NMR-Based Metabolomic Analysis and Microbial Composition of Soil Supporting Burkea africana Growth
Abstract
1. Introduction
2. Results
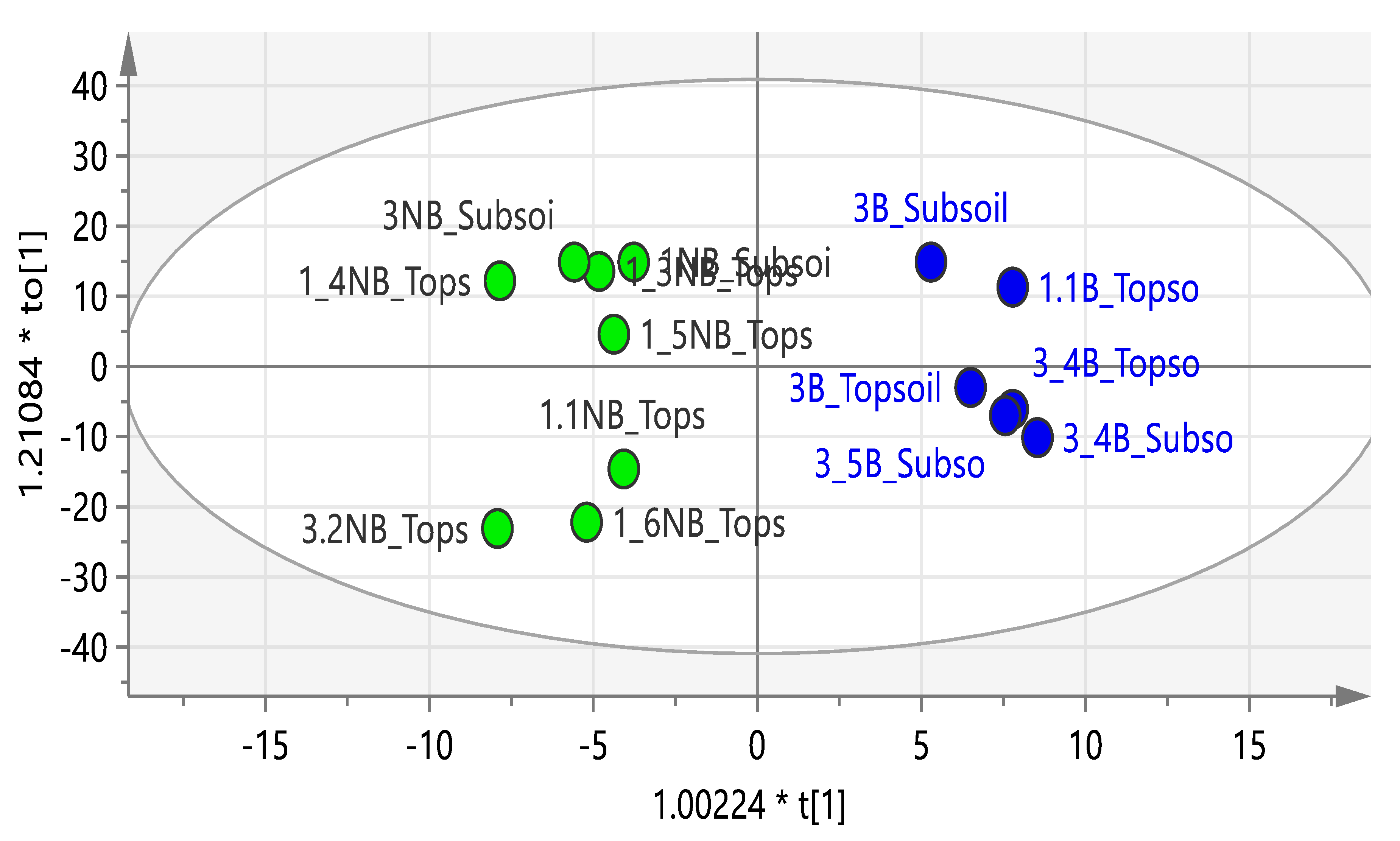
2.1. Composition of Soils
2.2. Annotation of Compounds
2.3. Identification of Annotated Metabolites
2.4. Bacterial Community Composition
Order and Family and Species Classification
2.5. Taxonomical Kingdom and Phylum Classification of Fungal Composition
2.5.1. Order and Family Classification
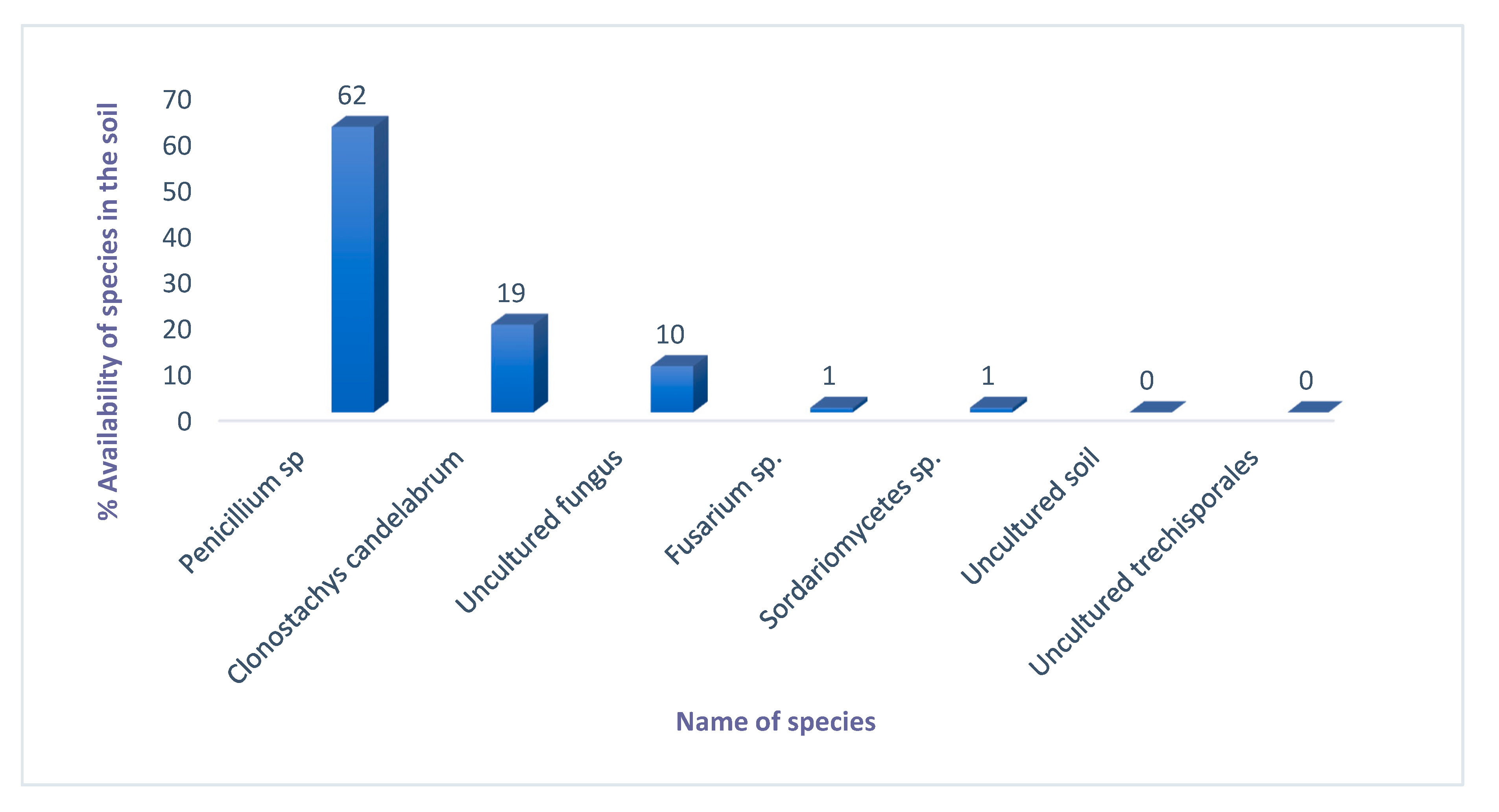
2.5.2. Identification of Fungal Species
3. Discussion
4. Materials and Methods
4.1. Sampling Site
4.2. Soil Collection
4.3. Soil Nutrient Analysis
4.4. Statistical Analysis
4.5. NMR Metabolomics Analysis
4.6. Identification of Compounds
4.7. LC-MS Analysis
4.8. Soil Genome and Microbial Community Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Marschner, H. Mineral Nutrition of Higher Plants; Academic Press: San Diego, CA, USA, 1995. [Google Scholar]
- Witkowski, E.T.F.; Lamont, B.B. Disproportionate allocation of mineral nutrients and carbon between vegetative and reproductive structures in Banksia hookeriana. Oecologia 1996, 105, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.G.; Witkowski, E.T.F. Seed banks, bark thickness and change in age and size structure (1978–1999) of the African savannah tree, Burkea africana. Plant Ecol. 2003, 167, 151–162. [Google Scholar] [CrossRef]
- Malterud, K.E. Ethnopharmacology, Chemistry and Biological Properties of Four Malian Medicinal Plants. Plants 2017, 6, 11. [Google Scholar] [CrossRef]
- Mathisen, E.; Diallo, D.; Malterud, K.E. Antioxidants from the bark of Burkea africana, an African medicinal plant. Phytother. Res. 2002, 16, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef]
- Gowda, G.A.N.; Zhang, S.; Gu, H.; Asiago, V.; Shanaiah, N.; Raftery, D. Metabolomics-based methods for early disease diagnostics. Expert Rev. Mol. Diagn. 2008, 8, 617–633. [Google Scholar] [CrossRef]
- Wardle, D.A.; Bardgett, R.D.; Klironomos, J.N.; Setälä, H.; Van Der Putten, W.H.; Wall, D.H. Ecological Linkages Between Aboveground and Belowground Biota. Science 2004, 304, 1629–1633. [Google Scholar] [CrossRef]
- Wagg, C.; Bender, S.F.; Widmer, F.; Van Der Heijden, M.G.A. Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Natl. Acad. Sci. USA 2014, 111, 5266–5270. [Google Scholar] [CrossRef]
- Wardle, D.A.; Bonner, K.I.; Barker, G.M. Linkages between plant litter decomposition, litter quality, and vegetation responses to herbivores. Funct. Ecol. 2002, 16, 585–595. [Google Scholar] [CrossRef]
- Adl, M.S.; Gupta, V.S. Protists in soil ecology and forest nutrient cycling. Can. J. For. Res. 2006, 36, 1805–1817. [Google Scholar] [CrossRef]
- Bennett, J.A.; Cahill, J.F. Fungal effects on plant-plant interactions contribute to grassland plant abundances: Evidence from the field. J. Ecol. 2016, 104, 755–764. [Google Scholar] [CrossRef]
- Hiiesalu, I.; Partel, M.; Dvison, J.; Gerhold, P.; Metsis, M.; Moora, M. Species richness of arbuscular mycorhizal fungi: Associations with grassland plant richness and biomass. New Phytol. 2014, 203, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Pellissier, L.; Niculita-Hirzel, H.; Dubuis, A.; Pagni, M.; Guex, N.; Ndiribe, C.; Salamin, N.; Xenarios, I.; Goudet, J.; Sanders, I.R.; et al. Soil fungal communities of grasslands are environmentally structured at a regional scale in the Alps. Mol. Ecol. 2014, 23, 4274–4290. [Google Scholar] [CrossRef] [PubMed]
- Bahram, M.; Põlme, S.; Kõljalg, U.; Zarre, S.; Tedersoo, L. Regional and local patterns of ectomycorrhizal fungal diversity and community structure along an altitudinal gradient in the Hyrcanian forests of northern Iran. New Phytol. 2011, 193, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Peay, K.G.; Baraloto, C.; Fine, P.V.A. Strong coupling of plant and fungal community structure across western Amazonian rainforests. ISME J. 2013, 7, 1852–1861. [Google Scholar] [CrossRef]
- Jumpponen, A.; Jones, K.L. Massively parallel 454 sequencing indicates hyperdiverse fungal communities in temperateQuercus macrocarpaphyllosphere. New Phytol. 2009, 184, 438–448. [Google Scholar] [CrossRef]
- Zheng, C.; Zhang, S.; Ragg, S.; Raftery, D.; Vitek, O. Identification and quantification of metabolites in 1H NMR spectra by Bayesian model selection. J Bioinform. Adv. 2011, 27, 1637–1644. [Google Scholar] [CrossRef]
- Nicholson, J.; Lindon, J.; Holmes, E. “Metabolomics”: Understanding the metabolomics response of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef]
- Lu, W.; Clasquin, M.F.; Melamud, E.; Amador-Noguez, D.; Caudy, A.A.; Rabinowitz, J.D. Metabolomic Analysis via Reversed-Phase Ion-Pairing Liquid Chromatography Coupled to a Stand Alone Orbitrap Mass Spectrometer. Anal. Chem. 2010, 82, 3212–3221. [Google Scholar] [CrossRef]
- Figueira, J.; Gouveia-Figueira, S.; Öhman, C.; Holgerson, P.L.; Nording, M.L.; Öhman, A. Metabolite quantification by NMR and LC-MS/MS reveals differences between unstimulated, stimulated, and pure parotid saliva. J. Pharm. Biomed. Anal. 2017, 140, 295–300. [Google Scholar] [CrossRef]
- Ulrich, E.L.; Akutsu, H.; Doreleijiers, J.F.; Harano, Y.; Ioannidis, Y.E.; Lin, J.; Mading, S.; Maziuk, D.; Miller, Z.; Nakatani, E.; et al. BioMagResBank. Nucleic Acids Res. 2008, 36, D402–D408. [Google Scholar] [CrossRef]
- Weljie, A.M.; Newton, J.; Mercier, P.; Carlson, A.E.; Slupsky, C.M. Targeted Profiling: Quantitative Analysis of1H NMR Metabolomics Data. Anal. Chem. 2006, 78, 4430–4442. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Zaidi, A.; Ahemad, M.; Oves, M.; Wani, P.A. Plant growth promotion by phosphate solubilizing fungi—Current perspective. Arch. Agron. Soil Sci. 2010, 56, 73–98. [Google Scholar] [CrossRef]
- Britto, D.T.; Kronzucker, H.J. NH4+ toxicity in higher plants: A critical review. J. Plant Physiol. 2002, 159, 567–584. [Google Scholar] [CrossRef]
- Cedergren, N.; Madsen, T.V. Nitrogen uptake by the floating macrophyte Lemma minor. J. New Phytol. 2002, 155, 285–292. [Google Scholar] [CrossRef]
- Tylová, E.; Lorenzen, B.; Brix, H.; Votrubova, O. The effects of NH4+ and NO3− on growth, resource allocation and nitrogen uptake kinetics of Phragmites australis and Glyceria maxima. Aquat. Bot. 2005, 81, 326–342. [Google Scholar] [CrossRef]
- Fang, Y.Y.; Babourina, O.; Rengel, Z.; Yang, X.E.; Pu, P.M. Ammonium and nitrate uptake by the floating plant Landoltia punctate. Ann. Bot. 2007, 99, 365–370. [Google Scholar] [CrossRef]
- Ashraf, M.; Foolad, M. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Matassini, C.; Parmeggiani, C.; Cardona, F. New Frontiers on Human Safe Insecticides and Fungicides: An Opinion on Trehalase Inhibitors. Molecules 2020, 25, 3013. [Google Scholar] [CrossRef]
- Elbein, A.D. The metabolism of α,α-trehalose. Adv. Carbohyd. Chem. Biochem. 1974, 30, 227–256. [Google Scholar]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; Van Dijck, P.; Stitt, M. Trehalose metabolism in plants. Plant J. 2014, 79, 544–567. [Google Scholar] [CrossRef] [PubMed]
- Crowe, J.H. Trehalose as a “chemical champerone”. Fact and fantasy. In Molecular Aspects of the Stress Response; Springer: Berlin/Heidelberg, Germany, 2007; Volume 594, pp. 143–158. [Google Scholar]
- Gancedo, C.; Flores, C.-L. The importance of a functional trehalose biosynthetic pathway for the life of yeasts and fungi. FEMS Yeast Res. 2004, 4, 351–359. [Google Scholar] [CrossRef]
- Valliyodan, B.; Nguyen, H.T. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol. 2006, 9, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Purvis, J.E.; Yomano, L.P.; Ingram, L.O. Enhanced Trehalose Production Improves Growth of Escherichia coli under Osmotic Stress. Appl. Environ. Microbiol. 2005, 71, 3761–3769. [Google Scholar] [CrossRef]
- Altamirano-Hernández, J.; Farías-Rodríguez, R.; Jaramillo, V.; Peña-Cabriales, J. Seasonal variation in trehalose contents of roots and nodules of leguminous trees in a tropical deciduous forest in Mexico. Soil Biol. Biochem. 2004, 36, 869–871. [Google Scholar] [CrossRef]
- Reina-Bueno, M.; Argandoña, M.; Nieto, J.J.; Garcia, A.H.; Iglesias-Guerra, F.; Delgado, M.J.; Vargas, C. Role of trehalose in heat and desiccation tolerance in the soil bacterium Rhizobium etli. BMC Microbiol. 2012, 12, 207. [Google Scholar] [CrossRef]
- Holmström, K.; Somersalo, S.; Mandal, A.; Palva, E.T.; Welin, B. Improved tolerance to salinity and low temperature in transgenic tobacco producing glycine betaine. J. Exp. Bot. 2000, 51, 177–185. [Google Scholar] [CrossRef]
- Ali, Q.; Ashraf, M. Induction of Drought Tolerance in Maize (Zea mays L.) due to Exogenous Application of Trehalose: Growth, Photosynthesis, Water Relations and Oxidative Defence Mechanism. J. Agron. Crop. Sci. 2011, 197, 258–271. [Google Scholar] [CrossRef]
- Holtmann, G.J.; Bremer, E. Thermoprotection of Bacillus subtilis by Exogenously Provided Glycine Betaine and Structurally Related Compatible Solutes: Involvement of Opu Transporters. J. Bacteriol. 2004, 186, 1683–1693. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, J.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0—The Human Metabolome Database. Nucleic Acids Res. 2003, 41, D801–D807. [Google Scholar] [CrossRef]
- Ichihashi, Y.; Date, Y.; Shino, A.; Shimizu, T.; Shibata, A.; Kumaishi, K.; Funahashi, F.; Wakayama, K.; Yamazaki, K.; Umezawa, A.; et al. Multi-omics analysis on an agroecosystem reveals the significant role of organic nitrogen to increase agricultural crop yield. Proc. Natl. Acad. Sci. USA 2020, 117, 14552–14560. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, Y.; Ogasawara, M.; Kim, S.; Konnai, M.; Takematsu, T.; Suzuki, A.; Hyeon, S.; Che, F.; Furushima, M. Promotive effect of Choline salts on groth of manila grass and bent grass. J. Jpn. Soc. Turf. Sci. 1990, 19, 15–21. [Google Scholar]
- Liu, H.; Cheng, S.; Logan, B.E. Production of Electricity from Acetate or Butyrate Using a Single-Chamber Microbial Fuel Cell. Environ. Sci. Technol. 2005, 39, 658–662. [Google Scholar] [CrossRef] [PubMed]
- Leroy, F.; De Vuyst, L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 2004, 15, 67–78. [Google Scholar] [CrossRef]
- Yamanaka, T. Corrosion by bacteria of concrete in sewerage systems and inhibitory effects of formates on their growth. Water Res. 2002, 36, 2636–2642. [Google Scholar] [CrossRef]
- Inoki, K.; Zhu, T.; Guan, K.-L. TSC2 NMediates cellular energy response to control cell growth and survival. J. Cell Sci. 2003, 115, 577–590. [Google Scholar]
- Kumari, K.; Singh, R.P.; Saxena, S.K. Effect of Different Factors on the Movement of Some Amino Acids in Soils Using Thin-Layer Chromatography. J. Liq. Chromatogr. 1987, 10, 1299–1325. [Google Scholar] [CrossRef]
- Zhang, H.; Jennings, A.; Barlow, P.W.; Forde, B.G. Dual pathways for regulation of root branching by nitrate. Proc. Natl. Acad. Sci. USA 1999, 96, 6529–6534. [Google Scholar] [CrossRef]
- Masclaux-Daubresse, C.; Reisdorf-Cren, M.; Pageau, K.; Lelandais, M.; Grandjean, O.; Kronenberger, J.; Valadier, M.-H.; Feraud, M.; Jouglet, T.; Suzuki, A. Glutamine sythetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source Nitrogen cycle in Tobacco. J. Plant Physiol. 2006, 140, 444–456. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef]
- Torsvik, V.; Goksøyr, J.; Daae, F.L. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 1990, 56, 782–787. [Google Scholar] [CrossRef] [PubMed]
- Nüsslein, K.; Tiedje, J.M. Soil Bacterial Community Shift Correlated with Change from Forest to Pasture Vegetation in a Tropical Soil. Appl. Environ. Microbiol. 1999, 65, 3622–3626. [Google Scholar] [CrossRef] [PubMed]
- Ward, N.L.; Challacombe, J.F.; Janssen, P.H.; Henrissat, B.; Coutinho, P.M.; Wu, M.; Xie, G.; Haft, D.H.; Sait, M.; Badger, J.; et al. Three Genomes from the Phylum Acidobacteria Provide Insight into the Lifestyles of These Microorganisms in Soils. Appl. Environ. Microbiol. 2009, 75, 2046–2056. [Google Scholar] [CrossRef] [PubMed]
- Borneman, J.; Triplett, E.W. Molecular microbial diversity in soils from eastern Amazonia: Evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 1997, 63, 2647–2653. [Google Scholar] [CrossRef] [PubMed]
- Visagie, C.M.; Seifert, K.A.; Houbraken, J. Diversity of Penicillium section Citrina within the fynbos biome of South Africa, including a new species from a Protea repens infructescence. J Mycol. 2014, 106, 537–552. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Stud. Mycol. 2004, 49, 1–174. [Google Scholar]
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage; Springer: New York, NY, USA, 2009. [Google Scholar]
- Samson, R.A.; Yilmaz, N.; Houbraken, J. Phylogeny and nomenculture of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud. Mycol. 2011, 70, 159–183. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Smedsgaard, J.; Larsen, T.O. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud. Mycol. 2004, 49, 201–241. [Google Scholar]
- Fleming, A. On the Antibacterial Action of Cultures of a Penicillium, with Special Reference to Their Use in the Isolation of B. influenzae. Clin. Infect. Dis. 1980, 2, 129–139. [Google Scholar] [CrossRef]
- Chain, E.; Florey, H.; Gardner, A.; Heatley, N.; Jennings, M.; Orr-Ewing, J.; Sanders, A. Penicillin As a Chemotherapeutic Agent. Lancet 1940, 236, 226–228. [Google Scholar] [CrossRef]
- Abraham, E.; Chain, E.; Fletcher, C.; Gardner, A.; Heatley, N.; Jennings, M.; Florey, H. Further Observations on Penicillin. Lancet 1941, 238, 177–189. [Google Scholar] [CrossRef]
- Thom, C. Mycology present penicillin. J. Mycol. 1945, 37, 460–475. [Google Scholar] [CrossRef]
- Van Der Heijden, E.W.; Kuyper, T.W. Ecological strategies of ectomycorrhizal fungi of Salix repens: Root manipulation versus root replacement. Oikos 2003, 103, 668–680. [Google Scholar] [CrossRef]
- Baxter, J.W.; Dighton, J. Phosphorus source alters host plant response to ectomycorrhizal diversity. Mycorrhiza 2005, 15, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Broeckling, C.D.; Broz, A.K.; Bergelson, J.; Manter, D.K.; Vivanco, J.M. Root Exudates Regulate Soil Fungal Community Composition and Diversity. Appl. Environ. Microbiol. 2007, 74, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Seastedt, I.T.; Callaway, R.M.; Pollock, J.L.; Kaur, J. Allelopathy and plant invasions: Traditional, congeneric, and bio-geographical approaches. Biol. Invasions 2008, 10, 875–890. [Google Scholar] [CrossRef]
- Waqas, M.; Khan, A.; Kang, S.-M.; Kim, Y.; Lee, S.-U. Phytohormone-producing fungal endophytes and hardwood-derived biochar interact to ameliorate heavy metal stress in soybeans. Biol. Fertil. Soils 2014, 50, 1155–1167. [Google Scholar] [CrossRef]
- Shivanna, M.B.; Meera, M.S.; Hyakumachi, M. Sterile fungi from zoysiagrass rhizosphere as plant growth promoters in spring wheat. Can. J. Microbiol. 1994, 40, 637–644. [Google Scholar] [CrossRef]
- Khan, S.A.; Hamayun, M.; Yoon, H.; Kim, H.-Y.; Suh, S.-J.; Hwang, S.-K.; Kim, J.-M.; Lee, S.-U.; Choo, Y.-S.; Yoon, U.-H.; et al. Plant growth promotion and Penicillium citrinum. BMC Microbiol. 2008, 8, 231. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Kang, S.-M.; Baek, I.-Y.; Lee, S.-U. Characterization of plant growth-promoting traits of Penicillium species against the effects of high soil salinity and root disease. J. Plant Interact. 2014, 9, 754–762. [Google Scholar] [CrossRef]
- Hossain, M.; Sultana, F.; Kubota, M.; Koyama, H.; Hyakumachi, M. The Plant Growth-Promoting Fungus Penicillium simplicissimum GP17-2 Induces Resistance in Arabidopsis thaliana by Activation of Multiple Defense Signals. Plant Cell Physiol. 2007, 48, 1724–1736. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.L.; Hamayun, M.; Ahmad, N.; Hussain, J.; Kang, S.-M.; Kim, Y.-H.; Adnan, M.; Tang, D.-S.; Wagas, M.; Radhakrishnan, R.; et al. Salinity Stress Resistance Offered by Endophytic Fungal Interaction Between Penicillium minioluteum LHL09 and Glycine max. J. Microbiol. Biotechnol. 2011, 21, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Swanepoel, B.; Bredenkamp, G. The Vegetation Ecology of Ezemvelo Nature Reserve, Bronkhorstspruit, South Africa. Master’s Thesis, University of Pretoria, Pretoria, South Africa, 2006. [Google Scholar]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-based metabolomic analysis of plants. Nature 2010, 5, 536–549. [Google Scholar] [CrossRef]
- Maree, J.; Viljoen, A. Phytochemical distinction between Pelargonium sidoides and Pelargonium reniforme—A quality control perspective. S. Afr. J. Bot. 2012, 82, 83–91. [Google Scholar] [CrossRef]
- Mediani, A.; Abas, F.; Khatib, A.; Maulidiani, M.; Shaari, K.; Choi, Y.H.; Lajis, N. 1H-NMR-based metabolomics approach to understanding the drying effects on the phytochemicals in Cosmos caudatus. Food Res. Int. 2012, 49, 763–770. [Google Scholar] [CrossRef]
- Fernie, A.R.; Aharoni, A.; Wilmitzer, L.; Stutt, M.; Tohge, T.; Kopka, J.; Carol, A.J.; Saito, K.; Fraser, P.D.; Deluca, V. Recommendations for reporting metabolite data. Plant Cell. 2011, 23, 2477–2482. [Google Scholar] [CrossRef]
- Suzuki, M.; Nishiumi, S.; Kobayashi, T.; Azuma, T.; Yoshida, M. LC–MS/MS-based metabolome analysis detected changes in the metabolic profiles of small and large intestinal adenomatous polyps in Apc Min/+ mice. Metabolomics 2016, 12. [Google Scholar] [CrossRef]
- Matsubara, A.; Fukusaki, E.; Bamba, T. Metabolite analysis by supercritical fluid chromatography. Bioanalysis 2010, 2, 27–34. [Google Scholar] [CrossRef]
- Xu, W.; Van Knegsel, A.; Saccenti, E.; Van Hoeij, R.; Kemp, B.; Vervoort, J. Metabolomics of Milk Reflects a Negative Energy Balance in Cows. J. Proteome Res. 2020, 19, 2942–2949. [Google Scholar] [CrossRef]
- Yang, J.; Schmelzer, K.; Georgi, K.; Hammock, B.D. Quantitative Profiling Method for Oxylipin Metabolome by Liquid Chromatography Electrospray Ionization Tandem Mass Spectrometry. Anal. Chem. 2009, 81, 8085–8093. [Google Scholar] [CrossRef]


| Soils | |||
|---|---|---|---|
| Nutrient (mg/kg) | Burkea | Non-Burkea | SEM |
| Fe | 30.26 | 20.27 | 6.070 |
| Mn | 27.38 | 24.76 | 5.964 |
| pH | 4.72 | 4.81 | 0.047 |
| P | 5.82 | 7.04 | 2.844 |
| C | 52.86 | 55.05 | 8.536 |
| Mg | 35.89 | 23.4 | 6.081 |
| K | 64.65 | 74.74 | 7.830 |
| Na | 1.78 | 2.56 | 0.917 |
| Total nitrogen | 0.13 | 0.16 | 0.001 |
| N-NO3− | 7.06 a | 5.35 b | 2.423 |
| N-NH4+ | 9.40 a | 7.11 b | 0.912 |
| Organic matter | 3.40 | 2.57 | 0.964 |
| Metabolite | NMR Region (ppm) | Chenomx | HMDB |
|---|---|---|---|
| Burkea Soil | |||
| Trehalose | 3.46 | 3.6 | 3.42 |
| 3.65 | 3.8 | 3.44 | |
| 3.83 | 3.9 | 3.65 | |
| 3.86 | 5.2 | 3.83 | |
| 3.88 | 3.87 | ||
| 5.2 | 5.18 | ||
| Betaine | 3.27 | 3.3 | 3.25 |
| 3.9 | 3.9 | 3.89 | |
| Choline-like | 3.10 | 3.1 | 3.1 |
| Carnitine-like | 3.1 | 3.1 | 3.1 |
| Non-Burkea Soil | |||
| Acetate | 1.92 | 1.9 | 1.91 |
| Formate | 8.46 | 8.4 | 8.44 |
| Lactate | 4.1 | 1.3 4.1 | 1.3 4.1 |
| Metabolites | Burkea | Non-Burkea | SEM |
|---|---|---|---|
| Aspartic acid | 54,572 a | 16,087 b | 1739.845 |
| Serine | 366,826 a | 170,977 b | 14,905.66 |
| 4-Hydroxyproline | 10,540 a | 7643 b | 447.987 |
| Glycine | 54,636 | 50,491 | 6662.825 |
| Glutamine | 130,176 a | 60,259 b | 6662.825 |
| Threonine | 66,867 a | 16,025 b | 1224.934 |
| Glutamic acid | 82,005 a | 13,149 b | 4022.601 |
| Citrulline | 22,565 a | 3089 b | 947.719 |
| Proline | 128,680 a | 26,096 b | 4530.320 |
| Lysine | 49075 a | 14,903 b | 2984.130 |
| Guanosine | 81,611 a | 36,711 b | 2731.467 |
| Cytidine | 35,296 | 34,131 | 1692.545 |
| Adenine | 134,064 | 125,639 | 19,526.543 |
| Tyrosine | 33,072 a | 16,720 b | 1379.953 |
| Adenosine | 768,010 a | 547,474 b | 18,855.132 |
| Isoleucine | 322,050 a | 72,972 b | 6094.5899 |
| Leucine | 282,818 | 257,661 | 7148.160 |
| Phenylalanine | 137,183 a | 35,000 b | 7738.684 |
| Acetylcarnitine | 75,057 | 63,587 | 366.627 |
| Tryptophan | 41,518 a | 15,248 b | 1480.327 |
| Fumaric acid | 336,355 | 335,903 | 4209.594 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nemadodzi, L.E.; Vervoort, J.; Prinsloo, G. NMR-Based Metabolomic Analysis and Microbial Composition of Soil Supporting Burkea africana Growth. Metabolites 2020, 10, 402. https://doi.org/10.3390/metabo10100402
Nemadodzi LE, Vervoort J, Prinsloo G. NMR-Based Metabolomic Analysis and Microbial Composition of Soil Supporting Burkea africana Growth. Metabolites. 2020; 10(10):402. https://doi.org/10.3390/metabo10100402
Chicago/Turabian StyleNemadodzi, Lufuno Ethel, Jacques Vervoort, and Gerhard Prinsloo. 2020. "NMR-Based Metabolomic Analysis and Microbial Composition of Soil Supporting Burkea africana Growth" Metabolites 10, no. 10: 402. https://doi.org/10.3390/metabo10100402
APA StyleNemadodzi, L. E., Vervoort, J., & Prinsloo, G. (2020). NMR-Based Metabolomic Analysis and Microbial Composition of Soil Supporting Burkea africana Growth. Metabolites, 10(10), 402. https://doi.org/10.3390/metabo10100402






