Metabolic Profiling of Glucose-Fed Metabolically Active Resting Zymomonas mobilis Strains
Abstract
1. Introduction
2. Results and Discussion
2.1. Shutdown of Cellular Growth Significantly Alters Zymomonas Mobilis Metabolome, Yet Partly Retains Its High Catabolic Rate
2.2. Aerobic to Anaerobic Transition Upregulates NADH/NAD, yet Downregulates ED Pathway Metabolite Pools
2.3. Metabolite Profiling of Mutants With Partially Impaired Ethanol Synthesis (Zm6 adhB) or Enhanced Respiratory Capacity (Zm6 cat) Exhibit Common Trends
3. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yang, S.; Fei, Q.; Zhang, Y.; Contreras, L.M.; Utturkar, S.M.; Brown, S.D.; Himmel, M.E.; Zhang, M. Zymomonas mobilis as a model system for production of biofuels and biochemicals. Microb. Biotechnol. 2016, 9, 699–717. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.-S.; Chong, H.; Park, H.S.; Yoon, K.-O.; Jung, C.; Kim, J.J.; Hong, J.H.; Kim, H.; Kim, J.-H.; Kil, J.-I.; et al. The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nat. Biotechnol. 2005, 23, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Kalnenieks, U. Physiology of Zymomonas mobilis: Some Unanswered Questions. In Advances in Microbial Physiology; Poole, R.K., Ed.; Academic Press: Cambridge, MA, USA, 2006; Volume 51, pp. 73–117. [Google Scholar]
- Rogers, P.L.; Jeon, Y.J.; Lee, K.J.; Lawford, H.G. Zymomonas mobilis for Fuel Ethanol and HigherValue Products. In Biofuels; Olsson, L., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 263–288. [Google Scholar]
- De Graaf, A.A.; Striegel, K.; Wittig, R.M.; Laufer, B.; Schmitz, G.; Wiechert, W.; Sprenger, G.A.; Sahm, H. Metabolic state of Zymomonas mobilis in glucose-, fructose-, and xylose-fed continuous cultures as analysed by 13 C- and 31 P-NMR spectroscopy. Arch. Microbiol. 1999, 171, 371–385. [Google Scholar] [CrossRef] [PubMed]
- Rutkis, R.; Galinina, N.; Strazdina, I.; Kalnenieks, U. The inefficient aerobic energetics of Zymomonas mobilis: Identifying the bottleneck. J. Basic Microbiol. 2014, 54, 1090–1097. [Google Scholar] [CrossRef] [PubMed]
- Belaïch, J.-P.; Senez, J.C. Influence of Aeration and of Pantothenate on Growth Yields of Zymomonas mobilis. J. Bacteriol. 1965, 89, 1195. [Google Scholar] [CrossRef]
- Rutkis, R.; Strazdina, I.; Balodite, E.; Lasa, Z.; Galinina, N.; Kalnenieks, U. The Low Energy-Coupling Respiration in Zymomonas mobilis Accelerates Flux in the Entner-Doudoroff Pathway. PLoS ONE 2016, 11, e0153866. [Google Scholar] [CrossRef]
- Kalnenieks, U.; Galinina, N.; Strazdina, I.; Kravale, Z.; Pickford, J.L.; Rutkis, R.; Poole, R.K. NADH dehydrogenase deficiency results in low respiration rate and improved aerobic growth of Zymomonas mobilis. Microbiology 2008, 154, 989–994. [Google Scholar] [CrossRef]
- Strazdina, I.; Balodite, E.; Lasa, Z.; Rutkis, R.; Galinina, N.; Kalnenieks, U. Aerobic catabolism and respiratory lactate bypass in Ndh-negative Zymomonas mobilis. Metab. Eng. Commun. 2018, 7, e00081. [Google Scholar] [CrossRef]
- Ishikawa, H.; Nobayashi, H.; Tanaka, H. Mechanism of fermentation performance of Zymomonas mobilis under oxygen supply in batch culture. J. Ferment. Bioeng. 1990, 70, 34–40. [Google Scholar] [CrossRef]
- Martien, J.I.; Hebert, A.S.; Stevenson, D.M.; Regner, M.R.; Khana, D.B.; Coon, J.J.; Amador-Noguez, D. Systems-Level Analysis of Oxygen Exposure in Zymomonas mobilis; Implications for Isoprenoid Production. mSystems 2019, 4, e00284-18. [Google Scholar] [CrossRef]
- Tatli, M.; Hebert, A.S.; Coon, J.J.; Amador-Noguez, D. Genome Wide Phosphoproteome Analysis of Zymomonas mobilis Under Anaerobic, Aerobic, and N2-Fixing Conditions. Front. Microbiol. 2019, 10, 1986. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, Q.; Yang, Y.; Wang, J.; Haning, K.; Hu, Y.; Wu, B.; He, M.; Zhang, Y.; Bao, J.; et al. Advances and prospects in metabolic engineering of Zymomonas mobilis. Metab. Eng. 2018, 50, 57–73. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Eddy, C.; Deanda, K.; Finkelstein, M.; Picataggio, S. Metabolic Engineering of a Pentose Metabolism Pathway in Ethanologenic Zymomonas mobilis. Science 1995, 267, 240. [Google Scholar] [CrossRef]
- Yang, S.; Tschaplinski, T.J.; Engle, N.L.; Carroll, S.L.; Martin, S.L.; Davison, B.H.; Palumbo, A.V.; Rodriguez, M., Jr.; Brown, S.D. Transcriptomic and metabolomic profiling of Zymomonas mobilis during aerobic and anaerobic fermentations. BMC Genom. 2009, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Kalnenieks, U.; Balodite, E.; Strähler, S.; Strazdina, I.; Rex, J.; Pentjuss, A.; Fuchino, K.; Bruheim, P.; Rutkis, R.; Pappas, K.M.; et al. Improvement of Acetaldehyde Production in Zymomonas mobilis by Engineering of Its Aerobic Metabolism. Front. Microbiol. 2019, 10, 2533. [Google Scholar] [CrossRef]
- Julsing, M.K.; Kuhn, D.; Schmid, A.; Bühler, B. Resting cells of recombinant E. coli show high epoxidation yields on energy source and high sensitivity to product inhibition. Biotechnol. Bioeng. 2012, 109, 1109–1119. [Google Scholar] [CrossRef]
- Kadisch, M.; Willrodt, C.; Hillen, M.; Bühler, B.; Schmid, A. Maximizing the stability of metabolic engineering-derived whole-cell biocatalysts. Biotechnol. J. 2017, 12, 1600170. [Google Scholar] [CrossRef]
- Bevers, J.; Verachtert, H. Synthesis of higher alcohols in the genus Zymomonas. J. Inst. Brew. 1976, 82, 35–40. [Google Scholar] [CrossRef]
- Fu, N.; Peiris, P.; Markham, J.; Bavor, J. A novel co-culture process with Zymomonas mobilis and Pichia stipitis for efficient ethanol production on glucose/xylose mixtures. Enzym. Microb. Technol. 2009, 45, 210–217. [Google Scholar] [CrossRef]
- Wirawan, F.; Cheng, C.-L.; Kao, W.-C.; Lee, D.-J.; Chang, J.-S. Cellulosic ethanol production performance with SSF and SHF processes using immobilized Zymomonas mobilis. Appl. Energy 2012, 100, 19–26. [Google Scholar] [CrossRef]
- Niu, X.; Wang, Z.; Li, Y.; Zhao, Z.; Liu, J.; Jiang, L.; Xu, H.; Li, Z. “Fish-in-Net”, a Novel Method for Cell Immobilization of Zymomonas mobilis. PLoS ONE 2013, 8, e79569. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pentjuss, A.; Odzina, I.; Kostromins, A.; Fell, D.A.; Stalidzans, E.; Kalnenieks, U. Biotechnological potential of respiring Zymomonas mobilis: A stoichiometric analysis of its central metabolism. J. Biotechnol. 2013, 165, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Michalowski, A.; Siemann-Herzberg, M.; Takors, R. Escherichia coli HGT: Engineered for high glucose throughput even under slowly growing or resting conditions. Metab. Eng. 2017, 40, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Jang, C.; Chen, L.; Rabinowitz, J.D. Metabolomics and Isotope Tracing. Cell 2018, 173, 822–837. [Google Scholar] [CrossRef]
- Rutkis, R.; Kalnenieks, U.; Stalidzans, E.; Fell, D.A. Kinetic modelling of the Zymomonas mobilis Entner–Doudoroff pathway: Insights into control and functionality. Microbiology 2013, 159, 2674–2689. [Google Scholar] [CrossRef]
- Hayashi, T.; Kato, T.; Watakabe, S.; Song, W.; Aikawa, S.; Furukawa, K. The respiratory chain provides salt stress tolerance by maintaining a low NADH/NAD+ ratio in Zymomonas mobilis. Microbiology 2015, 161, 2384–2394. [Google Scholar] [CrossRef]
- Yang, S.; Mohagheghi, A.; Franden, M.A.; Chou, Y.-C.; Chen, X.; Dowe, N.; Himmel, M.E.; Zhang, M. Metabolic engineering of Zymomonas mobilis for 2,3-butanediol production from lignocellulosic biomass sugars. Biotechnol. Biofuels 2016, 9, 189. [Google Scholar] [CrossRef]
- Wu, B.; Qin, H.; Yang, Y.; Duan, G.; Yang, S.; Xin, F.; Zhao, C.; Shao, H.; Wang, Y.; Zhu, Q.; et al. Engineered Zymomonas mobilis tolerant to acetic acid and low pH via multiplex atmospheric and room temperature plasma mutagenesis. Biotechnol. Biofuels 2019, 12, 10. [Google Scholar] [CrossRef]
- Wang, J.-L.; Wu, B.; Qin, H.; You, Y.; Liu, S.; Shui, Z.-X.; Tan, F.-R.; Wang, Y.-W.; Zhu, Q.-L.; Li, Y.-B.; et al. Engineered Zymomonas mobilis for salt tolerance using EZ-Tn5-based transposon insertion mutagenesis system. Microb. Cell Factories 2016, 15, 101. [Google Scholar] [CrossRef]
- Strazdina, I.; Kravale, Z.; Galinina, N.; Rutkis, R.; Poole, R.K.; Kalnenieks, U. Electron transport and oxidative stress in Zymomonas mobilis respiratory mutants. Arch. Microbiol. 2012, 194, 461–471. [Google Scholar] [CrossRef]
- Kalnenieks, U.; Galinina, N.; Toma, M.M.; Pickford, J.L.; Rutkis, R.; Poole, R.K. Respiratory behaviour of a Zymomonas mobilis adhB::kan r mutant supports the hypothesis of two alcohol dehydrogenase isoenzymes catalysing opposite reactions. FEBS Lett. 2006, 580, 5084–5088. [Google Scholar] [CrossRef]
- Galinina, N.; Lasa, Z.; Strazdina, I.; Rutkis, R.; Kalnenieks, U. Effect of ADH II deficiency on the intracellular redox homeostasis in Zymomonas mobilis. Sci. World J. 2012, 2012, 742610. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Grube, M.; Bekers, M.; Upite, D.; Kaminska, E. IR-spectroscopic studies of Zymomonas mobilis and levan precipitate. Vib. Spectrosc. 2002, 28, 277–285. [Google Scholar] [CrossRef]
- Low, K.S.; Rogers, P.L. The macromolecular composition and essential amino acid profiles of strains of Zymomonas mobilis. Appl. Microbiol. Biotechnol. 1984, 19, 75–78. [Google Scholar] [CrossRef]
- Jöbses, I.M.L.; Egberts, G.T.C.; Luyben, K.C.A.M.; Roels, J.A. Fermentation kinetics of Zymomonas mobilis at high ethanol concentrations: Oscillations in continuous cultures. Biotechnol. Bioeng. 1986, 28, 868–877. [Google Scholar] [CrossRef]
- Belenky, P.; Jonathan, D.Y.; Caroline, P.B.M.; Nadia, R.C.; Lobritz, M.A.; Ferrante, T.; Jain, S.; Korry, B.J.; Schwarz, E.G.; Walker, G.C.; et al. Bactericidal Antibiotics Induce Toxic Metabolic Perturbations that Lead to Cellular Damage. Cell Rep. 2015, 13, 968–980. [Google Scholar] [CrossRef] [PubMed]
- Kalnenieks, U.; de Graaf, A.A.; Bringer-Meyer, S.; Sahm, H. Oxidative phosphorylation in Zymomonas mobilis. Arch. Microbiol. 1993, 160, 74–79. [Google Scholar] [CrossRef]
- Kvitvang, H.F.N.; Kristiansen, K.A.; Bruheim, P. Assessment of capillary anion exchange ion chromatography tandem mass spectrometry for the quantitative profiling of the phosphometabolome and organic acids in biological extracts. J. Chromatogr. A 2014, 1370, 70–79. [Google Scholar] [CrossRef]
- Stafsnes, M.H.; Røst, L.M.; Bruheim, P. Improved phosphometabolome profiling applying isotope dilution strategy and capillary ion chromatography-tandem mass spectrometry. J. Chromatogr. B 2018, 1083, 278–283. [Google Scholar] [CrossRef]
- Cohen, S.A.; Strydom, D.J. Amino acid analysis utilizing phenylisothiocyanate derivatives. Anal. Biochem. 1988, 174, 1–16. [Google Scholar] [CrossRef]
- Tan, B.; Lu, Z.; Dong, S.; Zhao, G.; Kuo, M.-S. Derivatization of the tricarboxylic acid intermediates with O-benzylhydroxylamine for liquid chromatography–tandem mass spectrometry detection. Anal. Biochem. 2014, 465, 134–147. [Google Scholar] [CrossRef] [PubMed]

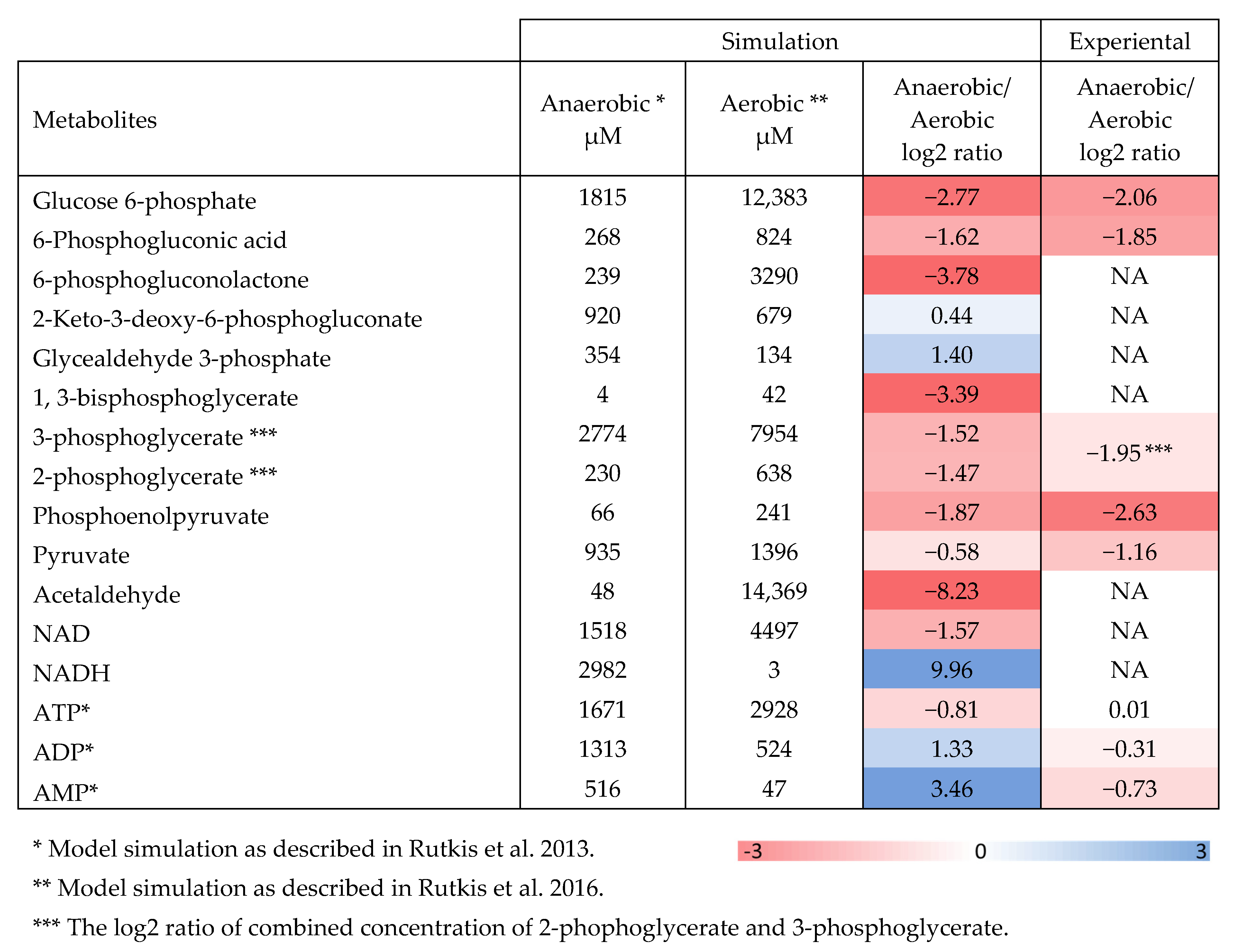
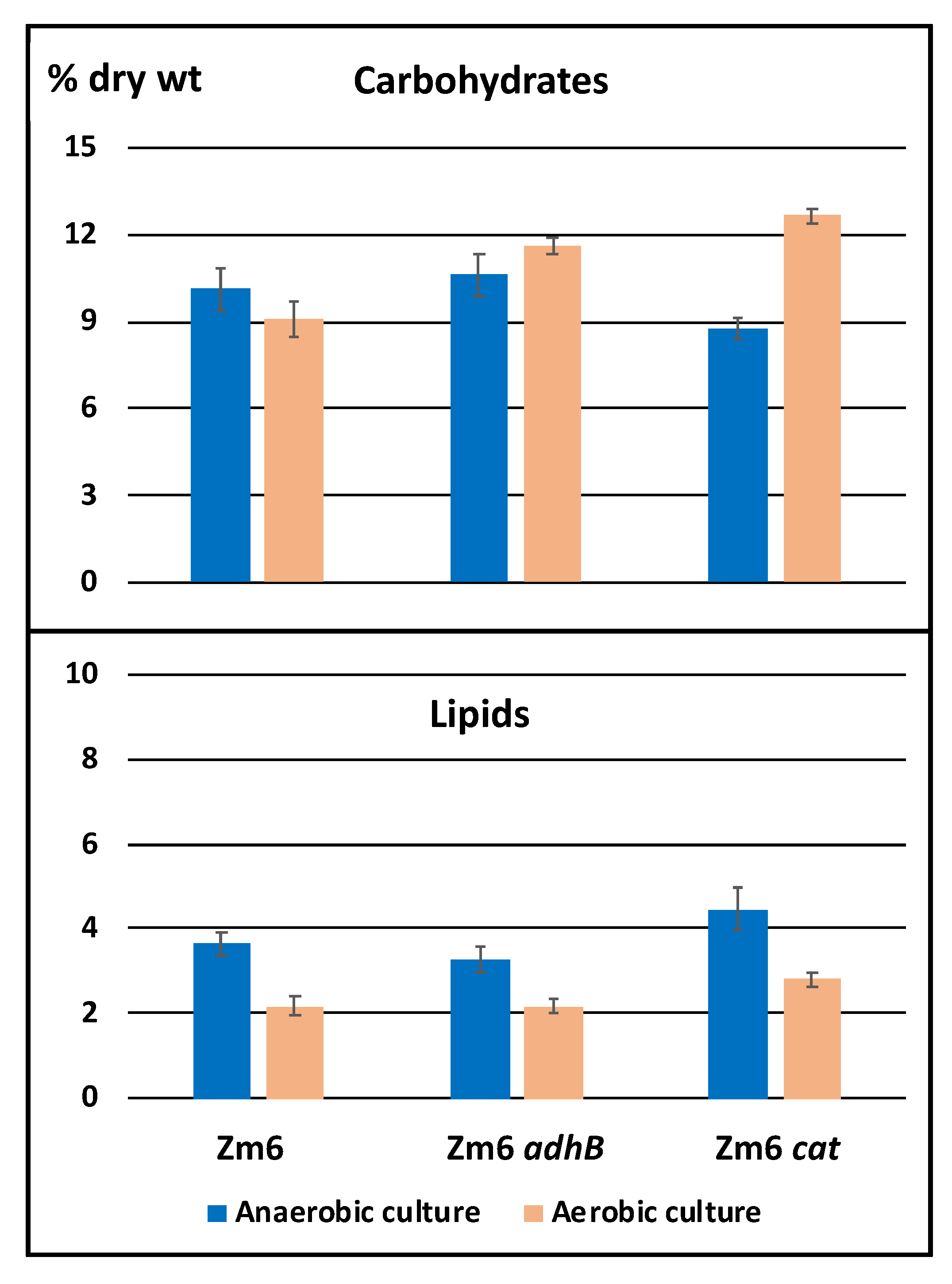
| Strain | Glucose mmol/CDW g/h | Y ethanol (g)/glucose (g) | ||
|---|---|---|---|---|
| Aerobic | Anaerobic | Aerobic | Anaerobic | |
| Zm6 | 12.13 ± 1.50 | 20.15 ± 0.89 | 0.22 ± 0.02 | 0.39 ± 0.02 |
| Zm6 cat | 11.27 ± 0.36 | 19.45 ± 0.15 | 0.18 ± 0.02 | 0.32 ± 0.01 |
| Zm6 adhB | 12.5 ± 1.92 | 15.6 ± 0.53 | 0.18 ± 0.03 | 0.33 ± 0.04 |
| Strains and Conditions | NADH/NAD | RSD (%) | |
|---|---|---|---|
| Zm6 resting | Aerobic | 0.51 | 38.11 |
| Anaerobic | 0.99 | 30.38 | |
| Zm6 cat resting | Aerobic | 0.57 | 15.1 |
| Anaerobic | 0.46 | 20.8 | |
| Zm6 adhB resting | Aerobic | 0.61 | 16.35 |
| Anaerobic | 0.74 | 1.97 | |
| Zm6 growing | Aerobic | 0.58 | 4.8 |
| Zm6 cat/ Zm6 | Zm6 cat/ Zm6 | Zm6 cat | Zm6 adh/ Zm6 | Zm6 adh/ Zm6 | Zm6 adh | ||
|---|---|---|---|---|---|---|---|
| Aerobic | Anaerobic | Anaerobic /Aerobic | Aerobic | Anaerobic | Anaerobic /Aerobic | ||
| G6P | −2.16 | 0.69 | 0.79 | G6P | −1.01 | 0.73 | −0.32 |
| 6PGA | 1.20 | 2.77 | −0.28 | 6PGA | 1.17 | 2.23 | −0.79 |
| P5P | 1.57 | 1.91 | 0.25 | P5P | 0.94 | 1.98 | 0.94 |
| Sedo-7p | 2.05 | 0.81 | −1.49 | Sedo-7p | 0.59 | 0.05 | −0.78 |
| Gly-3p | 0.98 | 1.85 | 0.52 | Gly-3p | 0.68 | 1.96 | 0.94 |
| 3PG-2PG | −1.04 | 0.88 | −0.04 | 3PG-2PG | −1.62 | 0.91 | 0.58 |
| PEP | −1.98 | 0.22 | −0.43 | PEP | −2.08 | 0.60 | 0.05 |
| Pyr | −2.20 | −0.80 | 0.24 | Pyr | −1.80 | −0.40 | 0.24 |
| Suc | 0.27 | 1.52 | 1.38 | Suc | 0.60 | 0.66 | 0.18 |
| Mal | 0.93 | 2.07 | 1.42 | Mal | −1.12 | −0.13 | 1.26 |
| AMP | 0.65 | −0.09 | −0.73 | AMP | 0.13 | −0.60 | −0.72 |
| ADP | 0.40 | 0.29 | −0.42 | ADP | 0.17 | 0.02 | −0.46 |
| ATP | 0.04 | 0.53 | −0.24 | ATP | 0.55 | 0.70 | −0.59 |
| GDP | 0.45 | 0.29 | 0.11 | GDP | 0.21 | −0.01 | 0.05 |
| GTP | 0.21 | 0.53 | 0.20 | GTP | 0.24 | 0.43 | 0.06 |
| CMP | −0.18 | 1.63 | 0.19 | CMP | −0.24 | −0.28 | −1.66 |
| CDP | 0.20 | 0.57 | −0.63 | CDP | −0.44 | −0.59 | −1.16 |
| CTP | 0.68 | 1.12 | −0.71 | CTP | 0.63 | 0.88 | −0.91 |
| UMP | 0.09 | 0.41 | 0.53 | UMP | 0.05 | −0.22 | −0.06 |
| UDP | 0.21 | 0.12 | 0.05 | UDP | −0.08 | −0.47 | −0.25 |
| UTP | 0.47 | 0.54 | −0.12 | UTP | 0.34 | 0.41 | −0.12 |
| dAMP | 0.85 | 0.56 | 0.05 | dAMP | −0.91 | −1.08 | 0.17 |
| dADP | 0.63 | 1.27 | 0.82 | dADP | −1.18 | −1.28 | 0.08 |
| dATP | −0.11 | 1.37 | 1.22 | dATP | −0.92 | −0.68 | −0.02 |
| dGDP | −0.09 | −1.86 | −2.01 | dGDP | 0.99 | −0.49 | −1.72 |
| dGTP | −0.15 | 1.50 | 0.74 | dGTP | 0.30 | −0.53 | −1.74 |
| dUMP | 1.22 | 0.00 | −0.81 | dUMP | 1.22 | 0.32 | −0.49 |
| dUTP | 0.83 | 0.19 | 0.00 | dUTP | 1.22 | 0.10 | −0.49 |
| dCTP | 0.33 | 0.66 | −0.45 | dCTP | 0.40 | 0.08 | −1.10 |
| Gly | −0.12 | 0.76 | 0.80 | Gly | −0.66 | −0.48 | 0.12 |
| Ala | 0.52 | 0.35 | −0.22 | Ala | 0.88 | −0.09 | −1.01 |
| Ser | 0.25 | 0.76 | −0.65 | Ser | 0.00 | 0.69 | −0.47 |
| Pro | 0.57 | 1.21 | 0.68 | Pro | 1.66 | 1.25 | −0.37 |
| Val | 0.12 | 0.27 | 0.70 | Val | 0.27 | −0.22 | 0.06 |
| Thr | 0.49 | 0.58 | 0.00 | Thr | 0.34 | 0.23 | −0.21 |
| Ile | 0.00 | 0.02 | 0.55 | Ile | 0.38 | −0.05 | 0.11 |
| Leu | −0.04 | 0.10 | 0.85 | Leu | 0.38 | −0.03 | 0.31 |
| Asp | 1.14 | 0.83 | 0.86 | Asp | 1.92 | 0.95 | 0.19 |
| Gln | 1.65 | 2.12 | −0.29 | Gln | 1.22 | 0.94 | −1.03 |
| Glu | 0.26 | 0.71 | −0.13 | Glu | −0.11 | −0.12 | −0.58 |
| His | 0.17 | 0.32 | 0.07 | His | −0.20 | 0.15 | 0.26 |
| Phe | −1.11 | −0.82 | 0.74 | Phe | −1.25 | −1.67 | 0.03 |
| Arg | −0.43 | 0.71 | −0.68 | Arg | −1.11 | −0.04 | −0.76 |
| Lys | −0.41 | 0.55 | −0.38 | Lys | −1.36 | −0.59 | −0.56 |
 | |||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuchino, K.; Kalnenieks, U.; Rutkis, R.; Grube, M.; Bruheim, P. Metabolic Profiling of Glucose-Fed Metabolically Active Resting Zymomonas mobilis Strains. Metabolites 2020, 10, 81. https://doi.org/10.3390/metabo10030081
Fuchino K, Kalnenieks U, Rutkis R, Grube M, Bruheim P. Metabolic Profiling of Glucose-Fed Metabolically Active Resting Zymomonas mobilis Strains. Metabolites. 2020; 10(3):81. https://doi.org/10.3390/metabo10030081
Chicago/Turabian StyleFuchino, Katsuya, Uldis Kalnenieks, Reinis Rutkis, Mara Grube, and Per Bruheim. 2020. "Metabolic Profiling of Glucose-Fed Metabolically Active Resting Zymomonas mobilis Strains" Metabolites 10, no. 3: 81. https://doi.org/10.3390/metabo10030081
APA StyleFuchino, K., Kalnenieks, U., Rutkis, R., Grube, M., & Bruheim, P. (2020). Metabolic Profiling of Glucose-Fed Metabolically Active Resting Zymomonas mobilis Strains. Metabolites, 10(3), 81. https://doi.org/10.3390/metabo10030081






