STAT3 Inhibitory Activities of Lignans Isolated from the Stems of Lindera obtusiloba Blume
Abstract
1. Introduction
2. Materials and Methods
2.1. General Experimental Procedures
2.2. Extraction and Isolation
2.3. Cell Culture
2.4. Cell Viability
2.5. IL-6-Induced STAT3 Luciferase Reporter Assay
2.5.1. Real-Time PCR
2.5.2. Western Blot Analysis
2.6. Statistical Analysis
3. Results and Discussion
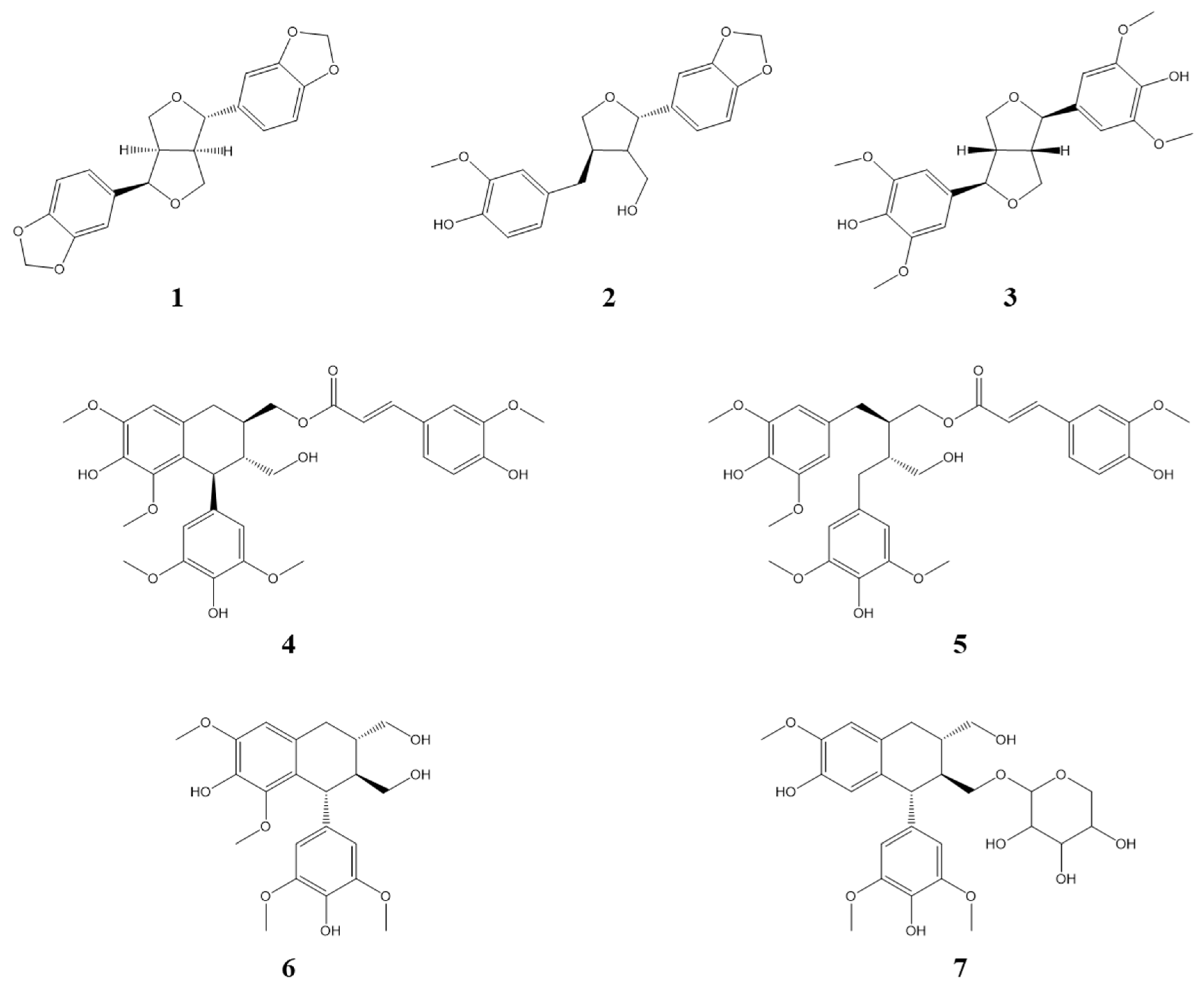
3.1. Identification of Compounds from Lindera obtusiloba
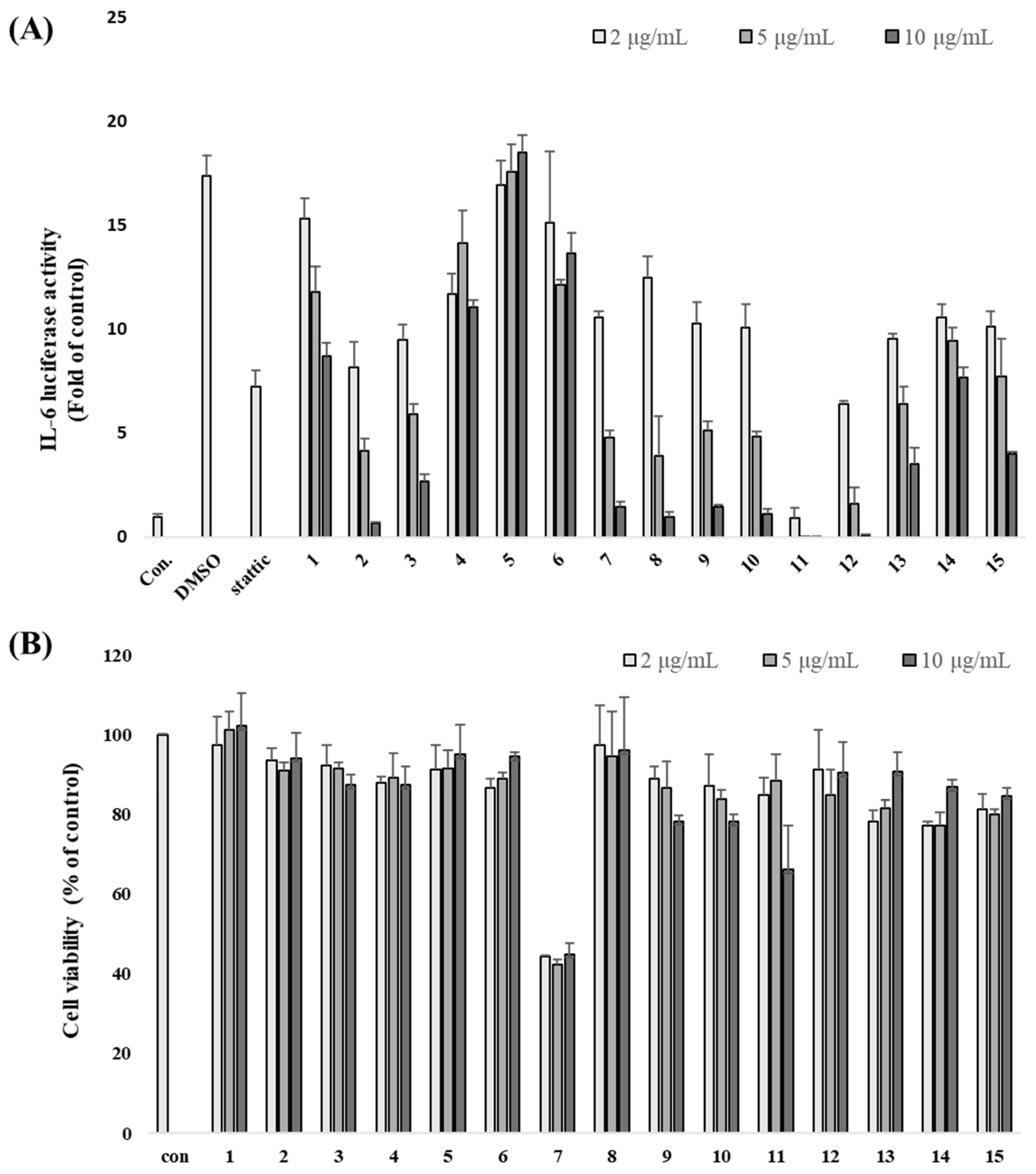
3.2. L. obtusiloba Blume Extract and Fractions Inhibit IL-6-Induced pSTAT3 Luciferase Activity
3.3. Inhibitory Effects of Isolated Compounds on IL-6-Induced pSTAT3 Luciferase Activity
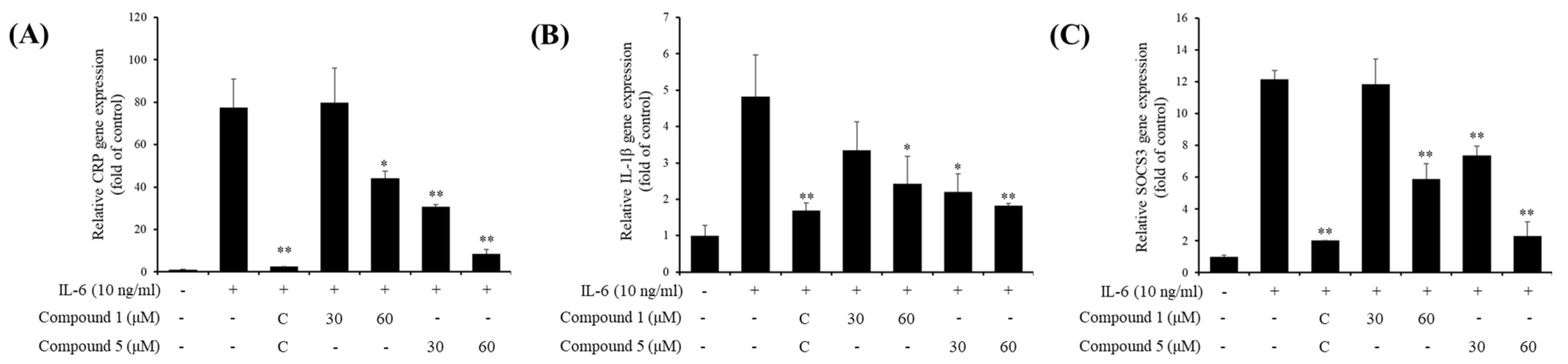
3.4. Inhibitory Effects of Active Compounds on IL-6-Induced Gene Expressions
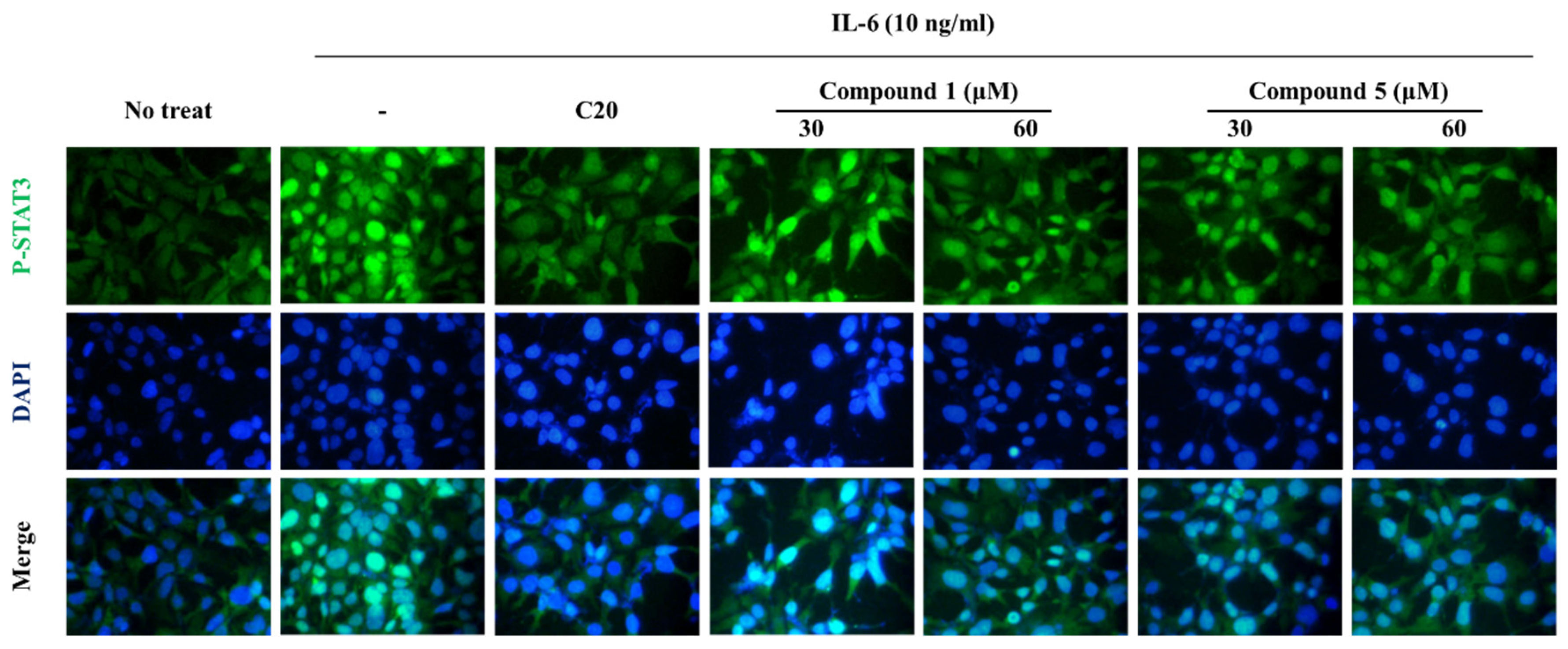
3.5. Effect of Compounds 1 and 5 on IL-6/STAT3 Signaling Molecules
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Haque, M.E.; Azam, S.; Balakrishnan, R.; Akther, M.; Kim, I.S. Therapeutic potential of Lindera obtusiloba: Focus on antioxidative and pharmacological properties. Plants 2020, 9, 1765. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.O.; Rhee, C.H.; Won, N.H.; Choi, H.D.; Lee, K.W. Protective effect of 70% ethanolic extract of Lindera obtusiloba Blume on tert-butyl hydroperoxide-induced oxidative hepatotoxicity in rats. Food Chem. Toxicol. 2013, 53, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Park, J.C.; Yu, Y.B.; Lee, J.H. Isolation and structure elucidation of flavonoid glycosides from Lindera obtusiloba BL. J. Korean Soc. Food Nutr. 1996, 25, 76–79. [Google Scholar]
- Kwon, H.C.; Choi, S.U.; Lee, J.O.; Kwon, H.C.; Choi, S.U.; Lee, J.O.; Bae, K.H.; Zee, O.P.; Lee, K.R. Two New Lignan from Lindera obtusiloba blume. Arch. Pharm. Res. 1999, 22, 417–422. [Google Scholar] [CrossRef]
- Kwon, H.C.; Baek, N.I.; Choi, S.U.; Lee, K.R. New cytotoxic butanolides from Lindera obtusiloba BLUME. Chem. Pharm. Bull. 2000, 48, 614–616. [Google Scholar] [CrossRef]
- Suh, W.M.; Park, S.B.; Lee, S.; Kim, H.H.; Suk, K.; Son, J.H.; Kwon, T.K.; Choi, H.G.; Lee, S.H.; Kim, S.H. Suppression of mast-cell-mediated allergic inflammation by Lindera obtusiloba. Exp. Biol. Med. 2011, 236, 240–246. [Google Scholar] [CrossRef]
- Park, K.J.; Park, S.H.; Kim, J.K. Anti-wrinkle activity of Lindera obtusiloba extract. J. Soc. Cosmet. Sci. Korea 2009, 35, 317–323. [Google Scholar]
- Bang, C.Y.; Won, E.K.; Park, K.W.; Lee, G.W.; Choung, S.Y. Antioxidant activites and whitening effect from Lindera obtusiloba BL. Extract. Yakhak Hoeji 2008, 52, 355–360. [Google Scholar]
- Kim, J.H.; Lee, J.; Kang, S.; Moon, H.; Chung, K.H.; Kim, K.R. Antiplatelet and antithrombotic effects of the extract of Lindera obtusiloba leaves. Biomol. Ther. 2016, 24, 659–664. [Google Scholar] [CrossRef]
- Lee, J.O.; Oak, M.H.; Jung, S.H.; Park, D.H.; Auger, C. An ethanolic extract of Lindera obtusiloba stems causes NO-mediated endothelium dependent relaxations in rat aortic rings and prevents angiotensin II-induced hypertension and endothelial dysfunction in rats. Naunyn Schmiedebergs Arch. Pharmacol. 2011, 383, 635–645. [Google Scholar] [CrossRef]
- Seo, K.H.; Baek, M.Y.; Lee, D.Y.; Cho, J.G. Isolation of Flavonoids and Lignans from the Stem Wood of Lindera Obtusioba Blume. J. Appl. Biol. Chem. 2011, 54, 178–183. [Google Scholar] [CrossRef][Green Version]
- Kwon, D.J.; Kim, J.K.; Bae, Y.S. Essential oils from leaves and twigs of Lindera obtusiloba. J. Korean For. Soc. 2007, 96, 65–69. [Google Scholar]
- Lee, K.Y.; Kim, S.H.; Jeong, E.J.; Park, J.H.; Kim, S.H.; Kim, Y.C.; Sung, S.H. New Secoisolariciresinol Derivatives from Lindera obtusiloba Stems and Their Neuroprotective Activities. Planta Med. 2010, 76, 294–297. [Google Scholar] [CrossRef] [PubMed]
- Freise, C.; Erben, U.; Neuman, U.; Kim, K.; Zeitz, M.; Somasundaram, R.; Ruehl, M. An active extract of Lindera obtusiloba inhibits adipogenesis via sustained Wnt signaling and exerts anti-inflammatory effects in the 3T3-L1 preadipocytes. J. Nutr. Biochem. 2011, 21, 1170–1177. [Google Scholar] [CrossRef] [PubMed]
- Mark, D.T.; Belinda, N.; Tara, H.; Daniel, J.P. Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta 2014, 1843, 2563–2582. [Google Scholar]
- Kumari, N.; Dwarakanath, B.S.; Das, A.; Bhatt, A.N. Role of interleukin-6 in cancer progression and therapeutic resistance. Tumour Biol. 2016, 37, 11553–11572. [Google Scholar] [CrossRef]
- Hunter, C.A.; Jones, S.A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 2015, 16, 448–457. [Google Scholar] [CrossRef]
- Mihara, M.; Hashizume, M.; Yoshida, H.; Suzuki, M.; Shiina, M. IL-6/IL-6 receptor system and its role in physiological and pathological conditions. Clin. Sci. 2012, 122, 143–159. [Google Scholar] [CrossRef]
- Jang, H.J.; Lim, H.J.; Park, E.J.; Lee, S.J.; Lee, S.; Lee, S.W.; Rho, M.C. STAT3-inhibitory activity of sesquiterpenoids and diterpenoids from Curcuma phaeocaulis. Bioorg. Chem. 2019, 93, 103267. [Google Scholar] [CrossRef]
- Lee, S.J.; Jang, H.J.; Kim, Y.; Oh, H.M.; Lee, S.; Jung, K.; Rho, M.C. Inhibitory effects of IL-6-induced STAT3 activation of bio-active compounds derived from Salvia plebeia R. Br. Process Biochem. 2016, 51, 2222–2229. [Google Scholar] [CrossRef]
- Liu, Z.; Saarinen, N.M.; Thompson, L.U. Sesamin is one of the major precursors of mammalian lignans in sesame seed (Sesamum indicum) as observed in vitro and in rats. J. Nutr. 2006, 136, 906–912. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xia, H.; Wang, L.; Xia, G.; Qu, Y.; Shang, X.; Lin, S. Lignans from the Twigs of Litsea cubeba and Their Bioactivities. Molecules 2019, 24, 306. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.H.; Huang, Y.H.; Lin, J.J. Cytotoxic lignan ester from cinnamomum osmophloeum. Planta Med. 2009, 76, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Sadhu, S.K.; Khatun, A.; Phattanawasin, P.; Ohtsuki, T.; Ishibashi, M. Lignan glycosides and flavonoids from Saraca asoca with antioxidant activity. J. Nat. Med. 2007, 61, 480–482. [Google Scholar] [CrossRef]
- Choi, H.G.; Choi, Y.H.; Kim, J.H.; Kim, H.H.; Kim, S.H.; Kim, J.A. A new neolignan and lignans from the stems of Lindera obtusiloba Blume and their anti-allergic inflammatory effects. Arch. Pharm. Res. 2014, 37, 467–472. [Google Scholar] [CrossRef]
- Rahman, M.A.; Katayama, T.; Suzuki, T.; Nakagawa, T. Stereochemistry and biosynthesis of (+)-lyoniresinol, a syringyl tetrahydronaphthalene lignan in Lyonia ovalifolia var. elliptica I: Isolation and stereochemistry of syringyl lignans and predicted precursors to (+)-lyoniresinol from wood. J. Wood Sci. 2007, 53, 161–167. [Google Scholar] [CrossRef]

| Compounds | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| IC50 (µM) | 14.3 ± 2.67 | >50 | >50 | 40.39 ± 3.71 | 10.83 ± 2.40 | >50 | >50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, E.-J.; Lim, H.J.; Lim, H.J.; Yun, B.-S.; Lee, S.; Lee, S.-J.; Lee, S.W. STAT3 Inhibitory Activities of Lignans Isolated from the Stems of Lindera obtusiloba Blume. Sci. Pharm. 2023, 91, 56. https://doi.org/10.3390/scipharm91040056
Park E-J, Lim HJ, Lim HJ, Yun B-S, Lee S, Lee S-J, Lee SW. STAT3 Inhibitory Activities of Lignans Isolated from the Stems of Lindera obtusiloba Blume. Scientia Pharmaceutica. 2023; 91(4):56. https://doi.org/10.3390/scipharm91040056
Chicago/Turabian StylePark, Eun-Jae, Hee Ju Lim, Hyung Jin Lim, Bong-Sik Yun, Soyoung Lee, Seung-Jae Lee, and Seung Woong Lee. 2023. "STAT3 Inhibitory Activities of Lignans Isolated from the Stems of Lindera obtusiloba Blume" Scientia Pharmaceutica 91, no. 4: 56. https://doi.org/10.3390/scipharm91040056
APA StylePark, E.-J., Lim, H. J., Lim, H. J., Yun, B.-S., Lee, S., Lee, S.-J., & Lee, S. W. (2023). STAT3 Inhibitory Activities of Lignans Isolated from the Stems of Lindera obtusiloba Blume. Scientia Pharmaceutica, 91(4), 56. https://doi.org/10.3390/scipharm91040056