Abstract
Semaglutide, a glucagon-like peptide-1 (GLP-1) receptor agonist, has gained considerable attention as a therapeutic agent for type 2 diabetes mellitus and obesity. Despite its clinical success, the precise mechanisms underlying its pharmacological effects remain incompletely understood. In this study, we employed ligand-based drug design strategies to investigate potential off-target interactions of semaglutide. Through a comprehensive in silico screening of semaglutide’s structural properties against a diverse panel of proteins, we have identified calmodulin (CaM) as a putative novel target of semaglutide. Molecular docking simulations revealed a strong interaction between semaglutide and CaM, characterized by favourable binding energies and a stable binding pose. Further molecular dynamics simulations confirmed the stability of the semaglutide–CaM complex, emphasizing the potential for a physiologically relevant interaction. In conclusion, our ligand-based drug design approach has uncovered calmodulin as a potential novel target of semaglutide. This discovery sheds light on the complex pharmacological profile of semaglutide and offers a promising direction for further research into the development of innovative therapeutic strategies for metabolic disorders. The CaM, and especially so the CaMKII, system is central in the experience of both drug- and natural-related reward. It is here hypothesized that, due to semaglutide binding, the reward pathway-based calmodulin system may be activated, and/or differently regulated. This may result in the positive semaglutide action on appetitive behaviour. Further studies are required to confirm these findings.
1. Introduction
Semaglutide (Sem), a potent glucagon-like peptide-1 (GLP-1) receptor agonist, has garnered significant attention in the field of metabolic research due to its remarkable effects on appetite regulation and weight loss. Emerging evidence suggests that the mechanisms underlying Sem’s effects on appetite and cravings may involve a broader spectrum of molecular targets than previously recognized [1,2]. While the well-established role of Sem involves activating GLP-1 receptors to enhance insulin secretion and reduce blood glucose levels, its ability to induce significant reductions in food cravings and body weight may be mediated by different receptors in humans [3]. As research continues to unravel the complex interplay of Sem within the intricate web of metabolic regulations, it becomes increasingly clear that its multifaceted effects on appetite and cravings may involve a network of receptors and signalling pathways beyond the conventional understanding of its mechanism of action. Further investigations are warranted to elucidate these novel molecular targets, shedding light on the potential for innovative therapeutic strategies for obesity and related metabolic disorders. In this article, we used a ligand-based drug design approach to identify novel targets for Sem. Ligand-based computational drug design focuses on the characteristics of a known or potential drug molecule that binds to the target protein. The main idea here is to analyse the chemical and physical properties of the drug molecule and understand how it interacts with the target protein [4]. With this information, new drug candidates that have similar chemical properties or structural features to the known ligands can be found [5,6]. PepBDB—a comprehensive structural database of all the biological peptide binding complexes with peptide lengths up to 50 residues in the Protein Data Bank was used for the research of similar peptides, and the research suggested Calmodulin (CaM), or calcium-modulated protein, as a possible target for Sem [7]. Hence, with the help of both molecular docking and dynamics simulations, we aimed here at assessing whether Sem may be tentatively considered as a calmodulin binder. However, it is crucial to acknowledge that our understanding is predominantly based on computational data, which inherently carries limitations in reflecting the complexities of biological systems. Nonetheless, recognizing that both computational methods and experimental models possess their inherent constraints, it is imperative to view this research as an initial step in the drug development process. Therefore, while the identification of potential interactions between Sem and novel targets like Calmodulin (CaM) offers valuable insights, it is pivotal to approach these findings with caution, understanding that they serve as suggestions for further experimental exploration rather than definitive conclusions regarding Sem’s pharmacological profile.
2. Results
The ligand-based approach was used here considering the following assumption: “if a molecule similar to Sem is known to interact with a particular target so should Sem itself”. To look for similar molecule to Sem, a database (PepBDB) of protein(ligand)–protein(receptor) interactions was screened to look for similar compounds. PepBDB is a comprehensive structural database (current number of structures: 13,299) of all the biological peptide binding complexes with peptide lengths up to 50 residues in the Protein Data Bank. The database was screened and interestingly several human protein ligands were identified as similar to the structure of the searched molecule. The whole set of identified proteins is included in Table S1 (Supplementary Material). Among all the investigated sequences, the recognition site for calmodulins appeared more than once and with a high similarity index (49.2 to 71.4% Peptide Sequence Identity for HEGTFTSDVSSYLEG and QAAKEFIAWLVRGRG). With a similar sequence to Sem, this means different structures were present on the PDB. This means that the structure of Sem is very similar to the recognition site for calmodulin (CaM). Calmodulin (CaM) is a multifunctional intermediate calcium-binding messenger protein expressed in all eukaryotic cells. It is an intracellular target of the secondary messenger Ca2+, and the binding of Ca2+ is required for the activation of calmodulin [8]. Once bound to Ca2+, calmodulin acts as part of a calcium signal transduction pathway by modifying its interactions with various target proteins such as kinases or phosphatases. While there are several classes of sequences that CaM can bind, calcium-loaded CaM preferentially binds to α-helical sequences, which are very similar to the sequence of Sem [9]. Intrigued by these findings, the structure of Sem (PDB ID: 4ZGM) was initially superposed on the structure of the recognition site from calcineurin (RSC) (PDB ID: 4Q5U) and the Root-mean-square deviation of atomic positions (RMSD), as reported in Figure 1, was calculated.
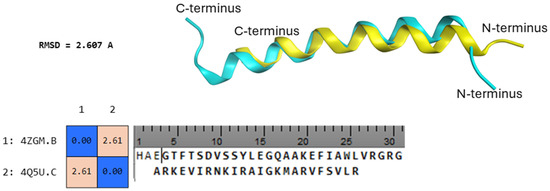
Figure 1.
(Down) Calculated RMSD between 4ZGM (Sem) and 4Q5U (RSC) and aligned sequences. (Up) In light blue, the structure of Sem superposed on the structure of the recognition site from calcineurin (yellow).
The calculated similarity after superposition was high and only 2.607 A of RMSD were calculated, which clearly is representative of high similarity with atomic positions. To further study the interaction between Sem and CaM, docking calculations (Figure 2) were performed and compared to docking calculations between the binding peptides RSC and CaM. Docking calculations were performed by flexible peptide–protein docking by fast modelling of peptide conformations and global/local sampling of binding orientations using HPEPDOCK 2.0 [10], and the results are reported in Table 1.
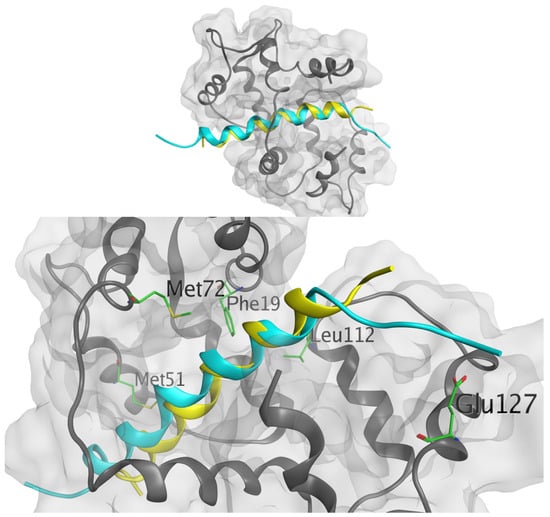
Figure 2.
(Up) Calculated docking poses by HPEPDOCK (rank 1 from Table 1) between Sem (light blue) and RSC (yellow) and CaM. (Down) Sem (light blue) and RSC (yellow) binding poses inside CaM with identified binding residues highlighted.

Table 1.
Docking score and ranking as calculated by HPEPDOCK for the complex of CaM/RSC and CaM/Sem. The values reported are the docking scores from HPEPDOCK; these do not reflect the real binding affinities, but are a relative ranking of the peptide binding.
As highlighted by the docking score of HPEPDOCK, the score between the two peptides is very similar, suggesting a similar interaction between the two studied peptides and the protein; moreover, the lowest score of −253.107 was calculated for Sem, i.e., the relative ranking for Sem seemed better than the native RSC binding peptide.
Hotspot residues, i.e., the residues that make a major contribution to the ligand binding free energy, were also calculated using fastDRH (http://cadd.zju.edu.cn/fastdrh/, accessed on 20 June 2020) and, as shown in Figure 3 and Figure 4, several residues are common—identified as hotspot residues for both ligands such as Met 51, Met 72, Leu 112 and Glu 127.
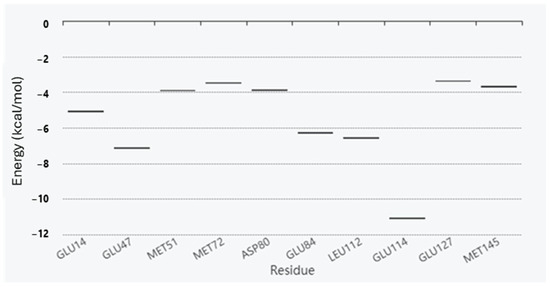
Figure 3.
Energy boxplot of the top 10 potential hotspot residues for the interaction between RSC/CaM.
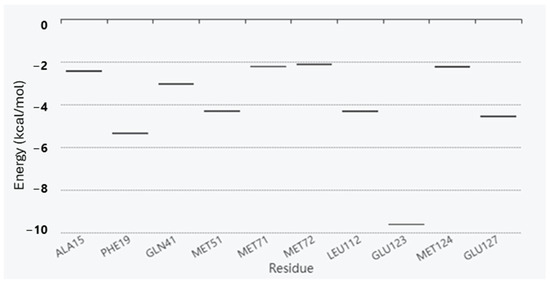
Figure 4.
Energy boxplot of the top 10 potential hotspot residues for the interaction between Sem/CaM.
To validate the anchoring point of Sem with CaM, we compared our results with the study of Ataman et al., who analysed the structures of 16 complexes of CaM bound to CaM binding peptides [11]. These authors determined that each of the residues in the FLMMN pocket (e.g., the binding pocket), Phe 19, Leu 32, Met 51, and Met 71, were in direct contact with bound peptide residues in ≥75% of the structures examined. In our structure, residues from the CaM binding peptide were in contact with Met 51 and Met 71, as already reported and discussed by Dunlap et al. [12]. Moreover, Sem is also able to interact with Phe 19 and Met 51, validating the binding poses of the molecule with CaM as consistent with the experimental observation [11].
As molecular dynamics simulations have emerged as a powerful tool in the field of structural biology and drug discovery, providing valuable insights into the dynamic behaviour of biomolecular systems at the atomic level, we employed molecular dynamics simulations to investigate the interaction between Sem and CaM. The objective of this investigation was to gain a comprehensive understanding of the stability of the Sem–CaM complex over an extended timescale. To achieve this, we conducted molecular dynamics simulations spanning a period of 300 nanoseconds (ns) using state-of-the-art computational techniques, as reported in the experimental section. Our results indicated that the Sem–CaM complex exhibited an overall stability throughout the simulation period, suggesting, by the calculated RMSD, a maintenance of the constant level (Figure 5) after the initial stabilization of the starting structure (0 ns).
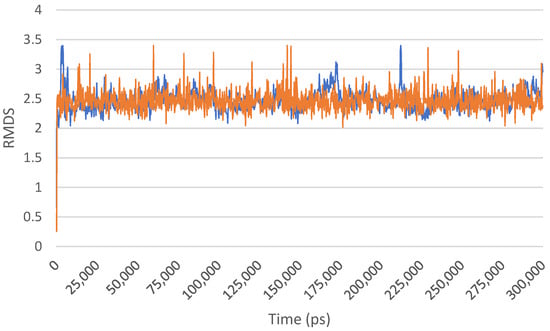
Figure 5.
RMSD of the Sem/Cam complex over the 300 ns of MD simulation. The different colours represent duplicate experiments.
During the 300 ns trajectory, we observed fluctuations in the intermolecular interactions between Sem and Cam, primarily driven by the inherent flexibility of both the peptide and the protein. However, these fluctuations remained within a physiologically acceptable range, and the complex maintained its overall structural integrity, as highlighted by the stability of the secondary structure of the simulated protein overtime, as shown Figure 6. Notably, the binding interface between Sem and CaM exhibited limited perturbation, underscoring the specificity of their interaction.
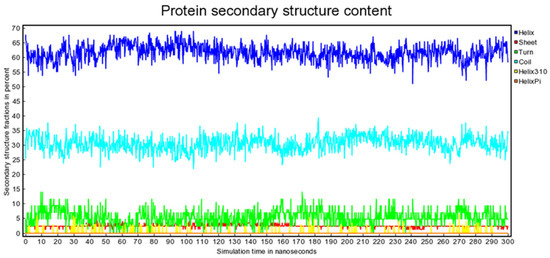
Figure 6.
Protein secondary structure content as a function of simulation time.
Furthermore, the simulations revealed critical insights into the dynamics of key residues involved in the Sem–CaM interaction, shedding light on the molecular mechanisms underpinning their association. Hotspot residues were calculated each 50 ns of MD and they are presented in Table 2. Interestingly, Met 124 and Met 144 were present among the hotspot residues; these residues, along with Phe 92, were found to be present in all the studied peptides by Ataman et al. [11], validating again the hypothesis of Sem binding to CaM. In conclusion, our molecular dynamics simulations provided levels of evidence of the overall stability of the Sem–CaM complex over a 300 ns timeframe. These findings expand our understanding of molecular interactions relating to the pharmacological activity of Sem and offer a solid foundation for future studies aimed at optimizing its therapeutic potential or developing novel drug candidates targeting this specific interaction. Such insights may well present the potential of advancing the field of diabetes research and drug discovery, ultimately benefitting patients worldwide.

Table 2.
Calculated hotspot residues over the time of MD.
3. Materials and Methods
To identify molecules similar to Sem, we screened the PepBDB database, a comprehensive structural repository containing 13,299 protein–ligand–protein complexes with peptide lengths of up to 50 residues sourced from the Protein Data Bank (PDB). Upon screening PepBDB, several human protein ligands with structural similarity to Sem were identified. The complete set of identified proteins is provided in Table S1 of the Supplementary Material. As only 20 standard amino acids are supported for sequence input in PepBDB, two different searches were performed using the following sequences: HEGTFTSDVSSYLEG and QAAKEFIAWLVRGRG. Details about the research input are reported in Figure S1. Notably, among the identified sequences, multiple instances of the recognition site for calmodulins (CaM) were observed. These sequences exhibited high similarity indices (ranging from 49.2% to 71.4% peptide sequence identity accordingly with the two different partial sequences of Sem studied) to Sem. Despite the fact that PKA, PUMA and BCL-xL actually appeared more frequently than CaM, this latter was chosen after a comparison of the secondary structure of the binding peptides. The 2D chemical structures were produced using Molecular Operating Environment (MOE), and each structure was subjected to molecular mechanics energy reduction using the MMFF94x force field of the same software [13]. PM3 Hamiltonian was then used to optimize the 3D geometry of all compounds [14], as implemented in the MOPAC 2016 package, assuming a pH of 7.0 [15]. Docking calculations were performed using HPEPDOCK 2.0 (http://huanglab.phys.hust.edu.cn/hpepdock/, accessed on 1 June 2023), with the default docking parameters [Search the database at 40% sequence identity, no Retrieve only representatives, no Search by peptide length, Include peptides with non-standard amino acids]. The CaM crystal structure was downloaded from the Protein Data Bank (www.rcsb.org, PDB ID: 4Q5U, accessed on 15 June 2023) and used for the calculations, and the structure of Sem was downloaded from the Protein Data Bank, PDB ID: 4ZGM. As the long alkyl side chain of Sem induces interaction with plasma albumin, resulting in extended half-life, and is not relevant to the interaction with the receptor, this was not considered in our study. The hotspots were calculated using fastDRH [16] using the default parameters [Hotspot prediction—Receptor force field: ff19SB (with OPC water model), Ligand force field: GAFF2, Method MM/GBSA (yield a energy decomposition on per-residues basis, Truncation radius: Retain protein residues within 20 Å of all ligand binding poses for rescoring]. The molecular dynamics simulations of the complexes were performed with YASARA [17]. A periodic simulation cell extending 10 Å from the protein surface was employed. The cell was filled with water, with a maximum sum of all water bumps of 1.0 Å and a density of 0.997 g/mL. The setup included optimizing the hydrogen bonding network [18] to increase the solute stability and a pKa prediction to fine-tune the protonation states of protein residues at the chosen pH of 7.4 [19]. With an excess of either Na or Cl to neutralize the cell, NaCl ions were supplied at a physiological concentration of 0.9%. The simulation was run using the ff14SB force field [20] for the solute, GAFF2 [21] and AM1BCC [22] for ligands and TIP3P for water. The cutoff was 10 Å for Van der Waals forces (the default used by AMBER) [23], and no cutoff was applied to electrostatic forces (using the Particle Mesh Ewald algorithm) [24]. The equations of motions were integrated with multiple time steps of 2.5 fs for bonded interactions and 5.0 fs for nonbonded interactions at a temperature of 298 K and a pressure of 1 atm using algorithms described in detail previously [25]. A short MD simulation was run on the solvent only to remove clashes. The entire system was then energy minimized using the steepest descent minimization to remove conformational stress, followed by a simulated annealing minimization until convergence (<0.01 kcal/mol Å). Finally, a 300 ns MD simulation without any restrictions was conducted, and the conformations were recorded every 200 ps, as proven effective by many other our studies [26,27].
4. Discussion
This is the first study providing levels of evidence relating to Sem posing as a calmodulin binder. Molecular docking simulations revealed here a strong interaction between Sem and CaM, characterized by favourable binding energies and a stable binding pose. Further molecular dynamics simulations confirmed the stability of the Sem–CaM complex, emphasizing the potential for a physiologically relevant interaction.
Sem, a widely used medication for the management of type 2 diabetes and obesity, exerts its therapeutic effects through several intricate mechanisms. While the primary mode of action of Sem is to mimic the activity of the naturally occurring hormone GLP-1, promoting glucose-dependent insulin secretion and reducing appetite [28], its ability to induce significant reductions in food cravings and body weight may be mediated by different receptors/proteins in humans such as specific subtypes of GLP-1 receptors or other, as yet unidentified, neural or hormonal pathways. Exploiting a ligand-based drug design approach, current research has uncovered CaM as a potential novel target of Sem.
The binding of Sem to CaM would physiologically result in a variety of events, but considering the intracellular localization of CaM, the issue of Sem internalization must be considered. In other words, one would argue if Sem can enter the cell and intracellularly exist in an active form to be able to affect CaM. Indeed, the remaining GLP-1 agonists (e.g., liraglutide and extendin-4 [29]) would be able to cross the blood–brain barrier (BBB) and to interact with GLP-r receptors in the central nervous system. Sem, a long-acting formulation derived from liraglutide’s structure, presents with a longer aliphatic chain with respect to its precursor and this improves its hydrophobicity/liphophilicity levels. Semaglutide has been structurally modified to prevent it from being metabolized by the dipeptidyl peptidase-4 (DPP-4) enzyme, thus enabling enhanced levels of binding with albumin (for an overview of the issue, see Mohapatra et al., 2022 [30]). Whilst one could argue that Sem may be unable to cross the BBB directly [31], Sem was suggested to bind with albumin and access, via the circumventricular organ, the brainstem, septal nucleus and hypothalamus [31]. It is of interest here that Sem appears to be able to cross those tanycytes (e.g., glial-like cells) which surround the ventricular wall or regions where the blood–brain barrier is absent [31]. Moreover, the GLP-1R is rapidly internalized when activated by its agonists, hence virtually localizing/repositioning the GLP-1R agonist into the cells [32].
In line with this, Sem could then arguably be able to virtually interact with any cells, with several possible results and biological consequences [33]. Conversely, one could think that Sem needs to be accumulated inside the cells and that the presence of a particular effect may reflect the level of its intracellular accumulation. Experimentally, this interaction has not been proven yet but there are several examples that may support both this hypothesis and the current findings, including the following: Sem is able to enter into tanycytes [31] and promote receptor internalization [34]; GLP-1 inhibits the KATP channel in mouse pancreatic beta-cells through a calmodulin-dependent mechanism [35]; a combined analysis study showed that calcium calmodulin-dependent kinase (CaMK) expression levels, possibly due to fear and stress, were significantly increased in the prefrontal cortex of suicide victims as compared to non-suicide subjects, [36]; Sem regulates the phosphorylation of several calmodulins [37]; and GLP-1 mediates changes to the phosphorylation of calcium/calmodulin-dependent protein kinase 1 (CaMKI) in the pituitary neuro-intermediate lobe [38]. Finally, the presence of an extracellular CaM (extCaM) in animals should also be considered and its presence is well documented in humans. extCaM appears to be required for cell division in the early preimplantation embryo and increases the proliferation of umbilical vein endothelial cells and acts as a mitogen in keratinocytes [39,40,41]. Considering this other localization, an extracellular interaction between Sem and extCaM could also be hypothesized.
4.1. Semaglutide Actions on Reward and Appetitive Behaviour; Clinical Relevance of Current Study Findings—Semaglutide; DAergic Reward Pathways; and Related Weight Loss
Sem intake is associated with significant levels of weight loss, levels which are comparable with those relating to the invasive bariatric surgery approach [42]. Indeed, obese non-diabetic patients receiving Sem lost and maintained about 15% of their body weight for over a year, with this result likely to be based on mechanisms occurring in the brain (for a commentary, see Gribble and O’Rahilly [43]), although these mechanisms are still a matter of debate.
The mesolimbic dopaminergic (DAergic) projections originating from the ventral tegmental area (VTA) and projected to both the nucleus accumbens (NAc) and the prefrontal cortex (PFC) represent the reward function neurochemical substrate (for a thorough review see Jia et al. [44]). DA levels in the NAc are elevated in association with both natural rewards (e.g., food and sex) but also by intake of a range of drugs, including nicotine, cocaine and amphetamine [44,45]. Consistent with this, responses to cues predicting food rewards in VTA DA axons and basal amygdala neurons in hungry mice were strongly attenuated following satiation [46]. Sem-related decreased levels of food cravings reported by subjects prescribed with this molecule have been associated with earlier attainment, in Sem-related subjects, of food satiety [47,48]. This may occur because GLP-1 mimetics show pro-dopaminergic efficacy [49], and as a result can modulate reward [50]. Hence, one could argue about the putative direct/indirect activities of Sem on the DA-mediated reward function.
Sem’s central effects on the brain’s centres affecting reward regulation could be driving this, as well the recently identified reported levels of Sem misuse [51]. This behaviour of misuse may arguably be occurring in both obese (e.g., through forged and/or diverted prescriptions) and healthy, non-obese individuals due to unrealistic ideals of physical attractiveness [51]. Although intake of Sem has not been associated with a stimulant-like ‘high’, a rebound syndrome may occur after abrupt Sem cessation [47].
4.2. Calmodulin Involvement in Both Drug and Natural Rewards
Calmodulin kinase II (CaMK II), an important [52] calmodulin, is activated when neuronal depolarization leads to Ca2+ entry into the cell [44]. Following activation, CaMKII translocates to the membrane where it regulates receptor activity. CaMKII is essential as well for neurotransmitter release, and thus it may mediate changes in DA function relevant to drug reward [53].
The stimulation of receptor-associated DA release in the NAc activates both cyclic adenosine monophosphate (cAMP) and protein kinase A (PKA), provoking cocaine-induced reinforcement and reinstatement [44]. Cocaine reinstatement behaviour induced by self-administration elevates CaMKII autophosphorylation in the rat NAc shell. This may suggest that CaMKII molecules may be critical in drug misuse/abuse/dependence issues relating to nicotine; morphine, alcohol; cocaine and amphetamine-type stimulants (for a review, see Jia et al. [44]).
Consistent with this, animals exhibiting elevated responses to methamphetamine relative to the vehicle showed increased D1R and CaMKII expression levels in the dorsal striatum [54]. CaMKII is involved in nicotine reward [55] and significant phosphorylated CaMKII level reductions in the NAc are observed after cessation of nicotine treatment in nicotine-dependent mice [56]. Additionally, endogenous CaMKII activity in the mPFC may exert inhibitory control over the positive reinforcing effects of alcohol [53]. Finally, calmodulines are primary molecular components of the aetiology of opiate/opioid [57] addiction. Conversely, nicotine reward, as measured by the conditioned place preference (CPP) test, is attenuated in CaMKIV knockout (−/−) mice [58]. Consistent with this, intra-VTA CaMKII inhibition via a specific αCaMKII inhibitor, as well as Ca2+-channel blockage via diltiazem (a calcium channel blocker), were shown to induce attenuated levels of cocaine-related behavioural sensitization [59]. In addition, αCaMKII-deficient mice showed reduced behavioural sensitization, as well as diminished locomotor response to amphetamine [60]. Hence, it has been suggested that CaMKII is important in facilitating both initiation and expression of drug addiction [44].
CaMKII has been suggested to play an essential role in the processing of natural reward as well [61]. CaMKII autophosphorylation-deficient mice showed impairments in the natural reward-related social interaction process [62]. Conversely, CaMKII activation/phosphorylation in the NAc core, but not shell, region was decreased during reinstatement of saccharine-(e.g., a form of natural reward) seeking activities [63]. Finally, with specific inhibition of CaMKII, sucrose-obtaining animals showed reduced behavioural response in comparison to naïve animals [53].
Overall, these considerations emphasize the relevance of CaMKII activity in the regulation of molecular pathways within both the drug- and natural-related reward system [61].
4.3. Semaglutide and Calmodulins
Several calmodulins were recently found to be upregulated or downregulated by Sem treatment [64]. Furthermore, calmodulin’s affinity for Ca2+ increases when it is bound to a calmodulin-binding protein [65], and current findings have suggested that Sem may act as a proper calmodulin binder.
As discussed here, CaMKII may represent a molecular point of convergence in the regulation of maladaptive behaviours associated with both natural [61] and drug [53] abuse.
Given the Sem-related food satiety and decrease in drug craving preclinical and clinical observations (for a commentary, see Leslie [28]), it is here hypothesized that, in the presence of Sem, the reward pathway-based calmodulin system may be better activated, and/or differently regulated. This process may, in turn, help explain Sem’s action on both natural and drug rewards. In line with this, following successful Sem obesity trials [43], nine phase 2 clinical trials are underway or being planned to test whether Sem can help patients to cut their use of cigarettes, alcohol, opioids or cocaine [28].
5. Conclusions
Calmodulin is a critical regulatory protein in cells, and its involvement in cellular signalling pathways, including those related to insulin release and metabolism, makes it an intriguing candidate for further investigation in the context of Sem’s action.
The CaM, and especially so the CaMKII, system is central in the experience of both drug- and natural-related reward. It is here hypothesized that, due to Sem binding, the reward pathway-based calmodulin system may be better activated, and/or differently regulated. This may result in positive action by Sem on appetitive behaviour. In conclusion, our study presents compelling evidence suggesting the potential interaction of semaglutide with Calmodulin (CaM), expanding the scope of its pharmacological targets beyond conventional GLP-1 receptors. However, it is paramount to recognize the inherent limitations of computational methodologies in fully capturing the intricacies of biological systems. As such, while our findings provide valuable insights, they should be interpreted as preliminary steps in the drug development journey, guiding future experimental endeavours rather than definitive determinants of Sem’s pharmacodynamics. We advocate for a prudent approach wherein these computational predictions serve as prompts for further in vitro and in vivo investigations, allowing for a comprehensive understanding of Sem’s molecular interactions and therapeutic potential. Furthermore, the dissemination of our findings stimulates scientific discourse, encouraging other research groups to explore and validate the speculated interactions, thereby collectively advancing our understanding of semaglutide’s multifaceted pharmacology and its implications for the management of metabolic disorders.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/scipharm92020017/s1, Figure S1: Input parameters used in PepBDB; Table S1: Identified proteins by PepBDB research.
Author Contributions
Conceptualization, G.F. and F.S.; Methodology, G.F. and V.C.; Validation, G.F. and V.C.; Formal Analysis, G.D.P.P., J.M.C. and A.G.; Resources, G.F. and F.S.; Data Curation, G.F.; Writing—Original Draft Preparation, G.F., V.C. and D.A.; Writing—Review and Editing, G.D.P.P., J.M.C., A.G., F.S. and G.F.; Supervision, F.S.; Project Administration, G.F. and F.S.; Funding Acquisition, F.S. All authors have read and agreed to the published version of the manuscript.
Funding
The study was carried out with internal support from the University of Hertfordshire (GL11101336—Centre Clinical and Health Services QR).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
F.S. was a member of the UK Advisory Council on the Misuse of Drugs (ACMD; 2011–2019) and is currently a member of the EMA Advisory Board (Psychiatry). J.M.C. is a co-opted member of the ACMD’s Technical and Novel Psychoactive Substances Committees. All the other authors declare no conflicts of interest.
References
- Gao, X.; Hua, X.; Wang, X.; Xu, W.; Zhang, Y.; Shi, C.; Gu, M. Efficacy and safety of semaglutide on weight loss in obese or overweight patients without diabetes: A systematic review and meta-analysis of randomized controlled trials. Front. Pharmacol. 2022, 13, 935823. [Google Scholar] [CrossRef]
- Xie, Z.; Yang, S.; Deng, W.; Li, J.; Chen, J. Efficacy and Safety of Liraglutide and Semaglutide on Weight Loss in People with Obesity or Overweight: A Systematic Review. Clin. Epidemiol. 2022, 14, 1463–1476. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, M.; Wen, Z.; Lu, Z.; Cui, L.; Fu, C.; Xue, H.; Liu, Y.; Zhang, Y. GLP-1 Receptor Agonists: Beyond Their Pancreatic Effects. Front. Endocrinol. 2021, 12, 721135. [Google Scholar] [CrossRef] [PubMed]
- Floresta, G.; Amata, E.; Gentile, D.; Romeo, G.; Marrazzo, A.; Pittalà, V.; Salerno, L.; Rescifina, A. Fourfold Filtered Statistical/Computational Approach for the Identification of Imidazole Compounds as HO-1 Inhibitors from Natural Products. Mar. Drugs 2019, 17, 113. [Google Scholar] [CrossRef]
- Floresta, G.; Gentile, D.; Perrini, G.; Patamia, V.; Rescifina, A. Computational Tools in the Discovery of FABP4 Ligands: A Statistical and Molecular Modeling Approach. Mar. Drugs 2019, 17, 624. [Google Scholar] [CrossRef]
- Floresta, G.; Patamia, V.; Gentile, D.; Molteni, F.; Santamato, A.; Rescifina, A.; Vecchio, M. Repurposing of FDA-Approved Drugs for Treating Iatrogenic Botulism: A Paired 3D-QSAR/Docking Approach(†). ChemMedChem 2020, 15, 256–262. [Google Scholar] [CrossRef]
- Wen, Z.; He, J.; Tao, H.; Huang, S.Y. PepBDB: A comprehensive structural database of biological peptide-protein interactions. Bioinformatics 2019, 35, 175–177. [Google Scholar] [CrossRef]
- Hoeflich, K.P.; Ikura, M. Calmodulin in action: Diversity in target recognition and activation mechanisms. Cell 2002, 108, 739–742. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, K.T.; Degrado, W.F. How calmodulin binds its targets: Sequence independent recognition of amphiphilic α-helices. Trends Biochem Sci. 1990, 15, 59–64. [Google Scholar] [CrossRef]
- Zhou, P.; Jin, B.; Li, H.; Huang, S.Y. HPEPDOCK: A web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res. 2018, 46, W443–W450. [Google Scholar] [CrossRef]
- Ataman, Z.A.; Gakhar, L.; Sorensen, B.R.; Hell, J.W.; Shea, M.A. The NMDA receptor NR1 C1 region bound to calmodulin: Structural insights into functional differences between homologous domains. Structure 2007, 15, 1603–1617. [Google Scholar] [CrossRef]
- Dunlap, T.B.; Guo, H.-F.; Cook, E.C.; Holbrook, E.; Rumi-Masante, J.; Lester, T.E.; Colbert, C.L.; Vander Kooi, C.W.; Creamer, T.P. Stoichiometry of the Calcineurin Regulatory Domain–Calmodulin Complex. Biochemistry 2014, 53, 5779–5790. [Google Scholar] [CrossRef]
- Cheng, A.; Best, S.A.; Merz, K.M., Jr.; Reynolds, C.H. GB/SA water model for the Merck molecular force field (MMFF). J. Mol. Graph. Model. 2000, 18, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.J. Optimization of parameters for semiempirical methods IV: Extension of MNDO, AM1, and PM3 to more main group elements. J. Mol. Model. 2004, 10, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.J. MOPAC: A semiempirical molecular orbital program. J. Comput. Aided Mol. Des. 1990, 4, 1–105. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Pan, H.; Sun, H.; Kang, Y.; Liu, H.; Cao, D.; Hou, T. fastDRH: A webserver to predict and analyze protein-ligand complexes based on molecular docking and MM/PB(GB)SA computation. Brief. Bioinform. 2022, 23, 201. [Google Scholar] [CrossRef]
- Cardullo, N.; Catinella, G.; Floresta, G.; Muccilli, V.; Rosselli, S.; Rescifina, A.; Bruno, M.; Tringali, C. Synthesis of Rosmarinic Acid Amides as Antioxidative and Hypoglycemic Agents. J. Nat. Prod. 2019, 82, 573–582. [Google Scholar] [CrossRef]
- Krieger, E.; Dunbrack, R.L.; Hooft, R.W.; Krieger, B. Assignment of protonation states in proteins and ligands: Combining pK a prediction with hydrogen bonding network optimization. In Computational Drug Discovery and Design; Springer: Berlin/Heidelberg, Germany, 2012; pp. 405–421. [Google Scholar]
- Krieger, E.; Nielsen, J.E.; Spronk, C.A.; Vriend, G. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model. 2006, 25, 481–486. [Google Scholar] [CrossRef]
- Maier, J.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins Struct. Funct. Bioinform. 2006, 65, 712–725. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef] [PubMed]
- Fuochi, V.; Floresta, G.; Emma, R.; Patamia, V.; Caruso, M.; Zagni, C.; Ronchi, F.; Ronchi, C.; Drago, F.; Rescifina, A.; et al. Heparan Sulfate and Enoxaparin Interact at the Interface of the Spike Protein of HCoV-229E but Not with HCoV-OC43. Viruses 2023, 15, 663. [Google Scholar] [CrossRef]
- Crocetti, L.; Floresta, G.; Zagni, C.; Merugu, D.; Mazzacuva, F.; de Oliveira Silva, R.R.; Vergelli, C.; Giovannoni, M.P.; Cilibrizzi, A. Ligand Growing Experiments Suggested 4-amino and 4-ureido pyridazin-3(2H)-one as Novel Scaffold for FABP4 Inhibition. Pharmaceuticals 2022, 15, 1335. [Google Scholar] [CrossRef] [PubMed]
- Leslie, M. Hot weight loss drugs tested against addiction. Science 2023, 381, 930–931. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Wen, S.; Zhou, L. The Relationship Between the Blood-Brain-Barrier and the Central Effects of Glucagon-Like Peptide-1 Receptor Agonists and Sodium-Glucose Cotransporter-2 Inhibitors. Diabetes Metab. Syndr. Obes. 2022, 15, 2583–2597. [Google Scholar] [CrossRef] [PubMed]
- Mahapatra, M.K.; Karuppasamy, M.; Sahoo, B.M. Semaglutide, a glucagon like peptide-1 receptor agonist with cardiovascular benefits for management of type 2 diabetes. Rev. Endocr. Metab. Disord. 2022, 23, 521–539. [Google Scholar] [CrossRef]
- Gabery, S.; Salinas, C.G.; Paulsen, S.J.; Ahnfelt-Ronne, J.; Alanentalo, T.; Baquero, A.F.; Buckley, S.T.; Farkas, E.; Fekete, C.; Frederiksen, K.S.; et al. Semaglutide lowers body weight in rodents via distributed neural pathways. JCI Insight 2020, 5, 133429. [Google Scholar] [CrossRef]
- Jones, B.; Buenaventura, T.; Kanda, N.; Chabosseau, P.; Owen, B.M.; Scott, R.; Goldin, R.; Angkathunyakul, N.; Corrêa, I.R., Jr.; Bosco, D.; et al. Targeting GLP-1 receptor trafficking to improve agonist efficacy. Nat. Commun. 2018, 9, 1602. [Google Scholar] [CrossRef] [PubMed]
- Rusnak, F.; Mertz, P. Calcineurin: Form and function. Physiol. Rev. 2000, 80, 1483–1521. [Google Scholar] [CrossRef]
- Novikoff, A.; O’Brien, S.L.; Bernecker, M.; Grandl, G.; Kleinert, M.; Knerr, P.J.; Stemmer, K.; Klingenspor, M.; Zeigerer, A.; DiMarchi, R.; et al. Spatiotemporal GLP-1 and GIP receptor signaling and trafficking/recycling dynamics induced by selected receptor mono- and dual-agonists. Mol. Metab. 2021, 49, 101181. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.G.; Kitasato, H.; Matsuura, H. Involvement of calmodulin in glucagon-like peptide 1(7-36) amide-induced inhibition of the ATP-sensitive K+ channel in mouse pancreatic beta-cells. Exp. Physiol. 2001, 86, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Le, T.; Xing, G.; Johnson, L.R.; Ursano, R.J. Analysis of kinase gene expression in the frontal cortex of suicide victims: Implications of fear and stress. Front. Behav. Neurosci. 2011, 5, 46. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ma, L.; Gan, K.; Pan, X.; Chen, S. Phosphorylated proteomics-based analysis of the effects of semaglutide on hippocampi of high-fat diet-induced-obese mice. Diabetol. Metab. Syndr. 2023, 15, 63. [Google Scholar] [CrossRef]
- Greenwood, M.P.; Greenwood, M.; Barez-Lopez, S.; Hawkins, J.W.; Short, K.; Tatovic, D.; Murphy, D. Osmoadaptive GLP-1R signalling in hypothalamic neurones inhibits antidiuretic hormone synthesis and release. Mol. Metab. 2023, 70, 101692. [Google Scholar] [CrossRef]
- Dawson, R.A.; Mac Neil, S. Mitogenic role for extracellular calmodulin-like activity in normal human umbilical vein endothelial cells. Br. J. Haematol. 1992, 82, 151–160. [Google Scholar] [CrossRef]
- Goberdhan, N.J.; Dawson, R.A.; Freedlander, E.; Mac Neil, S. A calmodulin-like protein as an extracellular mitogen for the keratinocyte. Br. J. Dermatol. 1993, 129, 678–688. [Google Scholar] [CrossRef]
- Woodward, B.J.; Lenton, E.A.; Mac Neil, S. Requirement of preimplantation human embryos for extracellular calmodulin for development. Hum. Reprod. 1993, 8, 272–276. [Google Scholar] [CrossRef]
- Patel, P.N.; Fox, C.K.; Bensignor, M.O.; Bomberg, E.M. Weight Loss from Combination Anti-Obesity Medication Regimens Can Approach that Achieved from Bariatric Surgery. JCEM Case Rep. 2023, 1, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Gribble, F.M.; O’Rahilly, S. Obesity therapeutics: The end of the beginning. Cell Metab. 2021, 33, 705–706. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Kawahata, I.; Cheng, A.; Fukunaga, K. The Role of CaMKII and ERK Signaling in Addiction. Int. J. Mol. Sci. 2021, 22, 3189. [Google Scholar] [CrossRef] [PubMed]
- Di Chiara, G.; Acquas, E.; Tanda, G.; Cadoni, C. Drugs of abuse: Biochemical surrogates of specific aspects of natural reward? Biochem. Soc. Symp. 1993, 59, 65–81. [Google Scholar] [PubMed]
- Lutas, A.; Kucukdereli, H.; Alturkistani, O.; Carty, C.; Sugden, A.U.; Fernando, K.; Diaz, V.; Flores-Maldonado, V.; Andermann, M.L. State-specific gating of salient cues by midbrain dopaminergic input to basal amygdala. Nat. Neurosci. 2019, 22, 1820–1833. [Google Scholar] [CrossRef] [PubMed]
- Arillotta, D.; Floresta, G.; Guirguis, A.; Corkery, J.M.; Catalani, V.; Martinotti, G.; Sensi, S.L.; Schifano, F. GLP-1 Receptor Agonists and Related Mental Health Issues; Insights from a Range of Social Media Platforms Using a Mixed-Methods Approach. Brain Sci. 2023, 13, 1503. [Google Scholar] [CrossRef] [PubMed]
- Anderberg, R.H.; Anefors, C.; Bergquist, F.; Nissbrandt, H.; Skibicka, K.P. Dopamine signaling in the amygdala, increased by food ingestion and GLP-1, regulates feeding behavior. Physiol. Behav. 2014, 136, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Athauda, D.; Maclagan, K.; Skene, S.S.; Bajwa-Joseph, M.; Letchford, D.; Chowdhury, K.; Hibbert, S.; Budnik, N.; Zampedri, L.; Dickson, J.; et al. Exenatide once weekly versus placebo in Parkinson’s disease: A randomised, double-blind, placebo-controlled trial. Lancet 2017, 390, 1664–1675. [Google Scholar] [CrossRef]
- van Bloemendaal, L.; RG, I.J.; Ten Kulve, J.S.; Barkhof, F.; Konrad, R.J.; Drent, M.L.; Veltman, D.J.; Diamant, M. GLP-1 receptor activation modulates appetite- and reward-related brain areas in humans. Diabetes 2014, 63, 4186–4196. [Google Scholar] [CrossRef]
- Chiappini, S.; Vickers-Smith, R.; Harris, D.; Papanti Pelletier, G.D.; Corkery, J.M.; Guirguis, A.; Martinotti, G.; Sensi, S.L.; Schifano, F. Is There a Risk for Semaglutide Misuse? Focus on the Food and Drug Administration’s FDA Adverse Events Reporting System (FAERS) Pharmacovigilance Dataset. Pharmaceuticals 2023, 16, 994. [Google Scholar] [CrossRef]
- Hirst, N.L.; Lawton, S.P.; Walker, A.J. CaMKII regulates neuromuscular activity and survival of the human blood fluke Schistosoma mansoni. Sci. Rep. 2022, 12, 19831. [Google Scholar] [CrossRef] [PubMed]
- Faccidomo, S.; Reid, G.T.; Agoglia, A.E.; Ademola, S.A.; Hodge, C.W. CaMKII inhibition in the prefrontal cortex specifically increases the positive reinforcing effects of sweetened alcohol in C57BL/6J mice. Behav. Brain Res. 2016, 298, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Oliver, R.J.; Purohit, D.C.; Kharidia, K.M.; Mandyam, C.D. Transient Chemogenetic Inhibition of D1-MSNs in the Dorsal Striatum Enhances Methamphetamine Self-Administration. Brain Sci. 2019, 9, 330. [Google Scholar] [CrossRef]
- Jackson, K.J.; Muldoon, P.P.; Walters, C.; Damaj, M.I. Neuronal calcium/calmodulin-dependent protein kinase II mediates nicotine reward in the conditioned place preference test in mice. Behav. Pharmacol. 2016, 27, 50–56. [Google Scholar] [CrossRef][Green Version]
- Jackson, K.J.; Damaj, M.I. Calcium/calmodulin-dependent protein kinase IV mediates acute nicotine-induced antinociception in acute thermal pain tests. Behav. Pharmacol. 2013, 24, 689–692. [Google Scholar] [CrossRef]
- Rosen, L.G.; Zunder, J.; Renard, J.; Fu, J.; Rushlow, W.; Laviolette, S.R. Opiate Exposure State Controls a D2-CaMKIIα-Dependent Memory Switch in the Amygdala-Prefrontal Cortical Circuit. Neuropsychopharmacology 2016, 41, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Jackson, K.J.; Sanjakdar, S.S.; Chen, X.; Damaj, M.I. Nicotine reward and affective nicotine withdrawal signs are attenuated in calcium/calmodulin-dependent protein kinase IV knockout mice. PLoS ONE 2012, 7, e51154. [Google Scholar] [CrossRef]
- Licata, S.C.; Schmidt, H.D.; Pierce, R.C. Suppressing calcium/calmodulin-dependent protein kinase II activity in the ventral tegmental area enhances the acute behavioural response to cocaine but attenuates the initiation of cocaine-induced behavioural sensitization in rats. Eur. J. Neurosci. 2004, 19, 405–414. [Google Scholar] [CrossRef]
- Steinkellner, T.; Mus, L.; Eisenrauch, B.; Constantinescu, A.; Leo, D.; Konrad, L.; Rickhag, M.; Sørensen, G.; Efimova, E.V.; Kong, E.; et al. In vivo amphetamine action is contingent on αCaMKII. Neuropsychopharmacology 2014, 39, 2681–2693. [Google Scholar] [CrossRef]
- Amaral, I.M.; Scheffauer, L.; Hofer, A.; El Rawas, R. Protein kinases in natural versus drug reward. Pharmacol. Biochem. Behav. 2022, 221, 173472. [Google Scholar] [CrossRef]
- Harda, Z.; Dzik, J.M.; Nalberczak-Skóra, M.; Meyza, K.; Łukasiewicz, K.; Łęski, S.; Radwanska, K. Autophosphorylation of αCaMKII affects social interactions in mice. Genes. Brain Behav. 2018, 17, e12457. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, X.D.; Zhang, J.J.; Yu, L.C. Increases in αCaMKII phosphorylated on Thr286 in the nucleus accumbens shell but not the core during priming-induced reinstatement of morphine-seeking in rats. Neurosci. Lett. 2012, 526, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, S.; Li, Z.; Zhu, R.; Jia, Z.; Ban, J.; Zhen, R.; Chen, X.; Pan, X.; Ren, Q.; et al. Effect of semaglutide and empagliflozin on cognitive function and hippocampal phosphoproteomic in obese mice. Front. Pharmacol. 2023, 14, 975830. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.K.; Parameswaran, S. Calmodulin-binding proteins: A journey of 40 years. Cell Calcium 2018, 75, 89–100. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).