Metabolic Effects of Bovine Milk Oligosaccharides on Selected Commensals of the Infant Microbiome—Commensalism and Postbiotic Effects
Abstract
:1. Introduction
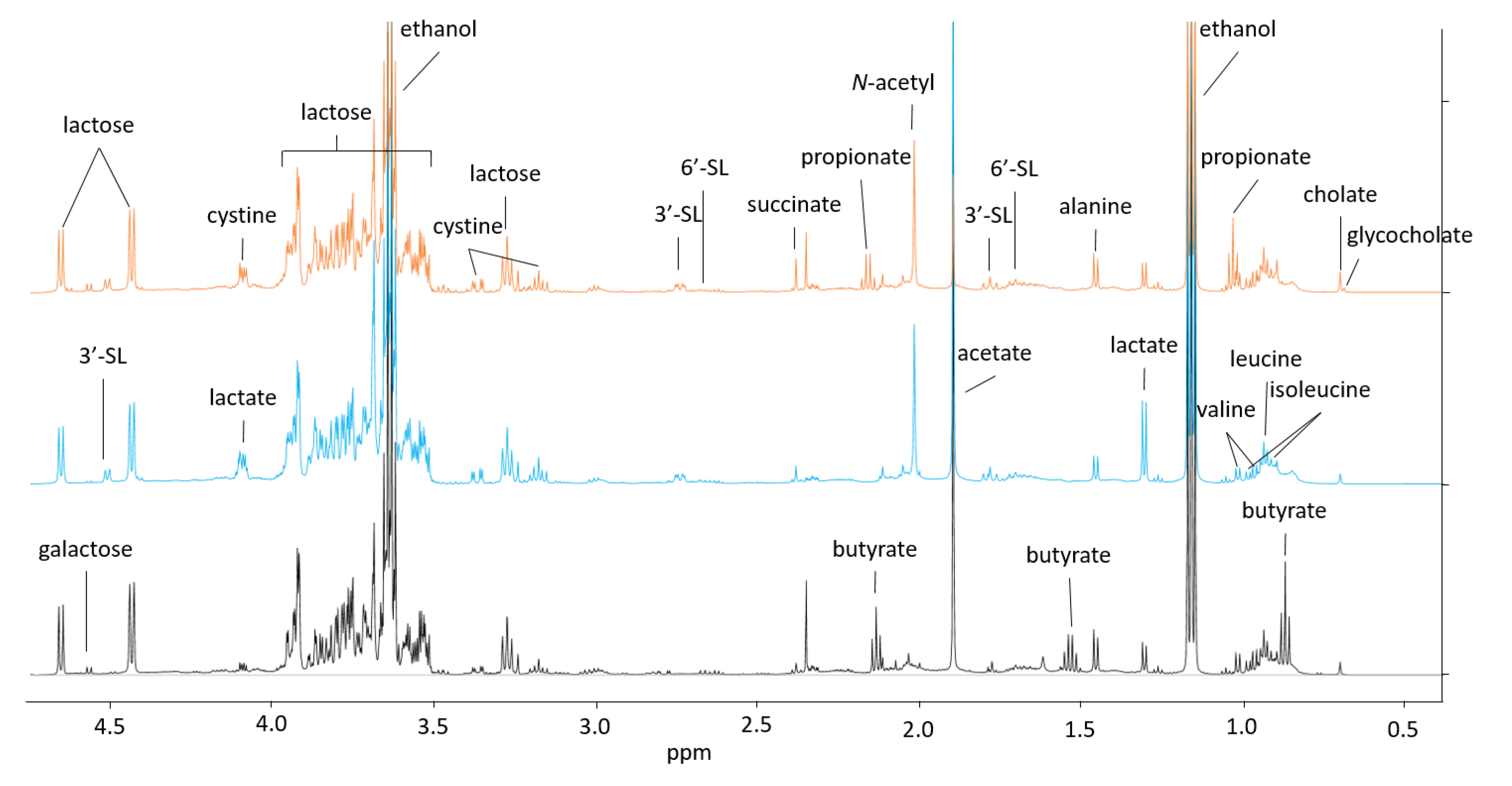
2. Results and Discussion
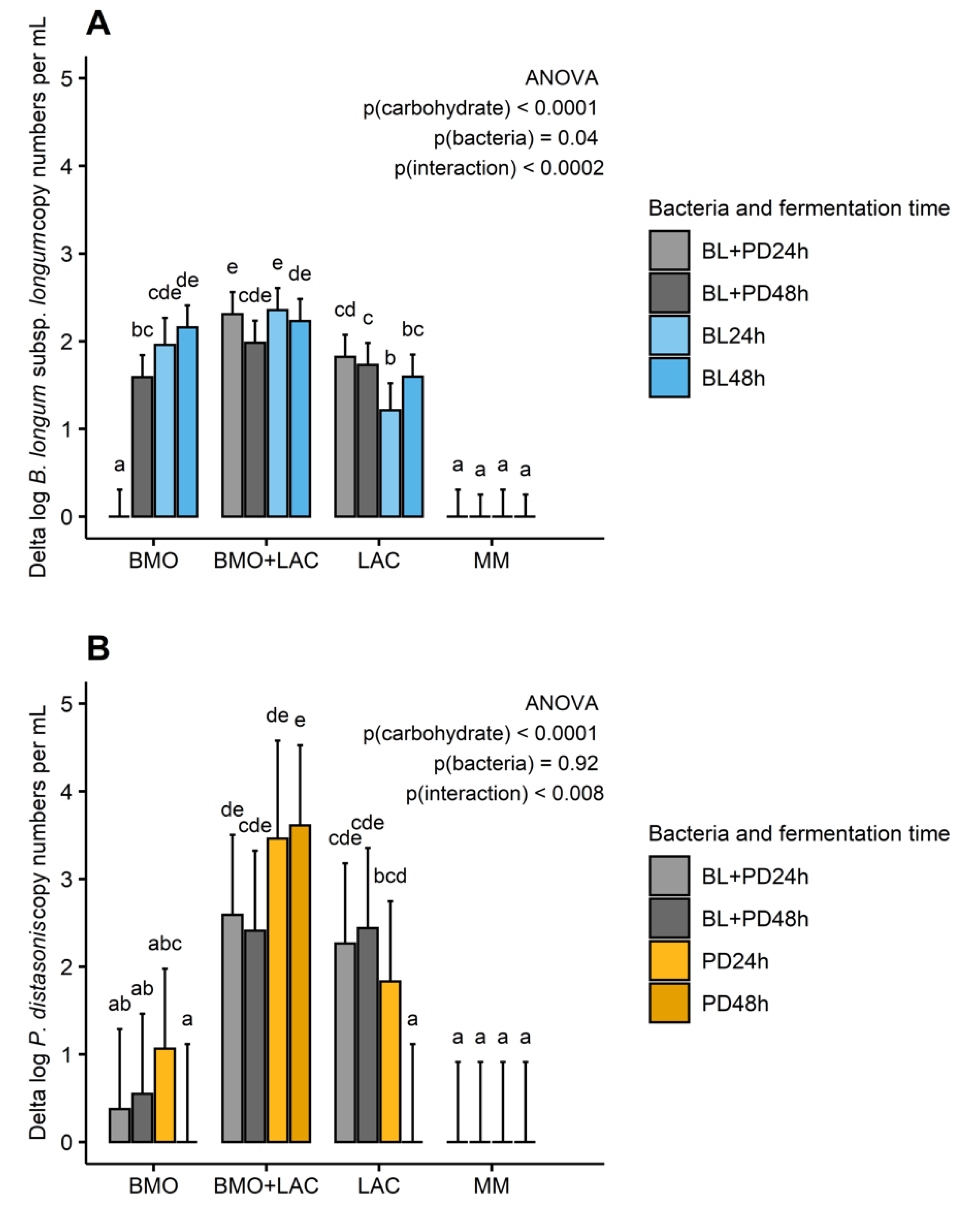
2.1. A Potential Synergistic Effect of BMO and Lactose on Stimulation of B. longum subsp. longum Metabolism
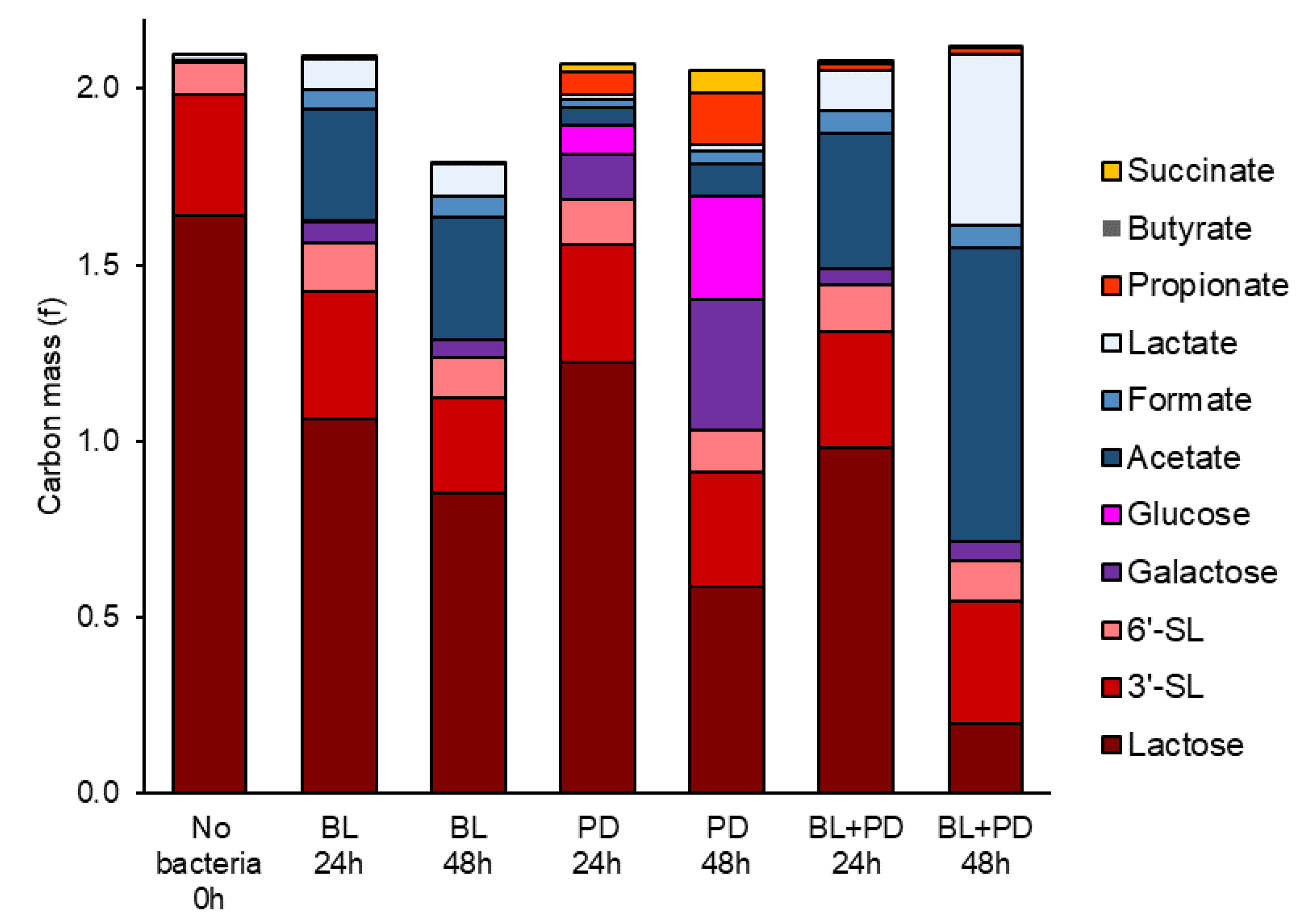
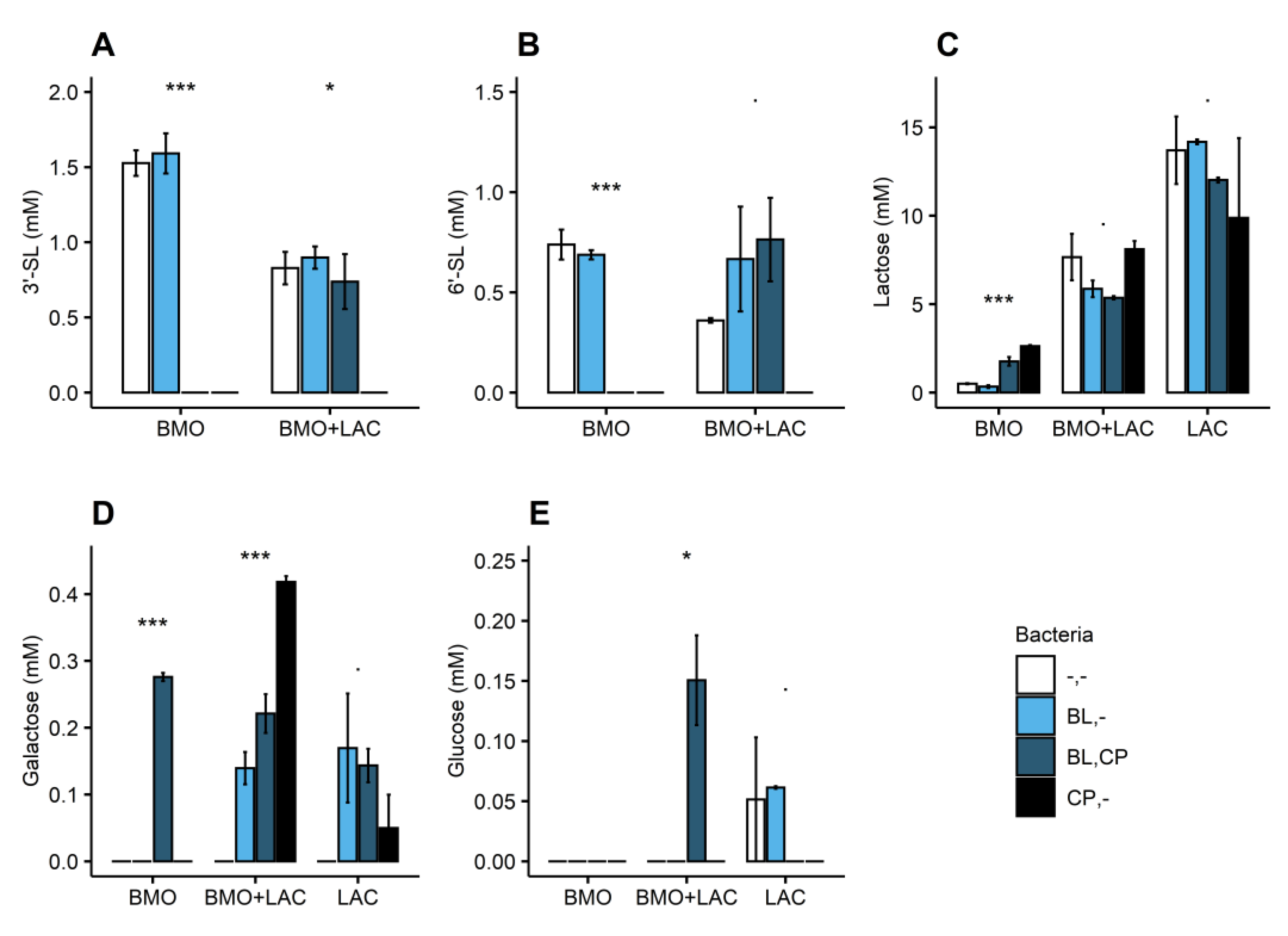
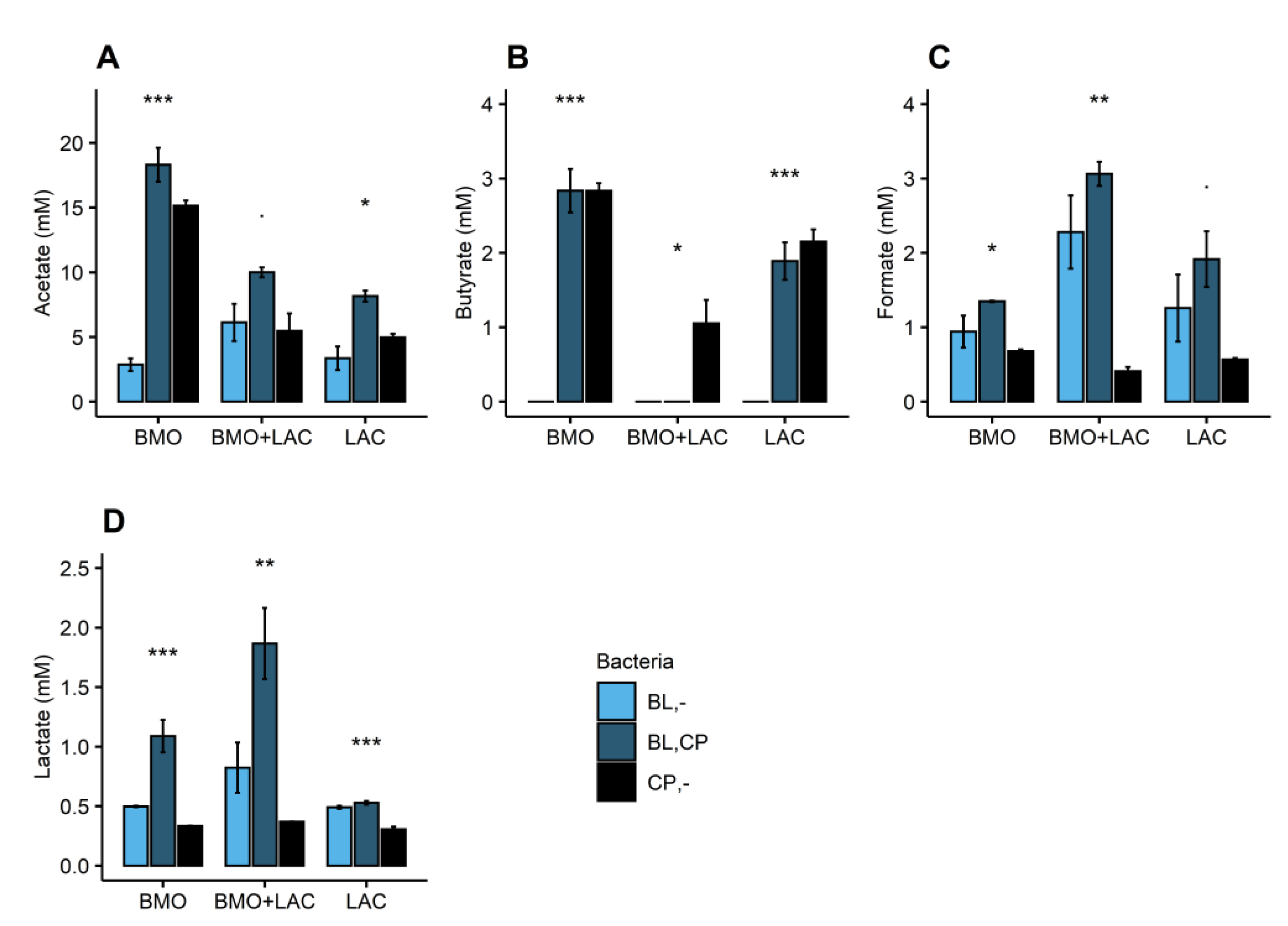
2.2. P. distasonis and B. longum subsp. longum Co-Culture Increase Utilization of Lactose
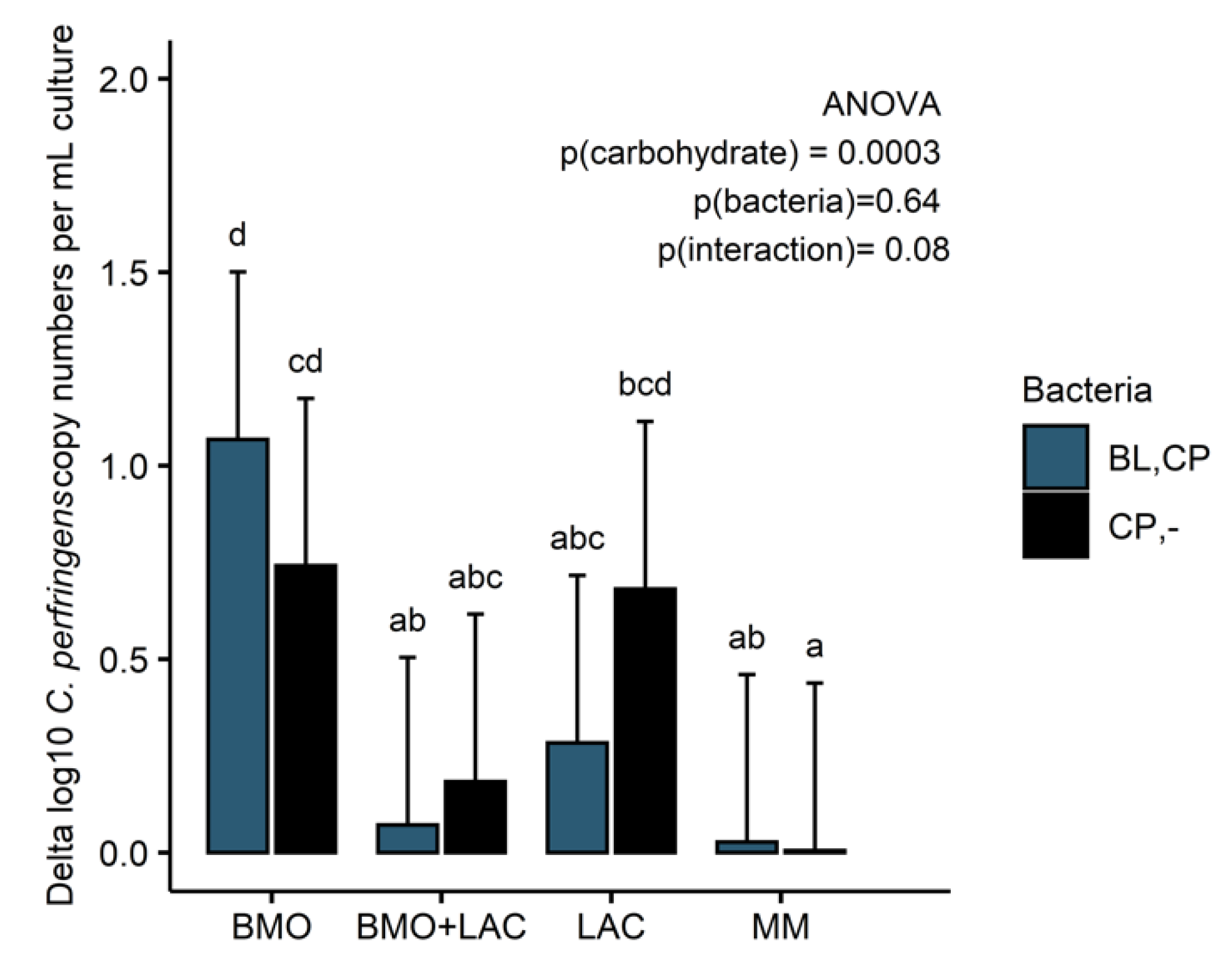
2.3. Through Metabolism of BMO + LAC B. longum subsp. longum Potentially Inhibits C. perfringens’ Ability to Metabolize Sialyllactose
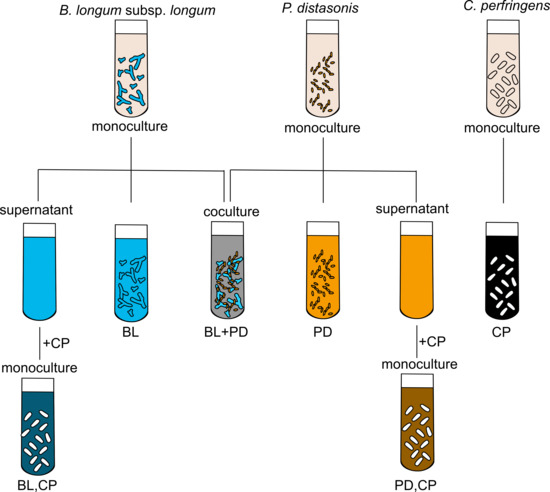
3. Materials and Methods
3.1. Strains and Propagation
3.2. Lactose and BMO Treatments
3.3. Standard Curves
3.4. Fermentation of Lactose and BMO in Mono- and Co-Culture
3.5. Proton Nuclear Magnetic Resonance (1H NMR) Metabolomics
3.6. Preprocessing of 1H NMR Data for Multivariate Analysis
3.7. Carbon Mass in Co-Culture Experiment
3.8. DNA Extraction
3.9. Quantitative PCR
3.10. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Newburg, D.S.; Walker, W.A. Protection of the neonate by the innate immune system of developing gut and of human milk. Pediatr. Res. 2007, 61, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Ten Bruggencate, S.J.M.; Bovee-Oudenhoven, I.M.J.; Feitsma, A.L.; van Hoffen, E.; Schoterman, M.H.C. Functional role and mechanisms of sialyllactose and other sialylated milk oligosaccharides. Nutr. Rev. 2014, 72, 377–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundekilde, U.K.; Downey, E.; O’Mahony, J.A.; O’Shea, C.-A.; Ryan, C.A.; Kelly, A.L.; Bertram, H.C. The effect of gestational and lactational age on the human milk metabolome. Nutrients 2016, 8, 304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, F.-P.J.; Sprenger, N.; Yap, I.K.; Wang, Y.; Bibiloni, R.; Rochat, F.; Rezzi, S.; Cherbut, C.; Kochhar, S.; Lindon, J.C.; et al. Panorganismal gut microbiome- host metabolic crosstalk. J. Proteome Res. 2009, 8, 2090–2105. [Google Scholar] [CrossRef] [Green Version]
- Martin, F.-P.J.; Wang, Y.; Sprenger, N.; Yap, I.K.S.; Lundstedt, T.; Lek, P.; Rezzi, S.; Ramadan, Z.; van Bladeren, P.; Fay, L.B.; et al. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol. Syst. Biol. 2008, 4, 157–171. [Google Scholar] [CrossRef] [Green Version]
- Yassour, M.; Vatanen, T.; Siljander, H.; Hämäläinen, A.-M.; Härkönen, T.; Ryhänen, S.J.; Franzosa, E.A.; Vlamakis, H.; Huttenhower, C.; Gevers, D.; et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 2016, 8, 343ra81. [Google Scholar] [CrossRef] [Green Version]
- Laursen, M.F.; Andersen, L.B.; Michaelsen, K.F.; Mølgaard, C.; Trolle, E.; Bahl, M.I.; Licht, T.R. Infant gut microbiota development is driven by transition to family foods independent of maternal obesity. Msphere 2016, 1, e00069-15. [Google Scholar] [CrossRef] [Green Version]
- Rios-Covian, D.; Arboleya, S.; Hernandez-Barranco, A.M.; Alvarez-Buylla, J.R.; Ruas-Madiedo, P.; Gueimonde, M.; Clara, G. Interactions between Bifidobacterium and Bacteroides species in cofermentations are affected by carbon sources, including exopolysaccharides produced by bifidobacteria. Appl. Environ. Microbiol. 2013, 79, 7518–7524. [Google Scholar] [CrossRef] [Green Version]
- Falony, G.; Calmeyn, T.; Leroy, F.; De Vuyst, L. Coculture fermentations of Bifidobacterium species and Bacteroides thetaiotaomicron reveal a mechanistic insight into the prebiotic effect of inulin-type fructans. Appl. Environ. Microbiol. 2009, 75, 2312–2319. [Google Scholar] [CrossRef] [Green Version]
- Sonnenburg, J.L.; Chen, C.T.; Gordon, J.I. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006, 4, e413. [Google Scholar] [CrossRef] [Green Version]
- Jakobsen, L.M.; Sundekilde, U.K.; Andersen, H.J.; Nielsen, D.S.; Bertram, H.C. Lactose and Bovine Milk Oligosaccharides Synergistically Stimulate B. longum subsp. longum Growth in a Simplified Model of the Infant Gut Microbiome. J. Proteome Res. 2019, 18, 3086–3098. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Toalá, J.; Garcia-Varela, R.; Garcia, H.; Mata-Haro, V.; González-Córdova, A.; Vallejo-Cordoba, B.; Hernández-Mendoza, A. Postbiotics: An evolving term within the functional foods field. Trends Food Sci. Technol. 2018, 75, 105–114. [Google Scholar] [CrossRef]
- Karav, S.; Le Parc, A.; de Moura Bell, J.M.L.N.; Frese, S.A.; Kirmiz, N.; Block, D.E.; Barile, D.; Mills, D.A. Oligosaccharides released from milk glycoproteins are selective growth substrates for infant-associated bifidobacteria. Appl. Environ. Microbiol. 2016, 3622–3630. [Google Scholar] [CrossRef] [Green Version]
- Sela, D.A.; Mills, D.A. Nursing our microbiota: Molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010, 18, 298–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawley, B.; Munro, K.; Hughes, A.; Hodgkinson, A.J.; Prosser, C.G.; Lowry, D.; Zhou, S.J.; Makrides, M.; Gibson, R.A.; Lay, C.; et al. Differentiation of Bifidobacterium longum subspecies longum and infantis by quantitative PCR using functional gene targets. PeerJ 2017, 5, e3375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrido, D.; Ruiz-Moyano, S.; Kirmiz, N.; Davis, J.C.; Totten, S.M.; Lemay, D.G.; Ugalde, J.A.; German, J.B.; Lebrilla, C.B.; Mills, D.A. A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp. longum SC596. Sci. Rep. 2016, 6, 35045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, N.; DePeters, E.; Freeman, S.; German, J.; Grimm, R.; Lebrilla, C.B. Bovine milk glycome. J. Dairy Sci. 2008, 91, 3768–3778. [Google Scholar] [CrossRef] [Green Version]
- Brigham, C.; Caughlan, R.; Gallegos, R.; Dallas, M.B.; Godoy, V.G.; Malamy, M.H. Sialic acid (N-acetyl neuraminic acid) utilization by Bacteroides fragilis requires a novel N-acetyl mannosamine epimerase. J. Bacteriol. 2009, 191, 3629–3638. [Google Scholar] [CrossRef] [Green Version]
- Parche, S.; Beleut, M.; Rezzonico, E.; Jacobs, D.; Arigoni, F.; Titgemeyer, F.; Jankovic, I. Lactose-over-glucose preference in Bifidobacterium longum NCC2705: glcP, encoding a glucose transporter, is subject to lactose repression. J. Bacteriol. Res. 2006, 188, 1260–1265. [Google Scholar] [CrossRef] [Green Version]
- Severi, E.; Hood, D.W.; Thomas, G.H. Sialic acid utilization by bacterial pathogens. Microbiology 2007, 153, 2817–2822. [Google Scholar] [CrossRef] [Green Version]
- Vimr, E.R.; Kalivoda, K.A.; Deszo, E.L.; Steenbergen, S.M. Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 2004, 68, 132–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, T.; Ohtani, K.; Hirakawa, H.; Ohshima, K.; Yamashita, A.; Shiba, T.; Ogasawara, N.; Hattori, M.; Kuhara, S.; Hayashi, H. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 2002, 99, 996–1001. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Uzal, F.A.; McClane, B.A. Clostridium perfringens sialidases: Potential contributors to intestinal pathogenesis and therapeutic targets. Toxins 2016, 8, 341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; McClane, B.A. The sialidases of Clostridium perfringens type D strain CN3718 differ in their properties and sensitivities to inhibitors. Appl. Environ. Microbiol. 2014, 80, 1701–1709. [Google Scholar] [CrossRef] [Green Version]
- Roggentin, P.; Kleineidam, R.G.; Schauer, R. Diversity in the properties of two sialidase isoenzymes produced by Clostridium perfringens spp. Biol. Chem. Hoppe-Seyler 1995, 376, 569–575. [Google Scholar] [PubMed]
- Stewart, D.B.; Hegarty, J.P. Correlation between virulence gene expression and proton pump inhibitors and ambient pH in Clostridium difficile: Results of an in vitro study. J. Med. Microbiol. 2013, 62, 1517–1523. [Google Scholar] [CrossRef] [PubMed]
- Sievers, M.; Lanini, C.; Weber, A.; Schuler-Schmid, U.; Teuber, M. Microbiology and fermentation balance in a kombucha beverage obtained from a tea fungus fermentation. Syst. Appl. Microbiol. 1995, 18, 590–594. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jakobsen, L.M.A.; Maldonado-Gómez, M.X.; Sundekilde, U.K.; Andersen, H.J.; Nielsen, D.S.; Bertram, H.C. Metabolic Effects of Bovine Milk Oligosaccharides on Selected Commensals of the Infant Microbiome—Commensalism and Postbiotic Effects. Metabolites 2020, 10, 167. https://doi.org/10.3390/metabo10040167
Jakobsen LMA, Maldonado-Gómez MX, Sundekilde UK, Andersen HJ, Nielsen DS, Bertram HC. Metabolic Effects of Bovine Milk Oligosaccharides on Selected Commensals of the Infant Microbiome—Commensalism and Postbiotic Effects. Metabolites. 2020; 10(4):167. https://doi.org/10.3390/metabo10040167
Chicago/Turabian StyleJakobsen, Louise M. A., Maria X. Maldonado-Gómez, Ulrik K. Sundekilde, Henrik J. Andersen, Dennis S. Nielsen, and Hanne C. Bertram. 2020. "Metabolic Effects of Bovine Milk Oligosaccharides on Selected Commensals of the Infant Microbiome—Commensalism and Postbiotic Effects" Metabolites 10, no. 4: 167. https://doi.org/10.3390/metabo10040167
APA StyleJakobsen, L. M. A., Maldonado-Gómez, M. X., Sundekilde, U. K., Andersen, H. J., Nielsen, D. S., & Bertram, H. C. (2020). Metabolic Effects of Bovine Milk Oligosaccharides on Selected Commensals of the Infant Microbiome—Commensalism and Postbiotic Effects. Metabolites, 10(4), 167. https://doi.org/10.3390/metabo10040167