Insights into the Natural Defenses of a Coral Reef Fish Against Gill Ectoparasites: Integrated Metabolome and Microbiome Approach
Abstract
1. Introduction
2. Results
2.1. Metabolomic Analysis of Chaetodon Gill Mucus
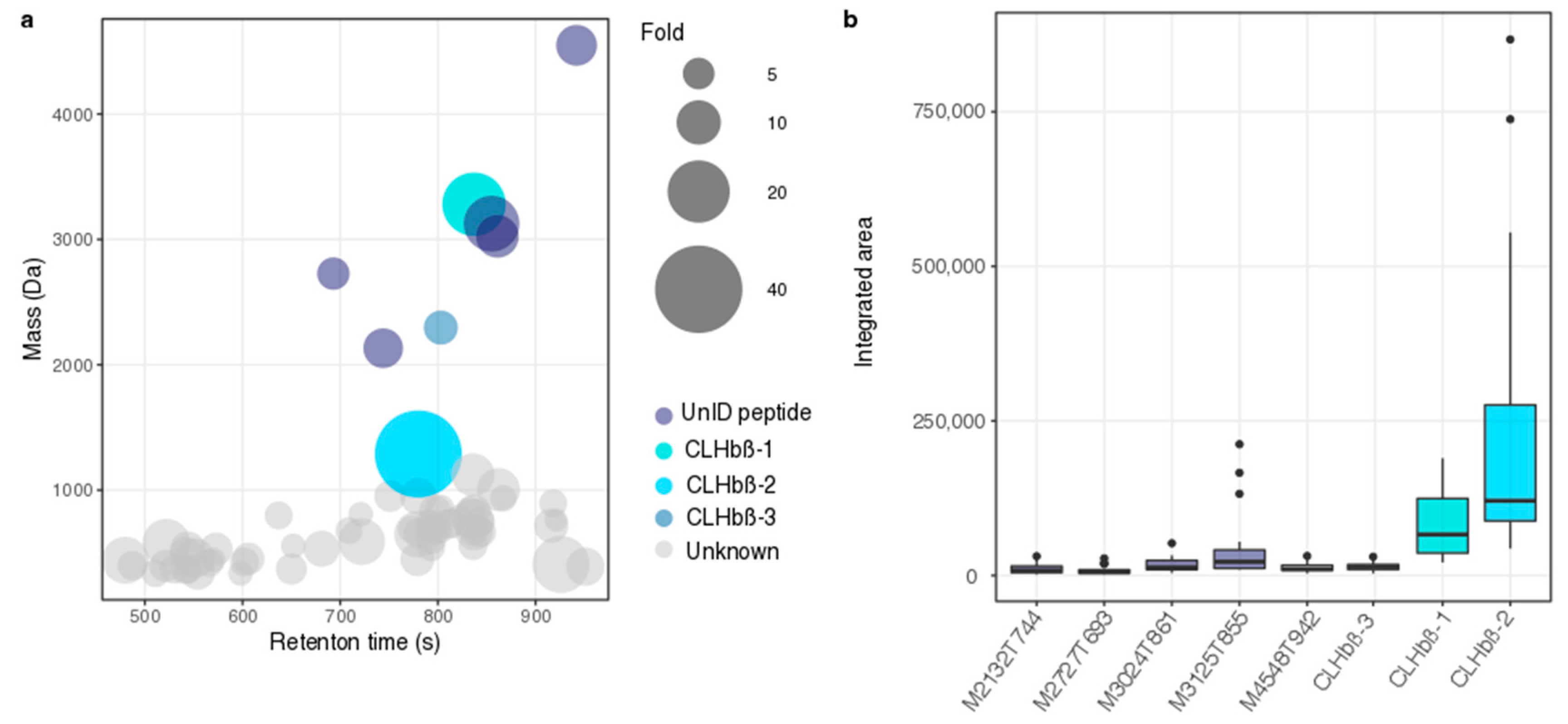
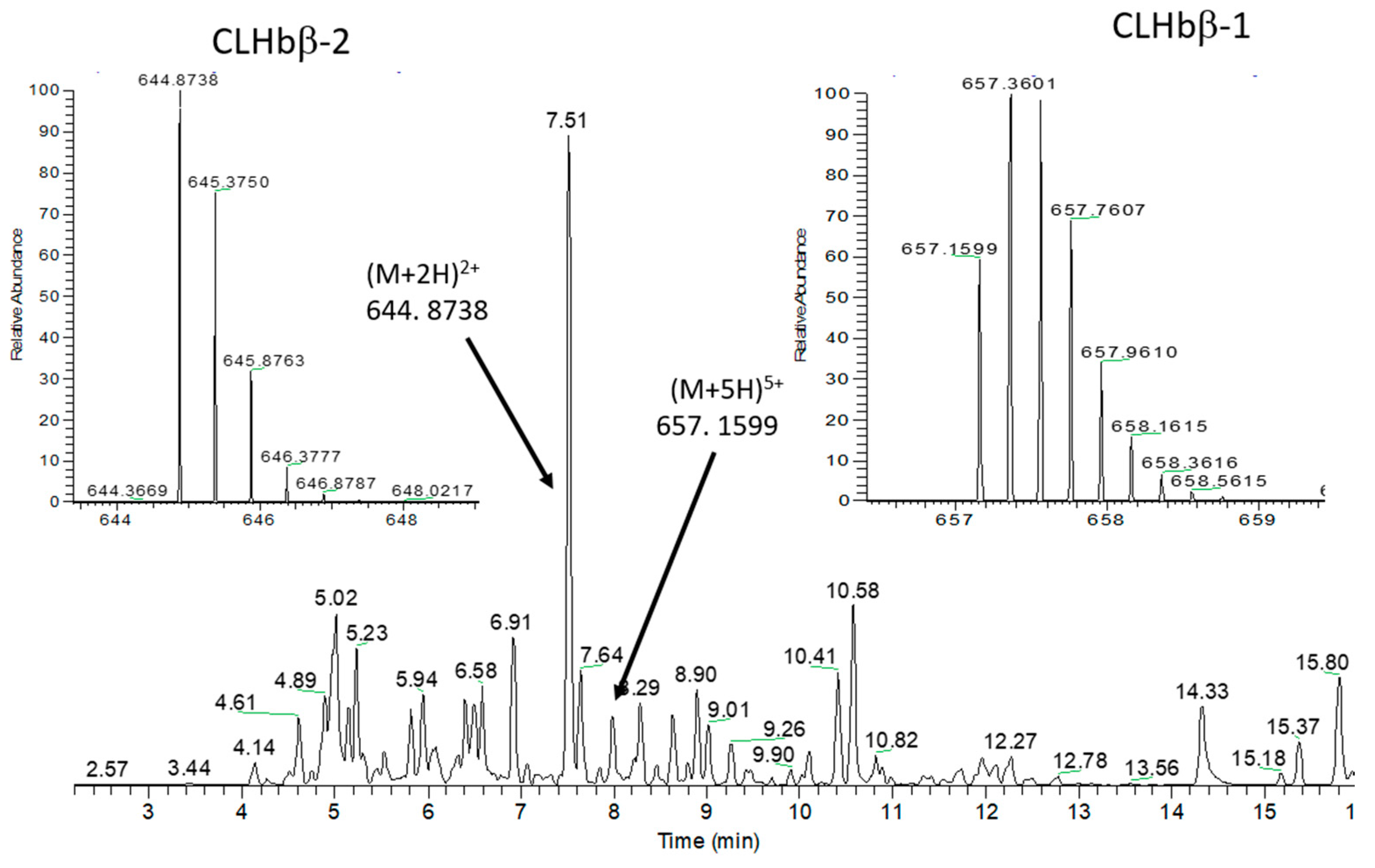
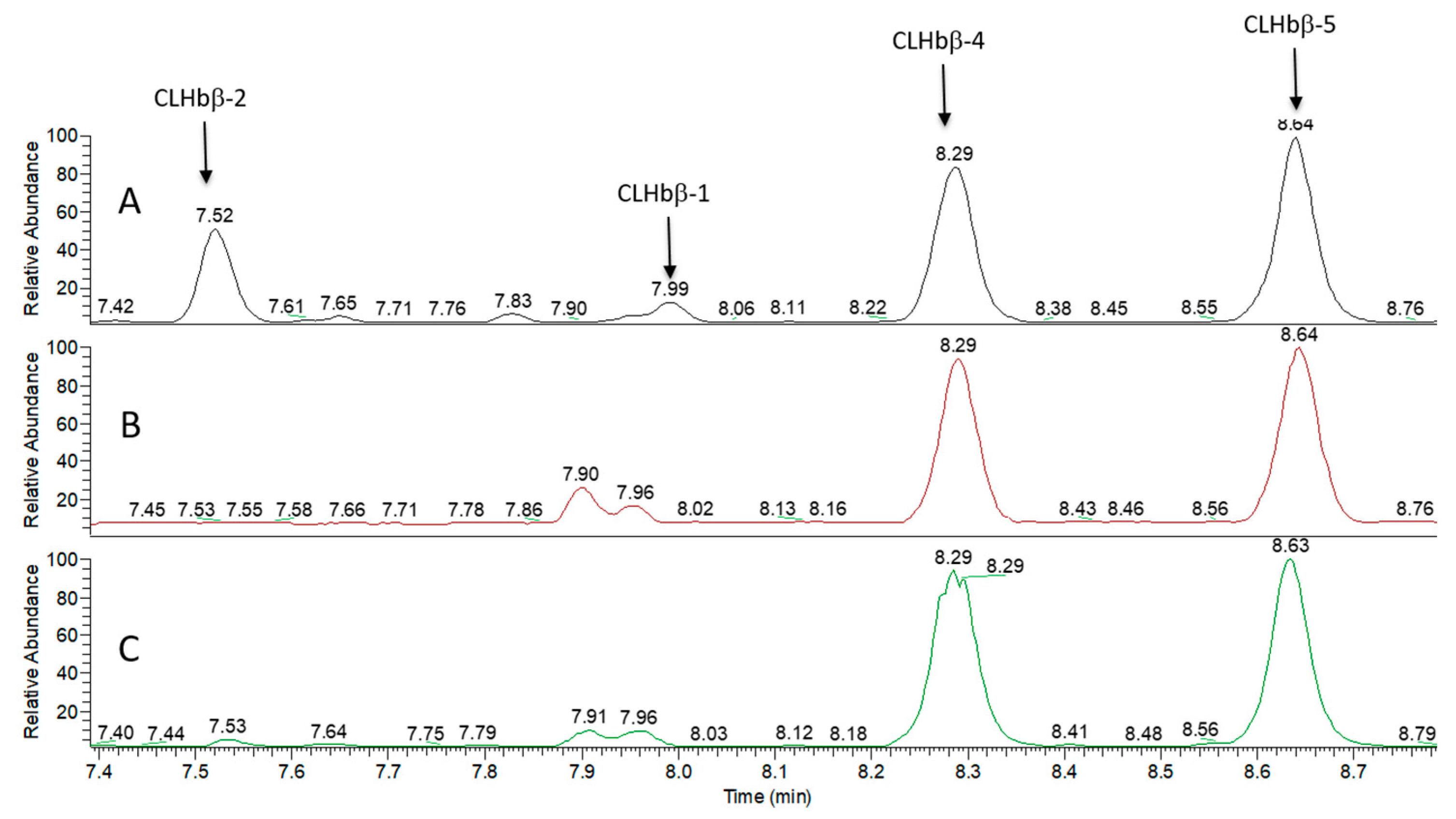
2.2. Peptide Characterisation and Synthesis
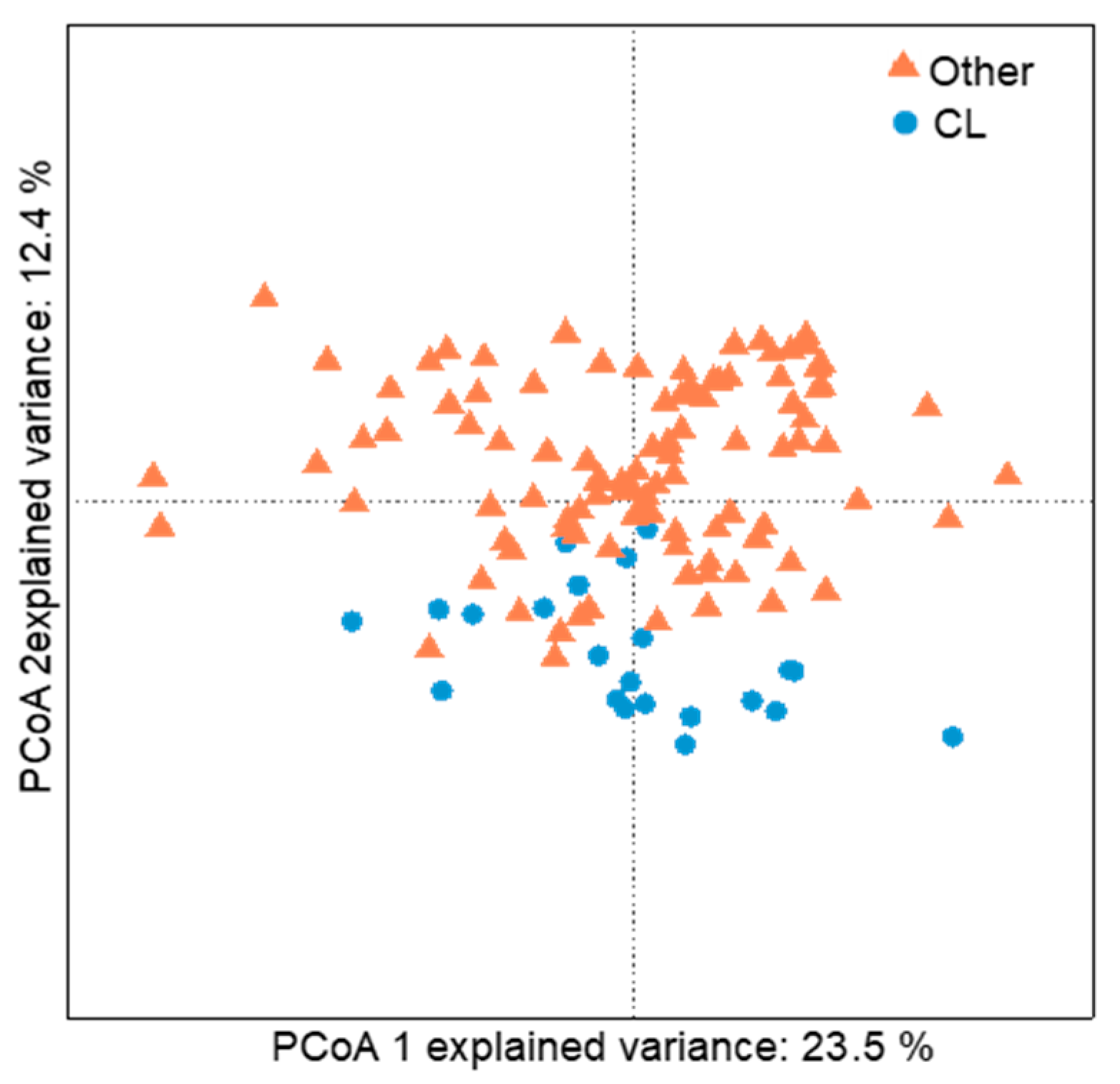
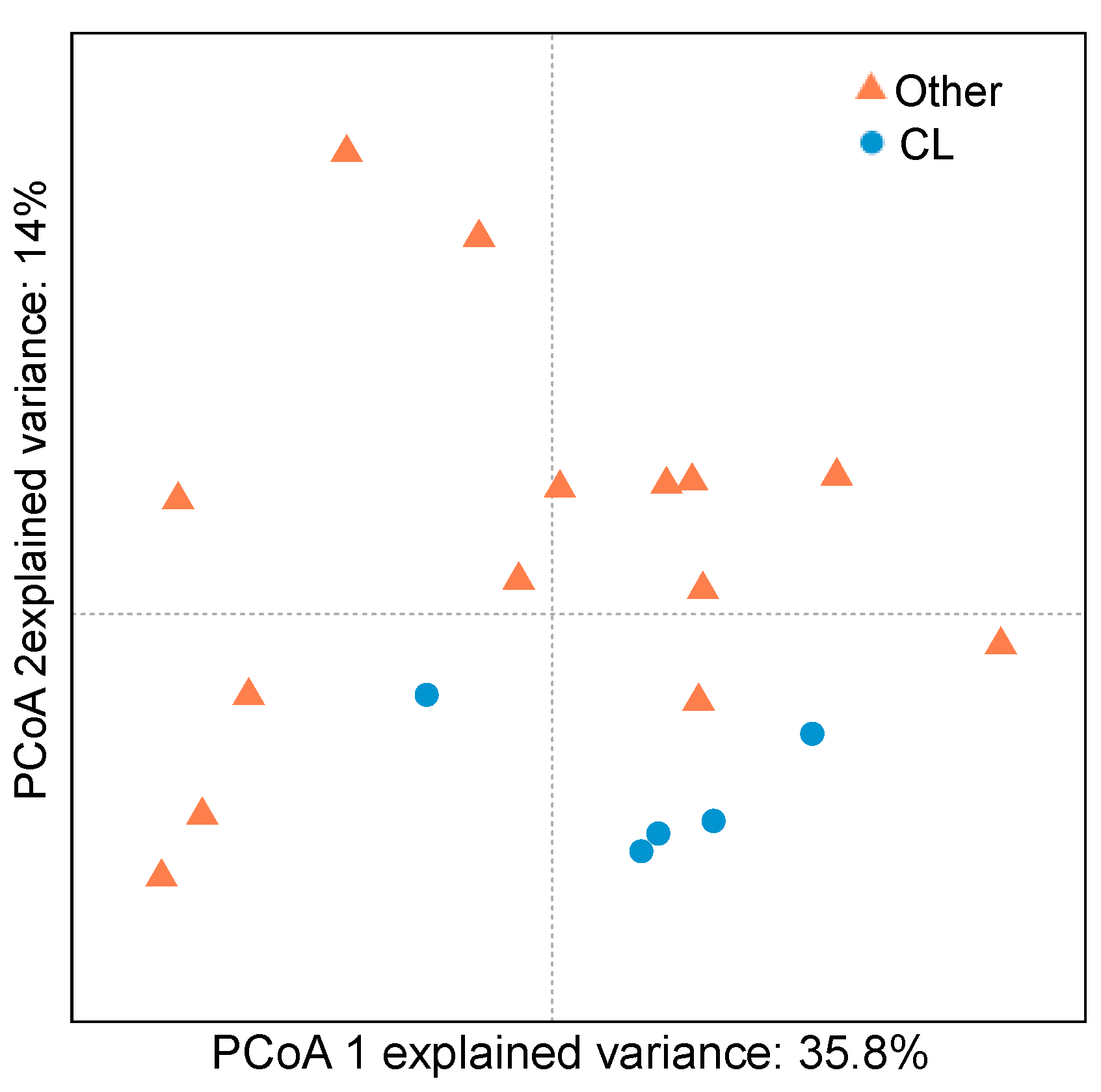
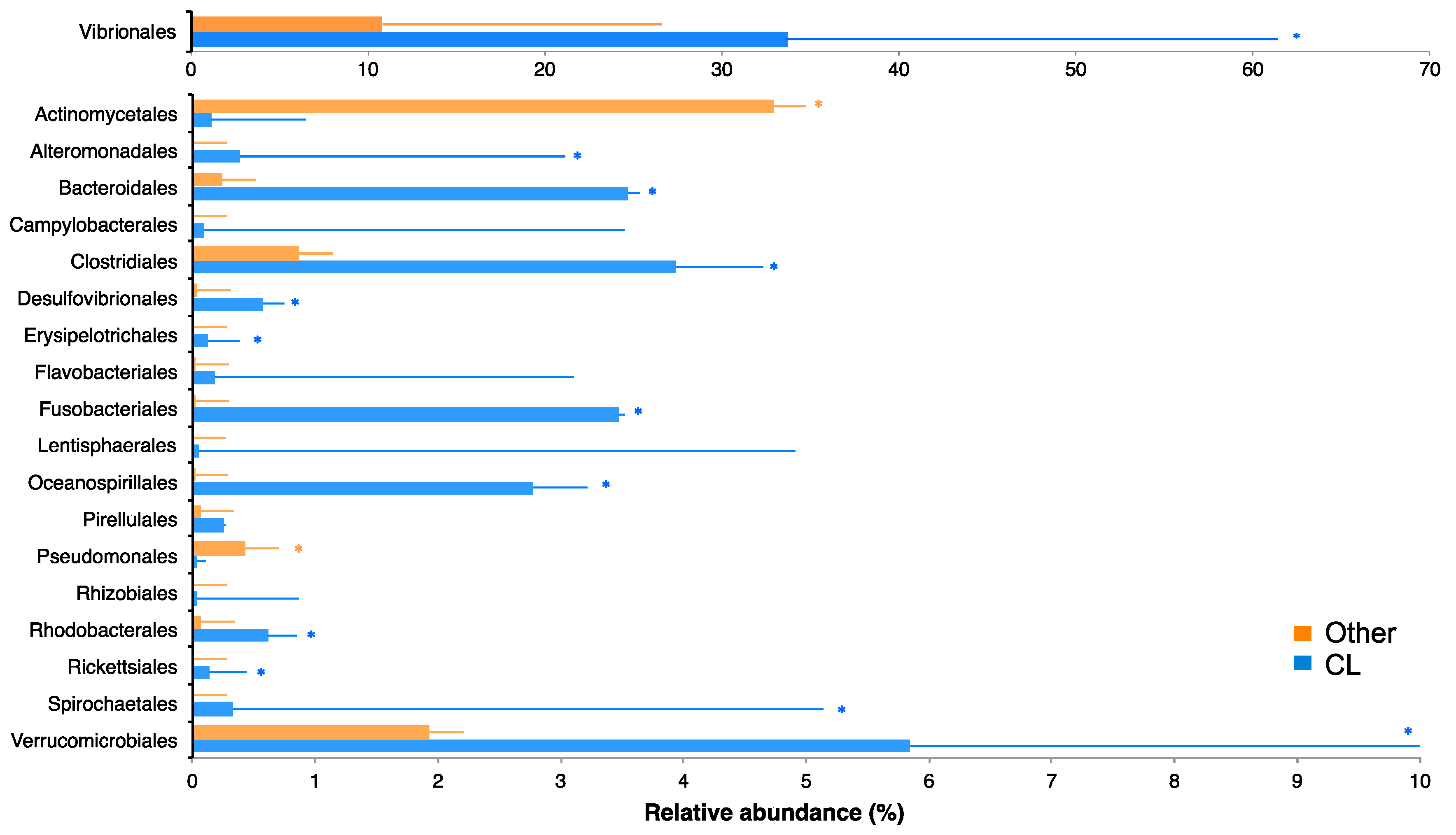
2.3. Bacterial Community of Chaetodon Gill Mucus
2.4. Correlation between VIP Metabolites and VIP OTUs
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Animal Ethics
4.3. Fish Mucus Sampling
4.4. Study of the Gill Mucus Metabolome
4.4.1. Untargeted Metabolomics
4.4.2. Optimized Peptide Extraction and Peptides Purification
4.4.3. Peptide Sequence Analysis
4.4.4. Solid-Phase Synthesis and Peptide Analysis
4.5. Study of the Gill Mucus Associated Bacterial Communities
4.6. Data analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hudson, P.J.; Dobson, A.P.; Lafferty, K.D. Is a healthy ecosystem one that is rich in parasites? Trends Ecol. Evol. 2006, 21, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Fox, N.J.; Marion, G.; Davidson, R.; White, P.C.L.; Hutchings, M. Climate-driven tipping-points could lead to sudden, high-intensity parasite outbreaks. R. Soc. Open Sci. 2015, 2, 140296. [Google Scholar] [CrossRef] [PubMed]
- Cable, J.; Barber, I.; Boag, B.; Ellison, A.R.; Morgan, E.R.; Murray, K.A.; Pascoe, E.L.; Sait, S.M.; Wilson, A.; Booth, M. Global change, parasite transmission and disease control: Lessons from ecology. Philos. Trans. R. Soc. B 2017, 372, 20160088. [Google Scholar] [CrossRef] [PubMed]
- Perry, B.D.; Grace, D.; Sones, K. Current drivers and future directions of global livestock disease dynamics. Proc. Natl. Acad. Sci. USA 2011, 110, 20871–20877. [Google Scholar] [CrossRef]
- Sana, S.; A Hardouin, E.; E Gozlan, R.; Ercan, D.; Tarkan, A.S.; Zhang, T.; Andreou, D. Origin and invasion of the emerging infectious pathogen Sphaerothecum destruens. Emerg. Microbes Infect. 2017, 6, e76–e78. [Google Scholar] [CrossRef]
- Cunningham, A.A.; Dobson, A.P.; Hudson, P.J. Disease invasion: Impacts on biodiversity and human health. Philos. Trans. R. Soc. B 2012, 367, 2804–2806. [Google Scholar] [CrossRef]
- Rakers, S.; Niklasson, L.; Steinhagen, D.; Kruse, C.; Schauber, J.; Sundell, K.; Paus, R. Antimicrobial Peptides (AMPs) from Fish Epidermis: Perspectives for Investigative Dermatology. J. Investig. Dermatol. 2013, 133, 1140–1149. [Google Scholar] [CrossRef]
- Hahn, M.A.; Dheilly, N.M. Experimental Models to Study the Role of Microbes in Host-Parasite Interactions. Front. Microbiol. 2016, 7, 3031. [Google Scholar] [CrossRef]
- Quigley, B.J.Z.; López, D.G.; Buckling, A.; McKane, A.J.; Brown, S. The mode of host–parasite interaction shapes coevolutionary dynamics and the fate of host cooperation. Proc. R. Soc. B 2012, 279, 3742–3748. [Google Scholar] [CrossRef]
- Dheilly, N.M.; Poulin, R.; Thomas, F. Biological warfare: Microorganisms as drivers of host–parasite interactions. Infect. Genet. Evol. 2015, 34, 251–259. [Google Scholar] [CrossRef]
- Cirimotich, C.; Dong, Y.; Clayton, A.M.; Sandiford, S.L.; Souza-Neto, J.A.; Mulenga, M.; Dimopoulos, G. Natural Microbe-Mediated Refractoriness to Plasmodium Infection in Anopheles Gambiae. Science 2011, 332, 855–858. [Google Scholar] [CrossRef] [PubMed]
- Lowrey, L.; Woodhams, D.C.; Tacchi, L.; Salinas, I. Topographical Mapping of the Rainbow Trout (Oncorhynchus mykiss) Microbiome Reveals a Diverse Bacterial Community with Antifungal Properties in the Skin. Appl. Environ. Microbiol. 2015, 81, 6915–6925. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.; Jackson, R.; Olson, J. Bacteria associated with lionfish (Pterois volitans/miles complex) exhibit antibacterial activity against known fish pathogens. Mar. Ecol. Prog. Ser. 2016, 558, 167–180. [Google Scholar] [CrossRef]
- Gómez, G.D.; Balcázar, J.L. A review on the interactions between gut microbiota and innate immunity of fish: Table 1. FEMS Immunol. Med. Microbiol. 2008, 52, 145–154. [Google Scholar] [CrossRef]
- Sepahi, A.; Cordero, H.; Goldfine, H.; Esteban, M. Ángeles; Salinas, I. Symbiont-derived sphingolipids modulate mucosal homeostasis and B cells in teleost fish. Sci. Rep. 2016, 6, 39054. [Google Scholar] [CrossRef] [PubMed]
- Reverter, M.; Tapissier-Bontemps, N.; Lecchini, D.; Banaigs, B.; Sasal, P. Biological and Ecological Roles of External Fish Mucus: A Review. Fishes 2018, 3, 41. [Google Scholar] [CrossRef]
- Hansson, G.C. Role of mucus layers in gut infection and inflammation. Curr. Opin. Microbiol. 2011, 15, 57–62. [Google Scholar] [CrossRef]
- Gómez, D.; Sunyer, J.O.; Salinas, I. The mucosal immune system of fish: The evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 2013, 35, 1729–1739. [Google Scholar] [CrossRef]
- Kallert, D.M.; Bauer, W.; Haas, W.; El-Matbouli, M. No shot in the dark: Myxozoans chemically detect fresh fish. Int. J. Parasitol. 2011, 41, 271–276. [Google Scholar] [CrossRef]
- Brooker, A.J.; Shinn, A.P.; Souissi, S.; Bron, J.E. Role of kairomones in host location of the pennellid copepod parasite, Lernaeocera branchialis (L. 1767). Parasitology 2013, 140, 756–770. [Google Scholar] [CrossRef]
- Ohashi, H.; Umeda, N.; Hirazawa, N.; Ozaki, Y.; Miura, C.; Miura, T. Purification and identification of a glycoprotein that induces the attachment of oncomiracidia of Neobenedenia girellae (Monogenea, Capsalidae). Int. J. Parasitol. 2007, 37, 1483–1490. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, K.; Matsunaga, R.; Hirakawa, S.; Hosoya, S.; Suetake, H.; Kikuchi, K.; Suzuki, Y.; Nakamura, O.; Miyadai, T.; Tasumi, S.; et al. Mucosal IgM Antibody with d-Mannose Affinity in Fugu Takifugu rubripes Is Utilized by a Monogenean Parasite Heterobothrium okamotoi for Host Recognition. J. Immunol. 2017, 198, 4107–4114. [Google Scholar] [CrossRef] [PubMed]
- Hellio, C.; Pons, A.M.; Beaupoil, C.; Bourgougnon, N.; Le Gal, Y. Antibacterial, antifungal and cytotoxic activities of extracts from fish epidermis and epidermal mucus. Int. J. Antimicrob. Agents 2002, 20, 214–219. [Google Scholar] [CrossRef]
- Wang, H.; Tang, W.; Zhang, R.; Ding, S. Analysis of enzyme activity, antibacterial activity, antiparasitic activity and physico-chemical stability of skin mucus derived from Amphiprion clarkii. Fish Shellfish Immunol. 2019, 86, 653–661. [Google Scholar] [CrossRef]
- Masso-Silva, J.A.; Diamond, G. Antimicrobial Peptides from Fish. Pharmaceuticals 2014, 7, 265–310. [Google Scholar] [CrossRef]
- Colorni, A.; Ullal, A.; Heinisch, G.; Noga, E.J. Activity of the antimicrobial polypeptide piscidin 2 against fish ectoparasites. J. Fish Dis. 2008, 31, 423–432. [Google Scholar] [CrossRef]
- Ullal, A.; Litaker, R.W.; Noga, E.J. Antimicrobial peptides derived from hemoglobin are expressed in epithelium of channel catfish (Ictalurus punctatus, Rafinesque). Dev. Comp. Immunol. 2008, 32, 1301–1312. [Google Scholar] [CrossRef]
- Ullal, A.J.; Noga, E.J. Antiparasitic activity of the antimicrobial peptide HbbetaP-1, a member of the beta-haemoglobin peptide family. J. Fish Dis. 2010, 33, 657–664. [Google Scholar] [CrossRef]
- Plaisance, L.; Bouamer, S.; Morand, S. Description and redescription of Haliotrema species (Monogenoidea: Poloyonchoinea: Dactylogyridae) parasitizing butterfly fishes (Teleostei: Chaetodontidae) in the Indo-West Pacific Ocean. Parasitol. Res. 2004, 93, 72–78. [Google Scholar] [CrossRef]
- Kearn, G. Evolutionary expansion of the Monogenea. Int. J. Parasitol. 1994, 24, 1227–1271. [Google Scholar] [CrossRef]
- Frantz, A.; Perga, M.-E.; Guillard, J. Parasitic versus nutritional regulation of natural fish populations. Ecol. Evol. 2018, 8, 8713–8725. [Google Scholar] [CrossRef] [PubMed]
- Dash, P.; Kar, B.; Mishra, A.; Sahoo, P.K. Effect of Dactylogyrus catlaius (Jain 1961) infection in Labeo rohita (Hamilton 1822): Innate immune responses and expression profile of some immune-related genes. Indian J. Exp. Biol. 2014, 52, 267–280. [Google Scholar] [PubMed]
- Li, J.-P.; Fu, Y.-W.; Zhang, Q.-Z.; Xu, D.-H.; Lin, Y.-M.; Zhou, S.-Y.; Lin, D.-J. Grass carp which survive Dactylogyrus ctenopharyngodonid infection also gain partial immunity against Ichthyophthirius multifili. Dis. Aquat. Org. 2018, 129, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Reverter, M.; Cutmore, S.C.; Bray, R.; Cribb, T.H.; Sasal, P. Gill monogenean communities (Platyhelminthes, Monogenea, Dactylogyridae) of butterflyfishes from tropical Indo-West Pacific Islands. Parasitology 2016, 143, 1580–1591. [Google Scholar] [CrossRef]
- Pratchett, M.S. Dietary overlap among coral-feeding butterflyfishes (Chaetodontidae) at Lizard Island, northern Great Barrier Reef. Mar. Biol. 2005, 148, 373–382. [Google Scholar] [CrossRef]
- Bellwood, D.R.; Klanten, S.; Cowman, P.F.; Pratchett, M.S.; Konow, N.; Van Herwerden, L. Evolutionary history of the butterflyfishes (f: Chaetodontidae) and the rise of coral feeding fishes. J. Evol. Biol. 2010, 23, 335–349. [Google Scholar] [CrossRef]
- Reverter, M.; Sasal, P.; Banaigs, B.; Lecchini, D.; Lecellier, G.; Tapissier-Bontemps, N. Fish mucus metabolome reveals fish life-history traits. Coral Reefs 2017, 36, 463–475. [Google Scholar] [CrossRef]
- Reverter, M.; Sasal, P.; Tapissier-Bontemps, N.; Lecchini, D.; Suzuki, M. Characterisation of the gill mucosal bacterial communities of four butterflyfish species: A reservoir of bacterial diversity in coral reef ecosystems. FEMS Microbiol. Ecol. 2017, 93, 051. [Google Scholar] [CrossRef]
- Dibattista, J.D.; Wang, X.; Saenz-Agudelo, P.; Piatek, M.J.; Aranda, M.; Berumen, M.L. Draft genome of an iconic Red Sea reef fish, the blacktail butterflyfish (Chaetodon austriacus): Current status and its characteristics. Mol. Ecol. Resour. 2016, 18, 347–355. [Google Scholar] [CrossRef]
- Shinn, A.; Pratoomyot, J.; Bron, J.; Paladini, G.; Brooker, E.; Brooker, A. Economic Impacts of Aquatic Parasites on Global Finfish Production. Available online: https://www.aquaculturealliance.org/advocate/economic-impacts-of-aquatic-parasites-on-global-finfish-production/ (accessed on 29 May 2020).
- Picón-Camacho, S.M.; Marcos-Lopez, M.; Bron, J.E.; Shinn, A. An assessment of the use of drug and non-drug interventions in the treatment of Ichthyophthirius multifiliis Fouquet, 1876, a protozoan parasite of freshwater fish. Parasitology 2011, 139, 149–190. [Google Scholar] [CrossRef]
- Forwood, J.M.; Harris, J.; DeVeney, M.R. Efficacy of current and alternative bath treatments for Lepidotrema bidyana infecting silver perch, Bidyanus bidyanus. Aquaculture 2013, 416, 65–71. [Google Scholar] [CrossRef]
- Reverter, M.; Sarter, S.; Caruso, D.; Avarre, J.-C.; Combe, M.; Pepey, E.; Pouyaud, L.; Vega-Heredía, S.; De Verdal, H.; Gozlan, R.E. Aquaculture at the crossroads of global warming and antimicrobial resistance. Nat. Commun. 2020, 11, 1870–1878. [Google Scholar] [CrossRef] [PubMed]
- Birkemo, G.A.; Lüders, T.; Andersen, Ø.; Nissen-Meyer, J. Hipposin, a histone-derived antimicrobial peptide in Atlantic halibut (Hippoglossus hippoglossus L.). Biochim. Biophys. Acta (BBA) - Bioenerg. 2003, 1646, 207–215. [Google Scholar] [CrossRef]
- Park, I.Y.; Park, C.B.; Kim, M.S.; Kim, S.C. Parasin I, an antimicrobial peptide derived from histone H2A in the catfish, Parasilurus asotus. FEBS Lett. 1998, 437, 258–262. [Google Scholar] [CrossRef]
- Salles, C.M.C.; Gagliano, P.; Leitão, S.A.T.; Salles, J.B.; Guedes, H.L.D.M.; Cassano, V.P.F.; De Simone, S.G. Identification and characterization of proteases from skin mucus of tambacu, a Neotropical hybrid fish. Fish Physiol. Biochem. 2007, 33, 173–179. [Google Scholar] [CrossRef]
- Fernandes, J.M.; Smith, V.J. A novel antimicrobial function for a ribosomal peptide from rainbow trout skin. Biochem. Biophys. Res. Commun. 2002, 296, 167–171. [Google Scholar] [CrossRef]
- Anthea, M. Human Biology and Health; Prentice Hall: Englewood Cliffs, NJ, USA, 1993. [Google Scholar]
- Zhang, D.L.; Rui-Zhang, G.; Huang, W.S.; Xiong, J. Isolation and characterization of a novel antibacterial peptide derived from hemoglobin alpha in the liver of Japanese eel, Anguilla japonica. Fish Shellfish Immunol. 2013, 35, 625–631. [Google Scholar] [CrossRef]
- Seo, J.-K.; Lee, M.J.; Jung, H.-G.; Go, H.-J.; Kim, Y.J.; Park, N.G. Antimicrobial function of SHβAP, a novel hemoglobin β chain-related antimicrobial peptide, isolated from the liver of skipjack tuna, Katsuwonus pelamis. Fish Shellfish. Immunol. 2014, 37, 173–183. [Google Scholar] [CrossRef]
- Wu, H.-J.; Wu, E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 2012, 3, 4–14. [Google Scholar] [CrossRef]
- Boutin, S.; Audet, C.; Derome, N. Probiotic treatment by indigenous bacteria decreases mortality without disturbing the natural microbiota of Salvelinus fontinalis. Can. J. Microbiol. 2013, 59, 662–670. [Google Scholar] [CrossRef]
- Boutin, S.; Sauvage, C.; Bernatchez, L.; Audet, C.; Derome, N. Inter Individual Variations of the Fish Skin Microbiota: Host Genetics Basis of Mutualism? PLoS ONE 2014, 9, e102649. [Google Scholar] [CrossRef] [PubMed]
- Gaulke, G.L.; Dennis, C.E.; Wahl, D.H.; Suski, C.D. Acclimation to a low oxygen environment alters the hematology of largemouth bass (Micropterus salmoides). Fish Physiol. Biochem. 2013, 40, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Heinicke, K.; Prommer, N.; Cajigal, J.; Viola, T.; Behn, C.; Schmidt, W. Long-term exposure to intermittent hypoxia results in increased hemoglobin mass, reduced plasma volume, and elevated erythropoietin plasma levels in man. Graefe’s Arch. Clin. Exp. Ophthalmol. 2003, 88, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Hunt, P.W.; Klok, E.J.; Trevaskis, B.; Watts, R.A.; Ellis, M.H.; Peacock, W.J.; Dennis, E.S. Increased level of hemoglobin 1 enhances survival of hypoxic stress and promotes early growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2002, 99, 17197–17202. [Google Scholar] [CrossRef]
- Sanchez, L.M.; Wong, W.R.; Riener, R.M.; Schulze, C.J.; Linington, R.G. Examining the Fish Microbiome: Vertebrate-Derived Bacteria as an Environmental Niche for the Discovery of Unique Marine Natural Products. PLoS ONE 2012, 7, e35398. [Google Scholar] [CrossRef]
- Bennett, K.W.; Eley, A. Fusobacteria: New taxonomy and related diseases. J. Med. Microbiol. 1993, 39, 246–254. [Google Scholar] [CrossRef]
- Von Engelhardt, W.; Bartels, J.; Kirschberger, S.; Zu Düttingdorf, H.M.; Busche, R. Role of short-chain fatty acids in the hind gut. Veter- Q. 1998, 20, 52–59. [Google Scholar] [CrossRef]
- Andoh, A.; Bamba, T.; Sasaki, M. Physiological and Anti-Inflammatory Roles of Dietary Fiber and Butyrate in Intestinal Functions. J. Parenter. Enter. Nutr. 1999, 23, S70–S73. [Google Scholar] [CrossRef]
- Hess, S.; Wenger, A.S.; Ainsworth, T.D.; Rummer, J.L. Exposure of clownfish larvae to suspended sediment levels found on the Great Barrier Reef: Impacts on gill structure and microbiome. Sci. Rep. 2015, 5, 10561. [Google Scholar] [CrossRef]
- Jiang, N.; Tan, N.S.; Ho, B.; Ding, J.L. Respiratory protein–generated reactive oxygen species as an antimicrobial strategy. Nat. Immunol. 2007, 8, 1114–1122. [Google Scholar] [CrossRef]
- Du, R.; Ho, B.; Ding, J.L. Rapid reprogramming of haemoglobin structure-function exposes multiple dual-antimicrobial potencies. EMBO J. 2009, 29, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Ding, J.L. A Perspective on the Role of Extracellular Hemoglobin on the Innate Immune System. DNA Cell Biol. 2013, 32, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Bakken, V.; Högh, B.T.; Jensen, H.B. Utilization of amino acids and peptides by Fusobacterium nucleatum. Eur. J. Oral Sci. 1989, 97, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Rogers, A.H.; Gully, N.J.; Pfennig, A.L.; Zilm, P.S. The breakdown and utilization of peptides by strains of Fusobacterium nucleatum. Oral Microbiol. Immunol. 1992, 7, 299–303. [Google Scholar] [CrossRef]
- Brokstad, K.A.; Jensen, H.B. Purification and characterization of a 65-kilodalton diisopropylfluorophosphate-binding protein in the outer membrane of Fusobacterium nucleatum Fev1. Eur. J. Oral Sci. 1991, 99, 20–29. [Google Scholar] [CrossRef]
- Doron, L.; Coppenhagen-Glazer, S.; Ibrahim, Y.; Eini, A.; Naor, R.; Rosen, G.; Bachrach, G. Identification and Characterization of Fusolisin, the Fusobacterium nucleatum Autotransporter Serine Protease. PLoS ONE 2014, 9, e111329. [Google Scholar] [CrossRef]
- Esteban, M. Ángeles An Overview of the Immunological Defenses in Fish Skin. ISRN Immunol. 2012, 2012, 1–29. [Google Scholar] [CrossRef]
- Albertsen, M.; Hugenholtz, P.; Skarshewski, A.; Nielsen, K.L.; Tyson, G.W.; Nielsen, P.H. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 2013, 31, 533–538. [Google Scholar] [CrossRef]
- Vidal-Dupiol, J.; Ladrière, O.; Destoumieux-Garzón, D.; Sautiere, P.-E.; Meistertzheim, A.-L.; Tambutté, E.; Tambutté, S.; Duval, D.; Fouré, L.; Adjeroud, M.; et al. Innate Immune Responses of a Scleractinian Coral to Vibriosis*. J. Biol. Chem. 2011, 286, 22688–22698. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
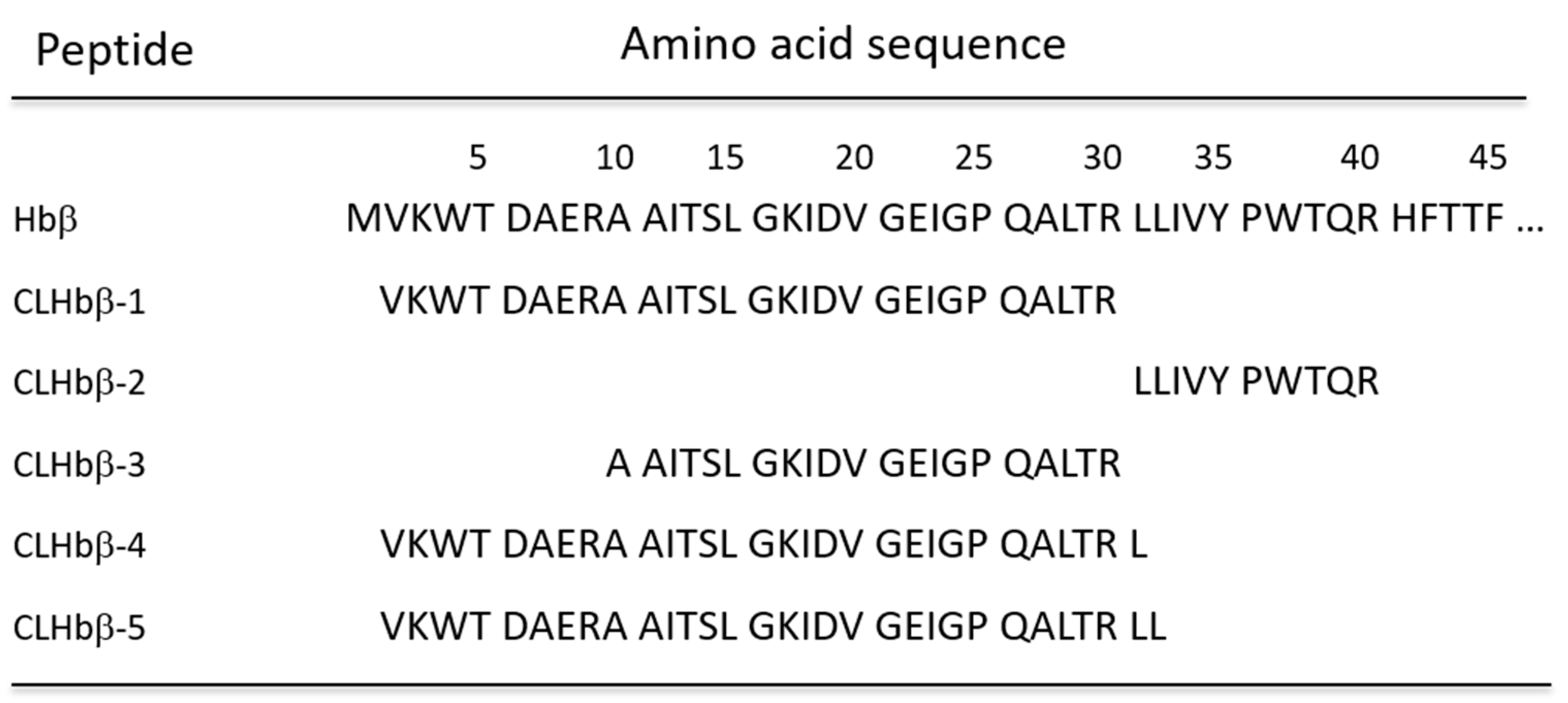

| Name | Sequence | Calculated MW (Monoisotopic Mass) | m/z LC-MS (Monoisotopic Mass) | m/z MALDI-MS (Average Value) | ||||
|---|---|---|---|---|---|---|---|---|
| Charge State | Calculated m/z LC-MS | Observed for Natural Peptides | Observed for Synthetic Peptides | Theoretical MH+ | Observed MH+ | |||
| CLHbβ-2 | LLIVYPWTQR | 1287.7339 | 2 | 644.8748 | 644.8738 | 644.8743 | 1289.562 | 1289.682 |
| CLHbβ-3 | AAITSLWGKIDVGEIGPQALTR | 2295.2586 | 3 | 766.0940 | 766.0947 | 766.0927 | 2297.659 | 2296.555 |
| CLHbβ-1 | VKWTDAERAAITSLWGKIDVGEIGPQALTR | 3280.7567 | 5 | 657.1592 | 657.1599 | 657.1589 | 3283.754 | 3283.980 |
| CLHbβ-4 | VKWTDAERAAITSLWGKIDVGEIGPQALTRL | 3393.8408 | 5 | 679.7760 | 679.7758 | 679.7773 | 3396.914 | 3396.935 |
| CLHbβ-5 | VKWTDAERAAITSLWGKIDVGEIGPQALTRLL | 3506.9248 | 5 | 702.3928 | 702.3929 | 702.3931 | 3510.073 | 3510.192 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reverter, M.; Sasal, P.; Suzuki, M.T.; Raviglione, D.; Inguimbert, N.; Pare, A.; Banaigs, B.; Voisin, S.N.; Bulet, P.; Tapissier-Bontemps, N. Insights into the Natural Defenses of a Coral Reef Fish Against Gill Ectoparasites: Integrated Metabolome and Microbiome Approach. Metabolites 2020, 10, 227. https://doi.org/10.3390/metabo10060227
Reverter M, Sasal P, Suzuki MT, Raviglione D, Inguimbert N, Pare A, Banaigs B, Voisin SN, Bulet P, Tapissier-Bontemps N. Insights into the Natural Defenses of a Coral Reef Fish Against Gill Ectoparasites: Integrated Metabolome and Microbiome Approach. Metabolites. 2020; 10(6):227. https://doi.org/10.3390/metabo10060227
Chicago/Turabian StyleReverter, Miriam, Pierre Sasal, Marcelino T. Suzuki, Delphine Raviglione, Nicolas Inguimbert, Alan Pare, Bernard Banaigs, Sébastien N. Voisin, Philippe Bulet, and Nathalie Tapissier-Bontemps. 2020. "Insights into the Natural Defenses of a Coral Reef Fish Against Gill Ectoparasites: Integrated Metabolome and Microbiome Approach" Metabolites 10, no. 6: 227. https://doi.org/10.3390/metabo10060227
APA StyleReverter, M., Sasal, P., Suzuki, M. T., Raviglione, D., Inguimbert, N., Pare, A., Banaigs, B., Voisin, S. N., Bulet, P., & Tapissier-Bontemps, N. (2020). Insights into the Natural Defenses of a Coral Reef Fish Against Gill Ectoparasites: Integrated Metabolome and Microbiome Approach. Metabolites, 10(6), 227. https://doi.org/10.3390/metabo10060227








