. Arch. Dermatol. Res. 1991, 283, 219–223. [Google Scholar] [CrossRef]
- Imokawa, G.; Abe, A.; Jin, K.; Higaki, Y.; Kawashima, M.; Hidano, A. Decreased Level of Ceramides in Stratum Corneum of Atopic Dermatitis: An Etiologic Factor in Atopic Dry Skin? J. Investig. Dermatol. 1991, 96, 523–526. [Google Scholar] [CrossRef] [Green Version]
- Van Smeden, J.; Janssens, M.; Kaye, E.C.J.; Caspers, P.J.; Lavrijsen, A.P.; Vreeken, R.J.; Bouwstra, J. The importance of free fatty acid chain length for the skin barrier function in atopic eczema patients. Exp. Dermatol. 2014, 23, 45–52. [Google Scholar] [CrossRef]
- Agrawal, K.; Hassoun, L.A.; Foolad, N.; Pedersen, T.L.; Sivamani, R.K.; Newman, J.W. Sweat lipid mediator profiling: A noninvasive approach for cutaneous research. J. Lipid Res. 2016, 58, 188–195. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, K.; Hassoun, L.A.; Foolad, N.; Borkowski, K.; Pedersen, T.L.; Sivamani, R.; Newman, J.W. Effects of atopic dermatitis and gender on sebum lipid mediator and fatty acid profiles. Prostaglandins Leukot. Essent. Fat. Acids 2018, 134, 7–16. [Google Scholar] [CrossRef]
- Schäfer, L.; Kragballe, K. Abnormalities in Epidermal Lipid Metabolism in Patients with Atopic Dermatitis. J. Investig. Dermatol. 1991, 96, 10–15. [Google Scholar] [CrossRef] [Green Version]
- Newell, L.; Polak, M.E.; Perera, J.; Owen, C.; Boyd, P.; Pickard, C.; Howarth, P.H.; Healy, E.; Holloway, J.W.; Friedmann, P.S.; et al. Sensitization via Healthy Skin Programs Th2 Responses in Individuals with Atopic Dermatitis. J. Investig. Dermatol. 2013, 133, 2372–2380. [Google Scholar] [CrossRef] [Green Version]
- Elias, P.M.; Schmuth, M. Abnormal skin barrier in the etiopathogenesis of atopic dermatitis. Curr. Opin. Allergy Clin. Immunol. 2009, 9, 437–446. [Google Scholar] [CrossRef] [Green Version]
- Danso, M.O.; Van Drongelen, V.; Mulder, A.A.; Van Esch, J.; Scott, H.; Van Smeden, J.; El Ghalbzouri, A.; Bouwstra, J.A. TNF-α and Th2 Cytokines Induce Atopic Dermatitis–Like Features on Epidermal Differentiation Proteins and Stratum Corneum Lipids in Human Skin Equivalents. J. Investig. Dermatol. 2014, 134, 1941–1950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danso, M.; Boiten, W.; Van Drongelen, V.; Meijling, K.G.; Gooris, G.; El Ghalbzouri, A.; Absalah, S.; Vreeken, R.J.; Kezic, S.; Van Smeden, J.; et al. Altered expression of epidermal lipid bio-synthesis enzymes in atopic dermatitis skin is accompanied by changes in stratum corneum lipid composition. J. Dermatol. Sci. 2017, 88, 57–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- HogenEsch, H.; Dunham, A.; Seymour, R.; Renninger, M.; Sundberg, J.P. Expression of chitinase-like proteins in the skin of chronic proliferative dermatitis (cpdm/cpdm) mice. Exp. Dermatol. 2006, 15, 808–814. [Google Scholar] [CrossRef]
- HogenEsch, H.; Torregrosa, S.E.; Boggess, D.; Sundberg, B.A.; Carroll, J.; Sundberg, J.P. Increased expression of type 2 cytokines in chronic proliferative dermatitis (cpdm) mutant mice and resolution of inflammation following treatment with IL-12. Eur. J. Immunol. 2001, 31, 734–742. [Google Scholar] [CrossRef]
- Franco, J.; Ferreira, C.; Sobreira, T.J.P.; Sundberg, J.P.; HogenEsch, H. Profiling of epidermal lipids in a mouse model of dermatitis: Identification of potential biomarkers. PLoS ONE 2018, 13, e0196595. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, D.J.; Hoeferlin, L.A.; Chalfant, C.E. Lipidomics in translational research and the clinical significance of lipid-based biomarkers. Transl. Res. 2017, 189, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Majd, T.M.; Kalantari, S.; Shahraki, H.R.; Nafar, M.; Almasi, A.; Samavat, S.; Parvin, M.; Hashemian, A. Application of Sparse Linear Discriminant Analysis and Elastic Net for Diagnosis of IgA Nephropathy: Statistical and Biological Viewpoints. Iran. Biomed. J. 2018, 22, 374–384. [Google Scholar] [CrossRef] [Green Version]
- Kirpich, A.; Ainsworth, E.A.; Wedow, J.M.; Newman, J.; Michailidis, G.; McIntyre, L.M. Variable selection in omics data: A practical evaluation of small sample sizes. PLOS ONE 2018, 13, e0197910. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, A.; Langs, G.; Denk, H.; Zatloukal, K.; Müller, H. Causability and explainability of artificial intelligence in medicine. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2019, 9, e1312. [Google Scholar] [CrossRef] [Green Version]
- Basu, A.; Mitra, R.; Liu, H.; Schreiber, S.L.; Clemons, P.A. RWEN: Response-weighted elastic net for prediction of chemosensitivity of cancer cell lines. Bioinformatics 2018, 34, 3332–3339. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, G.B.; De Saeger, S. Elastic net regularized regression for time-series analysis of plasma metabolome stability under sub-optimal freezing condition. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bujak, R.; Daghir-Wojtkowiak, E.; Kaliszan, R.; Markuszewski, M.J. PLS-Based and Regularization-Based Methods for the Selection of Relevant Variables in Non-targeted Metabolomics Data. Front. Mol. Biosci. 2016, 3, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furman, D.; Davis, M.M. New approaches to understanding the immune response to vaccination and infection. Vaccine 2015, 33, 5271–5281. [Google Scholar] [CrossRef] [Green Version]
- Joo, K.-M.; Hwang, J.-H.; Bae, S.; Nahm, D.-H.; Park, H.; Ye, Y.-M.; Lim, K.-M. Relationship of ceramide–, and free fatty acid–cholesterol ratios in the stratum corneum with skin barrier function of normal, atopic dermatitis lesional and non-lesional skins. J. Dermatol. Sci. 2015, 77, 71–74. [Google Scholar] [CrossRef]
- Agrawal, R.; Woodfolk, J.A. Skin barrier defects in atopic dermatitis. Curr. Allergy Asthma Rep. 2014, 14, 433. [Google Scholar] [CrossRef]
- Loiseau, N.; Obata, Y.; Moradian, S.; Sano, H.; Yoshino, S.; Aburai, K.; Takayama, K.; Sakamoto, K.; Holleran, W.M.; Elias, P.M.; et al. Altered sphingoid base profiles predict compromised membrane structure and permeability in atopic dermatitis. J. Dermatol. Sci. 2013, 72, 296–303. [Google Scholar] [CrossRef] [Green Version]
- Cotterill, J.A.; Cunliffe, W.J.; Williamson, B.; Bulusu, L. AGE AND SEX VARIATION IN SKIN SURFACE LIPID COMPOSITION AND SEBUM EXCRETION RATE. Br. J. Dermatol. 1972, 87, 333–340. [Google Scholar] [CrossRef]
- Norheim, F.; Bjellaas, T.; Hui, S.T.; Krishnan, K.C.; Lee, J.; Gupta, S.; Pan, C.; Hasin-Brumshtein, Y.; Parks, B.W.; Li, D.; et al. Genetic, dietary, and sex-specific regulation of hepatic ceramides and the relationship between hepatic ceramides and IR. J. Lipid Res. 2018, 59, 1164–1174. [Google Scholar] [CrossRef] [Green Version]
- Blaess, M.; Deigner, H.-P. Derailed Ceramide Metabolism in Atopic Dermatitis (AD): A Causal Starting Point for a Personalized (Basic) Therapy. Int. J. Mol. Sci. 2019, 20, 3967. [Google Scholar] [CrossRef] [Green Version]
- Rabionet, M.; Gorgas, K.; Sandhoff, R. Ceramide synthesis in the epidermis. Biochim. Biophys. Acta (BBA) Mol. Cell Boil. Lipids 2014, 1841, 422–434. [Google Scholar] [CrossRef]
- Borodzicz, S.; Rudnicka, L.; Mirowska-Guzel, D.; Cudnoch-Jędrzejewska, A. The role of epidermal sphingolipids in dermatologic diseases. Lipids Heal. Dis. 2016, 15, 13. [Google Scholar] [CrossRef] [Green Version]
- Sacotte, R.; Silverberg, J.I. Epidemiology of adult atopic dermatitis. Clin. Dermatol. 2018, 36, 595–605. [Google Scholar] [CrossRef]
- Elias, P.M. Lipid abnormalities and lipid-based repair strategies in atopic dermatitis. Biochim. Biophys. Acta. 2013, 1841, 323–330. [Google Scholar] [CrossRef] [Green Version]
- Elias, P.M.; Wakefield, J.S.; Man, M.-Q. Moisturizers versus Current and Next-Generation Barrier Repair Therapy for the Management of Atopic Dermatitis. Ski. Pharmacol. Physiol. 2018, 32, 1–7. [Google Scholar] [CrossRef]
- Di Nardo, A. Ceramide and cholesterol composition of the skin of patients with atopic dermatitis. Acta Derm. Venereol. 1998, 78, 27–30. [Google Scholar] [CrossRef] [Green Version]
- Berdyshev, E.; Goleva, E.; Bronova, I.A.; Dyjack, N.; Rios, C.; Jung, J.; Taylor, P.; Jeong, M.; Hall, C.F.; Richers, B.N.; et al. Lipid abnormalities in atopic skin are driven by type 2 cytokines. JCI Insight 2018, 3, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Fantini, J.; Yahi, N. Chemical Basis of Lipid Biochemistry. In Brain Lipids in Synaptic Function and Neurological Disease; Elsevier: Amsterdam, The Netherlands, 2015; pp. 1–28. ISBN 978-0-12-800111-0. [Google Scholar]
- Radner, F.; Fischer, J. The important role of epidermal triacylglycerol metabolism for maintenance of the skin permeability barrier function. Biochim. Biophys. Acta (BBA) Mol. Cell Boil. Lipids 2014, 1841, 409–415. [Google Scholar] [CrossRef]
- Han, X. Lipidomics for studying metabolism. Nat. Rev. Endocrinol. 2016, 12, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Piotto, S.; Trapani, A.; Bianchino, E.; Ibarguren, M.; Lopez, D.J.; Busquets, X.; Concilio, S. The effect of hydroxylated fatty acid-containing phospholipids in the remodeling of lipid membranes. Biochim. Biophys. Acta (BBA) Biomembr. 2014, 1838, 1509–1517. [Google Scholar] [CrossRef] [Green Version]
- Ottas, A.; Fishman, D.; Okas, T.-L.; Püssa, T.; Toomik, P.; Märtson, A.; Kingo, K.; Soomets, U. Blood serum metabolome of atopic dermatitis: Altered energy cycle and the markers of systemic inflammation. PLoS ONE 2017, 12, e0188580. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, N.; Suzuki, T.; Yamazaki, Y.; Sugiyama, K.; Koike, S.; Nishimukai, M. Supplemental feeding of phospholipid-enriched alkyl phospholipid from krill relieves spontaneous atopic dermatitis and strengthens skin intercellular lipid barriers in NC/Nga mice. Biosci. Biotechnol. Biochem. 2019, 83, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Mårtensson, C.U.; Doan, K.N.; Becker, T. Effects of lipids on mitochondrial functions. Biochim. Biophys. Acta (BBA) Mol. Cell Boil. Lipids 2017, 1862, 102–113. [Google Scholar] [CrossRef]
- Chegary, M.; Brinke, H.T.; Ruiter, J.P.; Wijburg, F.A.; Stoll, M.S.; Minkler, P.E.; Van Weeghel, M.; Schulz, H.; Hoppel, C.L.; Wanders, R.J.; et al. Mitochondrial long chain fatty acid β-oxidation in man and mouse. Biochim. Biophys. Acta (BBA) Mol. Cell Boil. Lipids 2009, 1791, 806–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutkowsky, J.M.; Knotts, T.A.; Ono-Moore, K.D.; McCoin, C.S.; Huang, S.; Schneider, D.; Singh, S.; Adams, S.H.; Hwang, D.H. Acylcarnitines activate proinflammatory signaling pathways. Am. J. Physiol. Metab. 2014, 306, E1378–E1387. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.-H.; Sviridov, D.; Miller, Y.I. Oxidized cholesteryl esters and inflammation. Biochim. Biophys. Acta (BBA) Mol. Cell Boil. Lipids 2017, 1862, 393–397. [Google Scholar] [CrossRef] [Green Version]
- Korber, M.; Klein, I.; Daum, G. Steryl ester synthesis, storage and hydrolysis: A contribution to sterol homeostasis. Biochim. Biophys. Acta (BBA) Mol. Cell Boil. Lipids 2017, 1862, 1534–1545. [Google Scholar] [CrossRef] [PubMed]
- Tachi, M.; Iwamori, M. Mass spectrometric characterization of cholesterol esters and wax esters in epidermis of fetal, adult and keloidal human skin. Exp. Dermatol. 2008, 17, 318–323. [Google Scholar] [CrossRef]
- Trieb, M.; Wolf, P.; Knuplez, E.; Weger, W.; Schuster, C.; Peinhaupt, M.; Holzer, M.; Trakaki, A.; Eichmann, T.; Lass, A.; et al. Abnormal composition and function of high-density lipoproteins in atopic dermatitis patients. Allergy 2019, 74, 398–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerl, M.J.; Vaz, W.L.C.; Domingues, N.; Klose, C.; Surma, M.A.; Sampaio, J.L.; Almeida, M.S.; Rodrigues, G.; Araújo-Gonçalves, P.; Ferreira, J.; et al. Cholesterol is Inefficiently Converted to Cholesteryl Esters in the Blood of Cardiovascular Disease Patients. Sci. Rep. 2018, 8, 14764. [Google Scholar] [CrossRef] [PubMed]
- Marsella, R.; Olivry, T.; Carlotti, D.-N. For the International Task Force on Canine Atopic Dermatitis Current evidence of skin barrier dysfunction in human and canine atopic dermatitis. Veter Dermatol. 2011, 22, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Tončić, R.J.; Jakasa, I.; Hadžavdić, S.L.; Goorden, S.M.; Der Vlugt, K.J.G.-V.; Stet, F.S.; Balic, A.; Petkovic, M.; Pavicic, B.; Žužul, K.; et al. Altered Levels of Sphingosine, Sphinganine and Their Ceramides in Atopic Dermatitis Are Related to Skin Barrier Function, Disease Severity and Local Cytokine Milieu. Int. J. Mol. Sci. 2020, 21, 1958. [Google Scholar] [CrossRef] [Green Version]
- Blagus, R.; Lusa, L. Class prediction for high-dimensional class-imbalanced data. BMC Bioinform. 2010, 11, 523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, C.R.; Yannell, K.E.; Mollenhauer, B.; Espy, R.D.; Cordeiro, F.B.; Ouyang, Z.; Cooks, R.G. Chemical profiling of cerebrospinal fluid by multiple reaction monitoring mass spectrometry. Anal. 2016, 141, 5252–5255. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, F.; Ferreira, C.R.; Sobreira, T.; Yannell, K.E.; Jarmusch, A.K.; Cedenho, A.P.; Turco, E.G.L.; Cooks, R.G. Multiple reaction monitoring (MRM)-profiling for biomarker discovery applied to human polycystic ovarian syndrome. Rapid Commun. Mass Spectrom. 2017, 31, 1462–1470. [Google Scholar] [CrossRef]
- Xie, Z.; Gonzalez, L.E.; Ferreira, C.R.; Vorsilak, A.; Frabutt, D.; Sobreira, T.J.P.; Pugia, M.; Cooks, R.G. Multiple Reaction Monitoring Profiling (MRM-Profiling) of Lipids To Distinguish Strain-Level Differences in Microbial Resistance in Escherichia coli. Anal. Chem. 2019, 91, 11349–11354. [Google Scholar] [CrossRef]
- Dipali, S.S.; Ferreira, C.R.; Zhou, L.T.; Pritchard, M.T.; Duncan, F.E. Histologic analysis and lipid profiling reveal reproductive age-associated changes in peri-ovarian adipose tissue. Reprod. Boil. Endocrinol. 2019, 17, 46. [Google Scholar] [CrossRef] [Green Version]
- Yannell, K.E.; Ferreira, C.R.; Tichy, S.E.; Cooks, R.G. Multiple reaction monitoring (MRM)-profiling with biomarker identification by LC-QTOF to characterize coronary artery disease. Analyst 2018, 143, 5014–5022. [Google Scholar] [CrossRef]
- Filzmoser, P.; Hron, K.; Reimann, C. Principal component analysis for compositional data with outliers. Environmetrics 2009, 20, 621–632. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B Stat. Methodol. 2005, 67, 301–320. [Google Scholar] [CrossRef] [Green Version]
- Chawla, N.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]




| Category | Tentative Attributions | MRM | Genotype (η2) | Sex (η2) |
|---|---|---|---|---|
| Phospholipid | PC(37:2), PC(O-38:2), PC(P-38:1) | 800.6→184.1 | 0.76 | 0.001 |
| Phospholipid | SM(d41:1) * | 801.6→184.1 | 0.72 | 0.002 |
| Sphingolipid | Cer[AS](d18:1/24:0)2OH | 666.4→264.3 | 0.63 | 0.025 |
| Phospholipid | PC(38:2), PC(P-39:1) | 814.6→184.1 | 0.63 | 0.018 |
| Phospholipid | SM(d36:0) * | 733.6→184.1 | 0.61 | 0.010 |
| Phospholipid | SM(d42:1) * | 815.6→184.1 | 0.60 | 0.001 |
| Phospholipid | PC(38:1), PC(P-38:1), PC(O-38:2) * | 816.6→184.1 | 0.60 | 0.022 |
| Phospholipid | PC (32:1), PC(O-33:1), PC(P-33:0) | 732.1→184.1 | 0.59 | 0.008 |
| Phospholipid | PC(40:8), PCo(40:1) | 830.1→184.1 | 0.57 | 0.014 |
| Category | Tentative Attributions | MRM | Importance Score |
|---|---|---|---|
| Phospholipid | PC(37:2), PC(O-38:2), PC(P-38:1) | 800.6→184.1 | 100.00 |
| Glycerolipid | Glycerolipid containing 22:5 residue | 627.1→280 | 50.58 |
| Sphingolipid | Cer[AS](d18:1/24:0)2OH | 666.4→264.3 | 48.40 |
| Phospholipid | PC(38:1), PC(P-38:1), PC(O-38:2) | 816.6→184.1 | 32.26 |
| Phospholipid | SM(d41:1) * | 801.6→184.1 | 29.57 |
| Phospholipid | PC(38:2), PC(P-39:1) | 814.6→184.1 | 19.86 |
| Glycerolipid | Glycerolipid containing 18:2 residue | 895.1→598 | 3.35 |
| Phospholipid | SM(d37:0) | 745.6→184.1 | 0.61 |
| Category | Tentative Attributions | MRM | Importance Scores | ||||
|---|---|---|---|---|---|---|---|
| Control | Non-Lesional | Established | Advanced | Overall | |||
| Phospholipid | PC(37:2), PC(O-38:2), PC(P-38:1) | 800.6→184.1 | 100.00 | 0.00 | 0.00 | 0.00 | 100.00 |
| Acylcarnitine | O-behenoylcarnitine | 484.4→85.1 | 0.00 | 0.00 | 77.26 | 0.00 | 77.26 |
| Sphingolipid | Sphingosine | 300.2→282.2 | 0.00 | 71.50 | 0.00 | 0.00 | 71.50 |
| Phospholipid | PC(31:0), PC(O-31:1), PC(P-31:0) | 720.4→184.1 | 0.00 | 0.00 | 0.00 | 66.55 | 66.55 |
| Cholesteryl Ester | 22:1 Cholesteryl ester | 725.4→369.1 | 0.00 | 0.00 | 0.00 | 60.59 | 60.59 |
| Sphingolipid | Cer(d27:2) | 438.2→ 266.2 | 0.00 | 0.00 | 44.09 | 0.00 | 44.09 |
| Glycerolipid | Glycerolipids containing 30:0 residue | 624.1→155.1 | 0.00 | 0.00 | 42.86 | 0.00 | 42.86 |
| Phospholipid | SM(d42:3) | 811.6→184.1 | 0.00 | 30.66 | 0.00 | 7.66 | 38.32 |
| Acylcarnitine | Non attributed | 837→85.1 | 0.00 | 36.96 | 0.00 | 0.00 | 36.96 |
| Phospholipid | PC (36:0), PCp(38:6) | 790.4→184.1 | 0.00 | 26.66 | 0.00 | 0.00 | 26.66 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franco, J.; Rajwa, B.; Ferreira, C.R.; Sundberg, J.P.; HogenEsch, H. Lipidomic Profiling of the Epidermis in a Mouse Model of Dermatitis Reveals Sexual Dimorphism and Changes in Lipid Composition before the Onset of Clinical Disease. Metabolites 2020, 10, 299. https://doi.org/10.3390/metabo10070299
Franco J, Rajwa B, Ferreira CR, Sundberg JP, HogenEsch H. Lipidomic Profiling of the Epidermis in a Mouse Model of Dermatitis Reveals Sexual Dimorphism and Changes in Lipid Composition before the Onset of Clinical Disease. Metabolites. 2020; 10(7):299. https://doi.org/10.3390/metabo10070299
Chicago/Turabian StyleFranco, Jackeline, Bartek Rajwa, Christina R. Ferreira, John P. Sundberg, and Harm HogenEsch. 2020. "Lipidomic Profiling of the Epidermis in a Mouse Model of Dermatitis Reveals Sexual Dimorphism and Changes in Lipid Composition before the Onset of Clinical Disease" Metabolites 10, no. 7: 299. https://doi.org/10.3390/metabo10070299
APA StyleFranco, J., Rajwa, B., Ferreira, C. R., Sundberg, J. P., & HogenEsch, H. (2020). Lipidomic Profiling of the Epidermis in a Mouse Model of Dermatitis Reveals Sexual Dimorphism and Changes in Lipid Composition before the Onset of Clinical Disease. Metabolites, 10(7), 299. https://doi.org/10.3390/metabo10070299