Metabolic Changes in Synaptosomes in an Animal Model of Schizophrenia Revealed by 1H and 1H,13C NMR Spectroscopy
Abstract
1. Introduction
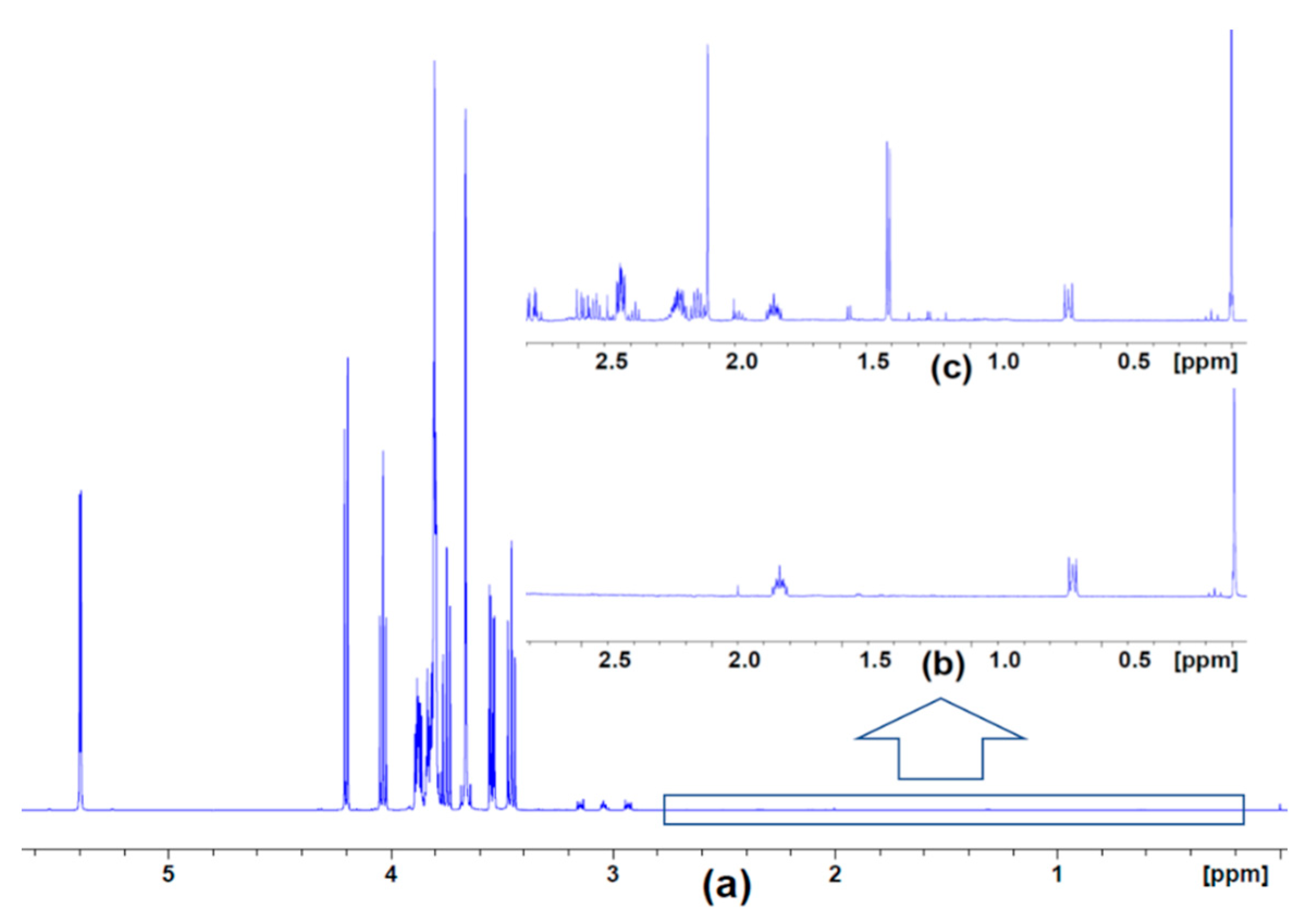
2. Results
3. Discussion
4. Materials and Methods
4.1. Animals, Tissue Collection, and Synaptosome Isolation
4.2. NMR Sample Preparation
4.3. NMR Data Collection and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Evans, G.J.O. The synaptosome as a model system for studying synaptic physiology. Cold Spring Harb. Protoc. 2015, 5, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, V.P.; Michaelson, I.A.; Kirkland, R.J.A. The separation of synaptic vesicles from nerve-ending particles (‘synaptosomes’). Biochem. J. 1964, 90, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Bai, F.; Witzmann, F.A. Synaptosome Proteomics; Bertrand, E., Faupel, M., Eds.; Subcellular Biochemistry Series 43; Springer: Dordrecht, The Netherlands, 2007; pp. 77–98. [Google Scholar] [CrossRef]
- Rohlff, C.; Hollis, K. Modern proteomic strategies in the study of complex neuropsychiatric disorders. Biol. Psychiatry 2003, 53, 847–853. [Google Scholar] [CrossRef]
- Chang, R.Y.K.; Nouwens, A.S.; Dodd, P.R.; Etheridge, N. The synaptic proteome in Alzheimer’s disease. Alzheimers Dement. 2013, 9, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Carroll, L.S.; Owen, M.J. Genetic overlap between autism, schizophrenia and bipolar disorder. Genome Med. 2009, 1, 102. [Google Scholar] [CrossRef] [PubMed]
- Forstner, A.J.; Hecker, J.; Hofmann, A.; Maaser, A.; Reinbold, C.S.; Muhleisen, T.W.; Leber, M.; Strohmaier, J.; Degenhardt, F.; Treutlein, J.; et al. Identification of shared risk loci and pathways for bipolar disorder and schizophrenia. PLoS ONE 2017, 12, e0171595. [Google Scholar] [CrossRef] [PubMed]
- Parikshak, N.N.; Swarup, V.; Belgard, T.G.; Irimia, M.; Ramaswami, G.; Gandal, M.J.; Hartl, C.; Leppa, V.; Ubieta, L.T.; Huang, J.; et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 2016, 540, 423–427. [Google Scholar] [CrossRef]
- Gandal, M.J.; Haney, J.R.; Parikshak, N.N.; Leppa, V.; Ramaswami, G.; Hartl, C.; Schork, A.J.; Appadurai, V.; Buil, A.; Werge, T.M.; et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 2018, 359, 693–697. [Google Scholar] [CrossRef]
- Gold, A.L.; Brotman, M.A.; Adleman, N.E.; Lever, S.N.; Steuber, E.R.; Fromm, S.J.; Mueller, S.C.; Pine, D.S.; Leibenluft, E. Comparing Brain Morphometry Across Multiple Childhood Psychiatric Disorders. J. Am. Acad. Child Adolesc. Psychiatry 2016, 55, 1027–1037. [Google Scholar] [CrossRef]
- Rowley, P.A.; Guerrero-Gonzalez, J.; Alexander, A.L.; Yu, J.J. Convergent microstructural brain changes across genetic models of autism spectrum disorder—A pilot study. Psychiatry Res. Neuroimaging 2019, 283, 83–91. [Google Scholar] [CrossRef]
- Albert, P.R.; Benkelfat, C. The neurobiology of depression—Revisiting the serotonin hypothesis. II. Genetic, epigenetic and clinical studies. Phil. Trans. R. Soc. B 2013, 368, 20120535. [Google Scholar] [CrossRef] [PubMed]
- Albert, P.R.; Benkelfat, C.; Descarries, L. The neurobiology of depression—Revisiting the serotonin hypothesis. I. Cellular and molecular mechanisms. Phil. Trans. R. Soc. B 2012, 367, 2378–2381. [Google Scholar] [CrossRef] [PubMed]
- Lanfumey, L.; Mongeau, R.; Cohen-Salmon, C.; Hamon, M. Corticosteroid-serotonin interactions in the neurobiological mechanisms of stress-related disorders. Neurosci. Biobehav. Rev. 2008, 32, 1174–1184. [Google Scholar] [CrossRef] [PubMed]
- Mahar, I.; Bambico, F.R.; Mechawar, N.; Nobrega, J.N. Stress, serotonin, and hippocampal neurogenesis in relation to depression and antidepressant effects. Neurosci. Biobehav. Rev. 2014, 38, 173–192. [Google Scholar] [CrossRef]
- Jacobsen, J.P.R.; Medvedev, I.O.; Caron, M.G. The 5-HT deficiency theory of depression: Perspectives from a naturalistic 5-HT deficiency model, the tryptophan hydroxylase 2Arg439His knockin mouse. Phil. Trans. R. Soc. B 2012, 367, 2444–2459. [Google Scholar] [CrossRef]
- Brown, R.P.; Mann, J.J. A clinical perspective on the role of neurotransmitters in mental disorders. Psychiatr. Serv. 1985, 36, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, B. Stress activation of glutamate neurotransmission in the prefrontal cortex: Implications for dopamine-associated psychiatric disorders. Biol. Psychiatry 2002, 51, 775–787. [Google Scholar] [CrossRef]
- Javitt, D.C. Glutamate as a therapeutic target in psychiatric disorders. Mol. Psychiatry 2004, 9, 984–997. [Google Scholar] [CrossRef]
- Lee, S.H.; Ripke, S.; Neale, B.M.; Faraone, S.V.; Purcell, S.M.; Perlis, R.H.; Mowry, B.J.; Thapar, A.; Goddard, M.E.; Witte, J.S.; et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet. 2013, 45, 984–994. [Google Scholar] [CrossRef]
- Miyoshi, K.; Asanuma, M.; Miyazaki, I.; Diaz-Corrales, F.J.; Katayama, T.; Tohyama, M.; Ogawa, N. DISC1 localizes to the centrosome by binding to kendrin. Biochem. Biophys. Res. Commun. 2004, 317, 1195–1199. [Google Scholar] [CrossRef]
- Morris, J.A.; Kandpal, G.; Ma, L.; Austin, C.P. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: Regulation and loss of interaction with mutation. Hum. Mol. Genet. 2003, 12, 1591–1608. [Google Scholar] [CrossRef] [PubMed]
- Higginbotham, H.R.; Gleeson, J.G. The centrosome in neuronal development. Trends Neurosci. 2007, 30, 276–283. [Google Scholar] [CrossRef]
- Clapcote, S.J.; Lipina, T.V.; Millar, J.K.; Mackie, S.; Christie, S.; Ogawa, F.; Lerch, J.P.; Trimble, K.; Uchiyama, M.; Sakuraba, Y.; et al. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron 2007, 54, 387–402. [Google Scholar] [CrossRef] [PubMed]
- Hikida, T.; Jaaro-Peled, H.; Seshadri, S.; Oishi, K.; Hookway, C.; Kong, S.; Wu, D.; Xue, R.; Andrade, M.; Tankou, S.; et al. Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc. Natl. Acad. Sci. USA 2007, 104, 14501–14506. [Google Scholar] [CrossRef] [PubMed]
- Barnett, B.R.; Torres-Velazquez, M.; Yi, S.Y.; Rowley, P.A.; Sawin, E.A.; Rubinstein, C.D.; Krentz, K.; Anderson, J.M.; Bakshi, V.P.; Yu, J.J. Sex-specific deficits in neurite density and white matter integrity are associated with targeted disruption of exon 2 of the Disc1 gene in the rat. Transl. Psychiatry 2019, 9, 82. [Google Scholar] [CrossRef] [PubMed]
- Newburn, E.N.; Hyde, T.M.; Ye, T.; Morita, Y.; Weinberger, D.R.; Kleinman, J.E.; Lipska, B.K. Interactions of human truncated DISC1 proteins: Implications for schizophrenia. Transl. Psychiatry 2011, 1, e30. [Google Scholar] [CrossRef] [PubMed]
- Forgacsova, A.; Galba, J.; Garruto, R.M.; Majerova, P.; Katina, S.; Kovac, A. A novel liquid chromatography/mass spectrometry method for determination of neurotransmitters in brain tissue: Application to human tauopathies. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2018, 1073, 154–162. [Google Scholar] [CrossRef]
- He, B.; Bi, K.; Jia, Y.; Wang, J.; Lv, C.; Liu, R.; Zhao, L.; Xu, H.; Chen, X.; Li, Q. Rapid analysis of neurotransmitters in rat brain using ultra-fast liquid chromatography and tandem mass spectrometry: Application to a comparative study in normal and insomnic rats. J. Mass Spectrom. 2013, 48, 969–978. [Google Scholar] [CrossRef]
- Kovac, A.; Somikova, Z.; Zilka, N.; Novak, M. Liquid chromatography-tandem mass spectrometry method for determination of panel of neurotransmitters in cerebrospinal fluid from the rat model for tauopathy. Talanta 2014, 119, 284–290. [Google Scholar] [CrossRef]
- Zhao, L.; Zheng, S.; Su, G.; Lu, X.; Yang, J.; Xiong, Z.; Wu, C. In vivo study on the neurotransmitters and their metabolites change in depressive disorder rat plasma by ultra high performance liquid chromatography coupled to tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2015, 988, 59–65. [Google Scholar] [CrossRef]
- Gemperline, E.; Chen, B.; Li, L. Challenges and recent advances in mass spectrometric imaging of neurotransmitters. Bioanalysis 2014, 6, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Fathi, F.; Brun, A.; Rott, K.H.; Falco Cobra, P.; Tonelli, M.; Eghbalnia, H.R.; Caviedes-Vidal, E.; Karasov, W.H.; Markley, J.L. NMR-based identification of metabolites in polar and non-polar extracts of avian liver. Metabolites 2017, 7, 61. [Google Scholar] [CrossRef] [PubMed]
- Nagana Gowda, G.A.; Raftery, D. Whole blood metabolomics by 1H NMR spectroscopy provides a new opportunity to evaluate coenzymes and antioxidants. Anal. Chem. 2017, 89, 4620–4627. [Google Scholar] [CrossRef] [PubMed]
- Nagana Gowda, G.A.; Raftery, D. Quantitating metabolites in protein precipitated serum using NMR spectroscopy. Anal. Chem. 2014, 86, 5433–5440. [Google Scholar] [CrossRef] [PubMed]
- Nagana Gowda, G.A.; Gowda, Y.N.; Raftery, D. Expanding the limits of human blood metabolite quantitation using NMR spectroscopy. Anal. Chem. 2015, 87, 706–715. [Google Scholar] [CrossRef] [PubMed]
- Callicott, J.H.; Straub, R.E.; Pezawas, L.; Egan, M.F.; Mattay, V.S.; Hariri, A.R.; Verchinski, B.A.; Meyer-Lindenberg, A.; Balkissoon, R.; Kolachana, B. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc. Natl. Acad. Sci. USA 2005, 102, 8627–8632. [Google Scholar] [CrossRef]
- Hamshere, M.L.; Bennett, P.; Williams, N.; Segurado, R.; Cardno, A.; Norton, N.; Lambert, D.; Williams, H.; Kirov, G.; Corvin, A. Genomewide linkage scan in schizoaffective disorder: Significant evidence for linkage at 1q42 close to DISC1, and suggestive evidence at 22q11 and 19p13. Arch. Gen. Psychiatry 2005, 62, 1081–1088. [Google Scholar] [CrossRef][Green Version]
- Hodgkinson, C.A.; Goldman, D.; Jaeger, J.; Persaud, S.; Kane, J.M.; Lipsky, R.H.; Malhotra, A.K. Disrupted in schizophrenia 1 (DISC1): Association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am. J. Hum. Genet. 2004, 75, 862–872. [Google Scholar] [CrossRef]
- Kilpinen, H.; Ylisaukko-oja, T.; Hennah, W.; Palo, O.M.; Varilo, T.; Vanhala, R.; Nieminen-von Wendt, T.; von Wendt, L.; Paunio, T.; Peltonen, L. Association of DISC1 with autism and Asperger syndrome. Mol. Psychiatry 2008, 13, 187–196. [Google Scholar] [CrossRef]
- Hashimoto, R.; Numakawa, T.; Ohnishi, T.; Kumamaru, E.; Yagasaki, Y.; Ishimoto, T.; Mori, T.; Nemoto, K.; Adachi, N.; Izumi, A. Impact of the DISC1 Ser704Cys polymorphism on risk for major depression, brain morphology and ERK signaling. Hum. Mol. Genet. 2006, 15, 3024–3033. [Google Scholar] [CrossRef]
- Zhou, Y.; Danbolt, N.C. Glutamate as a neurotransmitter in the healthy brain. J. Neural. Transm. 2014, 121, 799–817. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, B.; Javitt, D. From revolution to evolution: The glutamate hypothesis of schizophrenia and its implication for treatment. Neuropsychopharmacology 2012, 37, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Canitano, R.; Scandurra, V. Glutamatergic agents in Autism Spectrum disorders: Current trends. Res. Autism. Spectr. Disord. 2014, 8, 255–265. [Google Scholar] [CrossRef]
- Herring, B.E.; Silm, K.; Edwards, R.H.; Nicoll, R.A. Is Aspartate an Excitatory Neurotransmitter? J. Neurosci. 2015, 35, 10168–10171. [Google Scholar] [CrossRef] [PubMed]
- Errico, F.; Napolitano, F.; Squillace, M.; Vitucci, D.; Blasi, G.; de Bartolomeis, A.; Bertolino, A.; D’Aniello, A.; Usiello, A. Decreased levels of d-aspartate and NMDA in the prefrontal cortex and striatum of patients with schizophrenia. J. Psychiatr. Res. 2013, 47, 1432–1437. [Google Scholar] [CrossRef]
- Errico, F.; Mothet, J.P.; Usiello, A. D-Aspartate: An endogenous NMDA receptor agonist enriched in the developing brain with potential involvement in schizophrenia. J. Pharm. Biomed. Anal. 2015, 116, 7–17. [Google Scholar] [CrossRef]
- Ariyannur, P.S.; Arun, P.; Barry, E.S.; Andrews-Shigaki, B.; Bosomtwi, A.; Tang, H.; Selwyn, R.; Grunberg, N.E.; Moffett, J.R.; Namboodiri, A.M. Do reductions in brain N-acetylaspartate levels contribute to the etiology of some neuropsychiatric disorders? J. Neurosci. Res. 2013, 91, 934–942. [Google Scholar] [CrossRef]
- Nordengen, K.; Heuser, C.; Rinholm, J.E.; Matalon, R.; Gundersen, V. Localisation of N-acetylaspartate in oligodendrocytes/myelin. Brain Struct. Funct. 2015, 220, 899–917. [Google Scholar] [CrossRef]
- Singhal, N.K.; Huang, H.; Li, S.; Clements, R.; Gadd, J.; Daniels, A.; Kooijman, E.E.; Bannerman, P.; Burns, T.; Guo, F. The neuronal metabolite NAA regulates histone H3 methylation in oligodendrocytes and myelin lipid composition. Exp. Brain Res. 2017, 235, 279–292. [Google Scholar] [CrossRef]
- Jakaria, M.; Azam, S.; Haque, M.E.; Jo, S.H.; Uddin, M.S.; Kim, I.S.; Choi, D.K. Taurine and its analogs in neurological disorders: Focus on therapeutic potential and molecular mechanisms. Redox Biol. 2019, 24, 101223. [Google Scholar] [CrossRef]
- Norkett, R.; Modi, S.; Kittler, J.T. Mitochondrial roles of the psychiatric disease risk factor DISC1. Schizophr. Res. 2017, 187, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Van, Q.N.; Issaq, H.J.; Jiang, Q.; Li, Q.; Muschik, G.M.; Waybright, T.J.; Lou, H.; Dean, M.; Uitto, J.; Veenstra, T.D. Comparison of 1D and 2D NMR spectroscopy for metabolic profiling. J. Proteome Res. 2008, 7, 630–639. [Google Scholar] [CrossRef] [PubMed]
- Sturm, R.M.; Greer, T.; Woodards, N.; Gemperline, E.; Li, L. Mass spectrometric evaluation of neuropeptidomic profiles upon heat stabilization treatment of neuroendocrine tissues in crustaceans. J. Proteome Res. 2013, 12, 743–752. [Google Scholar] [CrossRef] [PubMed]
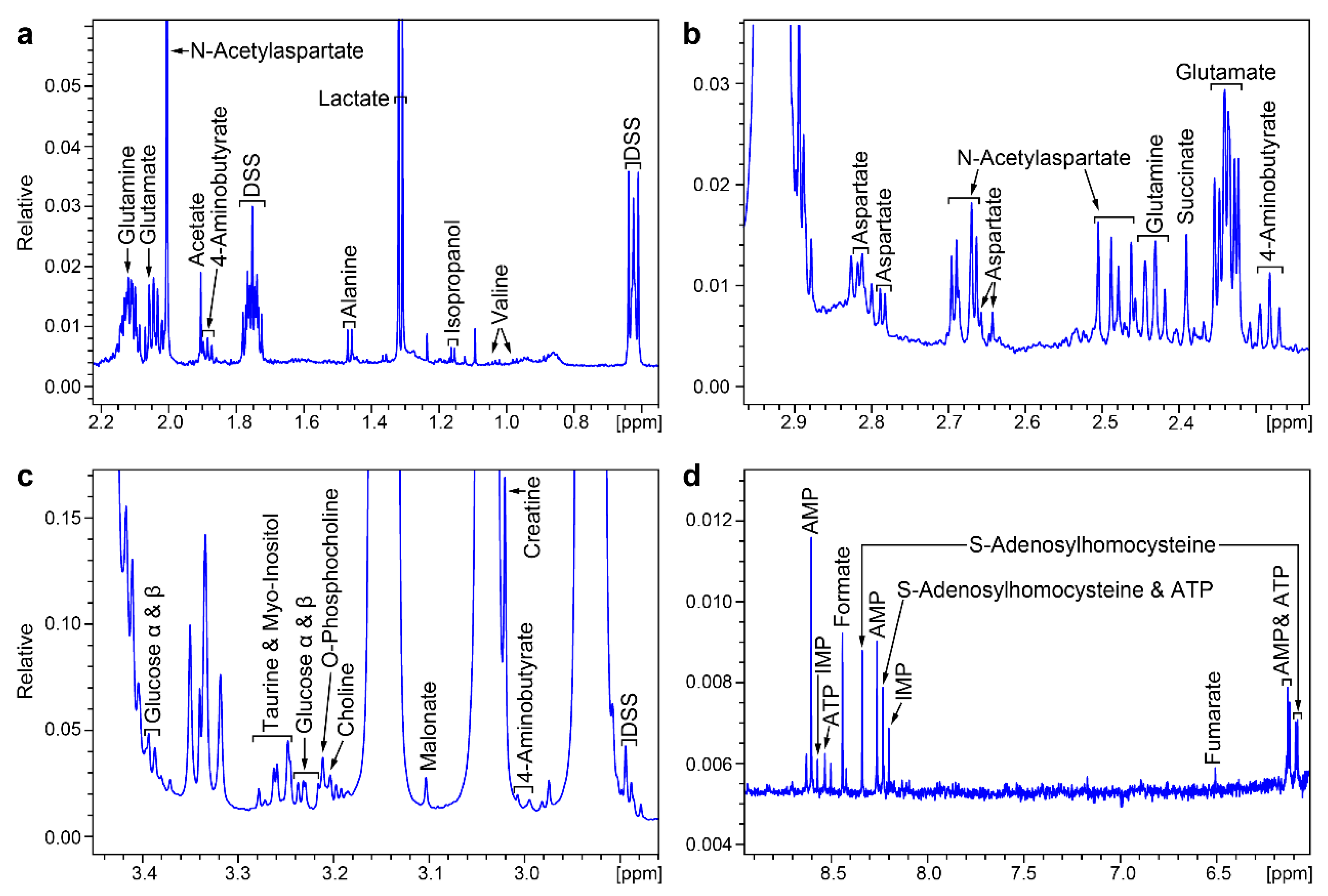
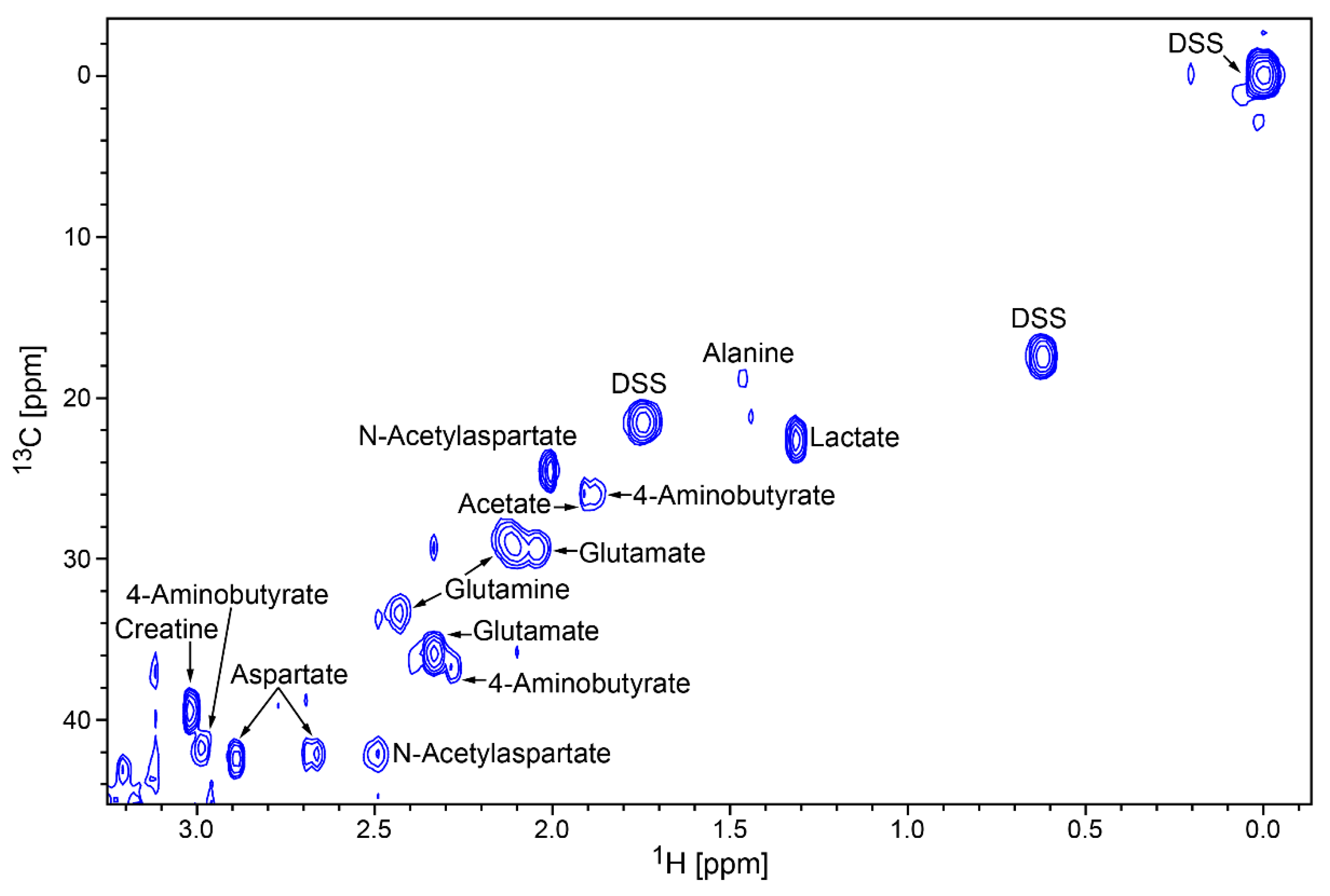
| Compound Name | Chemical Shifts and Multiplicities | Notes |
|---|---|---|
| 4-Aminobutyrate (GABA) | 1.9 m, 2.3 t, 3 t | Confirmed by HSQC |
| AMP | 4.01 m, 4.36 m, 4.50 q, 4.79 t, 6.12 d, 8.25 s, 8.58 s | Concentration too low to detect by HSQC |
| ATP | 4.23 m, 4.27 m, 4.56 t, 4.73 t, 6.12 d, 8.24 s, 8.49 s | Concentration too low to detect by HSQC |
| Acetate | 1.9 s | Confirmed by HSQC |
| Alanine | 1.47 d,3.78 q | Confirmed by HSQC |
| Aspartate | 2.66 dd, 2.80 dd, 3.91 dd | Confirmed by HSQC |
| Choline | 3.19 s, 3.50 dd, 4.05 t | Overlapped by SYN-PER peaks |
| Creatine | 3.02 s, 3.91 s | Confirmed by HSQC |
| Formate | 8.44 s | Concentration too low to detect by HSQC |
| Glucose | 3.25 m, 3.42 m, 3.49 m, 3.50 m, 3.54 m, 3.72 m, 3.73 m, 3.77 m, 3.87 m, 3.88 m, 4.66 d, 5.23 d | Confirmed by HSQC; close to SYN-PER area |
| Glutamate | 2.04 m, 2.12 m, 2.32 m, 2.32 m, 3.76 dd | Confirmed by HSQC |
| Glutamine | 2.15 m, 2.18 m, 2.42 m, 2.46 m, 3.76 t | Confirmed by HSQC |
| IMP | 8.53 s, 8.21 s, 6.13 d, 4.49 t, 4.36 m, 4.03 m | Concentration too low to detect by HSQC |
| Lactate | 1.31 d, 4.10 q | Confirmed by HSQC |
| N-Acetylaspartate | 2.1 s, 2.5 dd, 2.7 dd, 4.4 m | Confirmed by HSQC |
| O-Phosphocholine | 3.21 s, 3.58 m, 4.16 m | Confirmed by HSQC; close to SYN-PER area |
| S-Adenosyl-homocysteine | 2.1 m, 2.7 t, 3.0 q, 3.1 q, 3.8 q, 4.3 m, 4.4 t, 4.9 t, 8.10 d, 8.26 s, 8.33 s | Concentration too low to detect by HSQC |
| Succinate | 2.39 s | Concentration too low to detect by HSQC |
| Taurine | 3.26 t, 3.43 t | Confirmed by HSQC; close to SYN-PER area |
| Myo-Inositol | 3.3 t, 3.5 dd, 3.6 t, 4.1 t | Confirmed by HSQC; close to SYN-PER area |
| Metabolite 1 | Mean (mM) | p-Value | Mean (mM) | p-Value | ||
|---|---|---|---|---|---|---|
| Control Male | Disc1 svΔ2 Male | Male | Control Female | Disc1 svΔ2 Female | Female | |
| GABA | 0.08980 | 0.07940 | 0.66 | 0.06698 | 0.07175 | 0.79 |
| AMP | 0.01140 | 0.03762 | 0.01 | 0.01423 | 0.03068 | 0.02 |
| ATP | 0.00613 | 0.00757 | 0.46 | 0.00545 | 0.00638 | 0.38 |
| Acetate | 0.03620 | 0.03483 | 0.78 | 0.03195 | 0.03393 | 0.10 |
| Alanine | 0.02308 | 0.03030 | 0.38 | 0.01948 | 0.02625 | 0.23 |
| Aspartate | 0.09735 | 0.13747 | 0.19 | 0.07712 | 0.12468 | 0.03 |
| Choline | 0.01995 | 0.01900 | 0.87 | 0.01525 | 0.01517 | 0.98 |
| Creatine | 0.33330 | 0.49193 | 0.13 | 0.28313 | 0.41097 | 0.07 |
| Formate | 0.02252 | 0.02087 | 0.31 | 0.02390 | 0.02177 | 0.57 |
| Glucose | 0.40657 | 0.42493 | 0.30 | 0.42541 | 0.42785 | 0.94 |
| Glutamate | 0.37868 | 0.65835 | 0.07 | 0.34222 | 0.58792 | 0.04 |
| Glutamine | 0.18277 | 0.24677 | 0.24 | 0.14740 | 0.21837 | 0.11 |
| IMP | 0.00325 | 0.00815 | 0.04 | 0.00445 | 0.00673 | 0.10 |
| Lactate | 0.24647 | 0.34892 | 0.16 | 0.20898 | 0.29948 | 0.09 |
| N-Acetyl-aspartate | 0.21753 | 0.36463 | 0.08 | 0.19398 | 0.32667 | 0.04 |
| O-Phospho-choline | 0.01983 | 0.02862 | 0.14 | 0.01772 | 0.02702 | 0.14 |
| S-Adenosyl-homo-cysteine | 0.01528 | 0.01585 | 0.91 | 0.01328 | 0.01383 | 0.87 |
| Succinate | 0.01125 | 0.01980 | 0.06 | 0.01247 | 0.01703 | 0.14 |
| Taurine | 0.18560 | 0.29490 | 0.11 | 0.15967 | 0.25010 | 0.06 |
| myo-Inositol | 0.22202 | 0.33798 | 0.09 | 0.21327 | 0.27798 | 0.18 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barnett, B.R.; Fathi, F.; Falco Cobra, P.; Yi, S.Y.; Anderson, J.M.; Eghbalnia, H.R.; Markley, J.L.; Yu, J.-P.J. Metabolic Changes in Synaptosomes in an Animal Model of Schizophrenia Revealed by 1H and 1H,13C NMR Spectroscopy. Metabolites 2020, 10, 79. https://doi.org/10.3390/metabo10020079
Barnett BR, Fathi F, Falco Cobra P, Yi SY, Anderson JM, Eghbalnia HR, Markley JL, Yu J-PJ. Metabolic Changes in Synaptosomes in an Animal Model of Schizophrenia Revealed by 1H and 1H,13C NMR Spectroscopy. Metabolites. 2020; 10(2):79. https://doi.org/10.3390/metabo10020079
Chicago/Turabian StyleBarnett, Brian R., Fariba Fathi, Paulo Falco Cobra, Sue Y. Yi, Jacqueline M. Anderson, Hamid R. Eghbalnia, John L. Markley, and John-Paul J. Yu. 2020. "Metabolic Changes in Synaptosomes in an Animal Model of Schizophrenia Revealed by 1H and 1H,13C NMR Spectroscopy" Metabolites 10, no. 2: 79. https://doi.org/10.3390/metabo10020079
APA StyleBarnett, B. R., Fathi, F., Falco Cobra, P., Yi, S. Y., Anderson, J. M., Eghbalnia, H. R., Markley, J. L., & Yu, J.-P. J. (2020). Metabolic Changes in Synaptosomes in an Animal Model of Schizophrenia Revealed by 1H and 1H,13C NMR Spectroscopy. Metabolites, 10(2), 79. https://doi.org/10.3390/metabo10020079






