Chromobacterium Violaceum: A Model for Evaluating the Anti-Quorum Sensing Activities of Plant Substances
Abstract
1. Introduction
2. Quorum Sensing: Bacterial Communication Network
2.1. LuxR Receptors
2.2. Bicomponent Quorum-Sensing Receptors
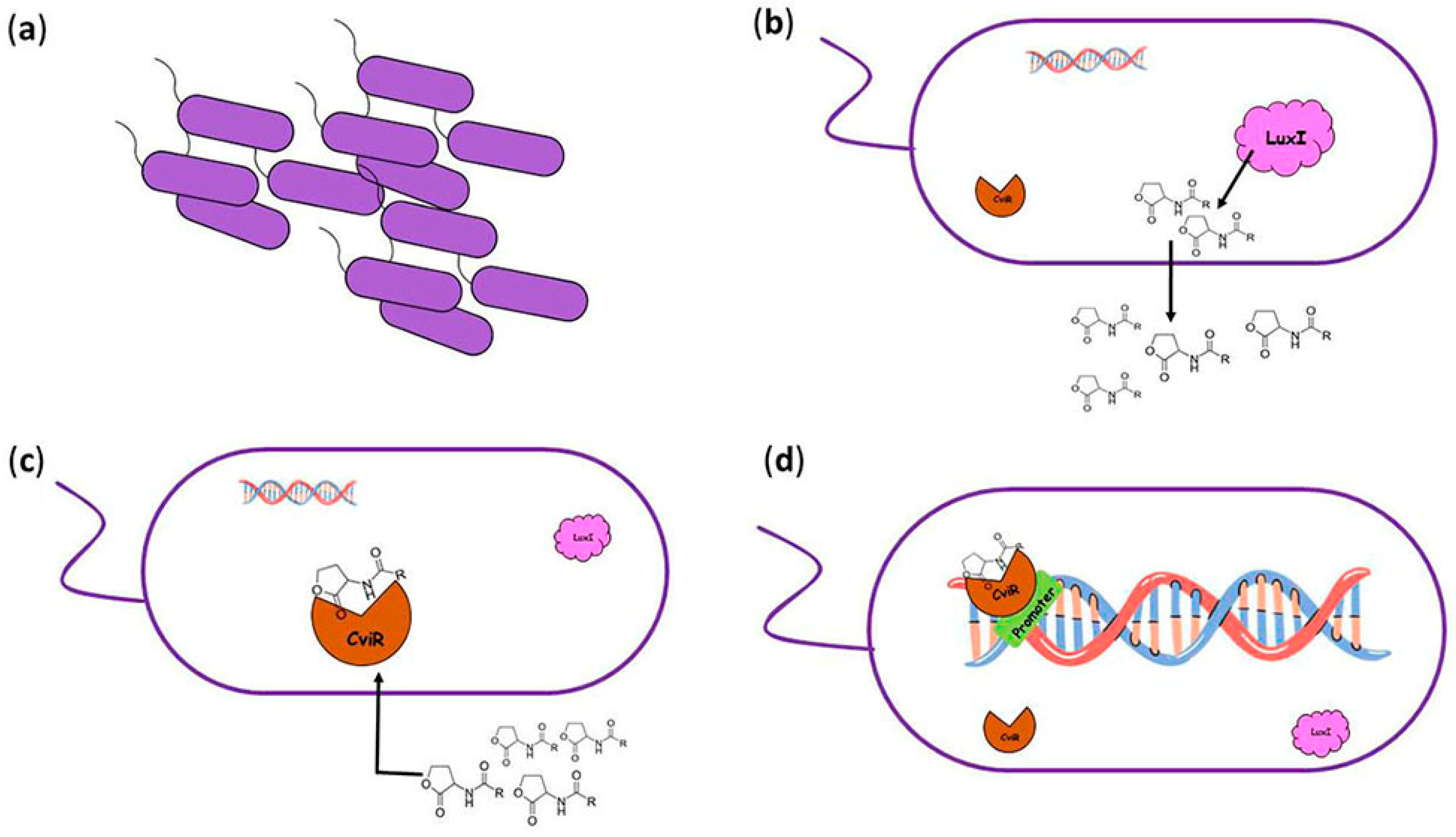
3. Quorum-Sensing System in Chromobacterium violaceum
3.1. Quorum-Sensing Mechanisms in Chromobacterium violaceum
3.2. Pigment Production
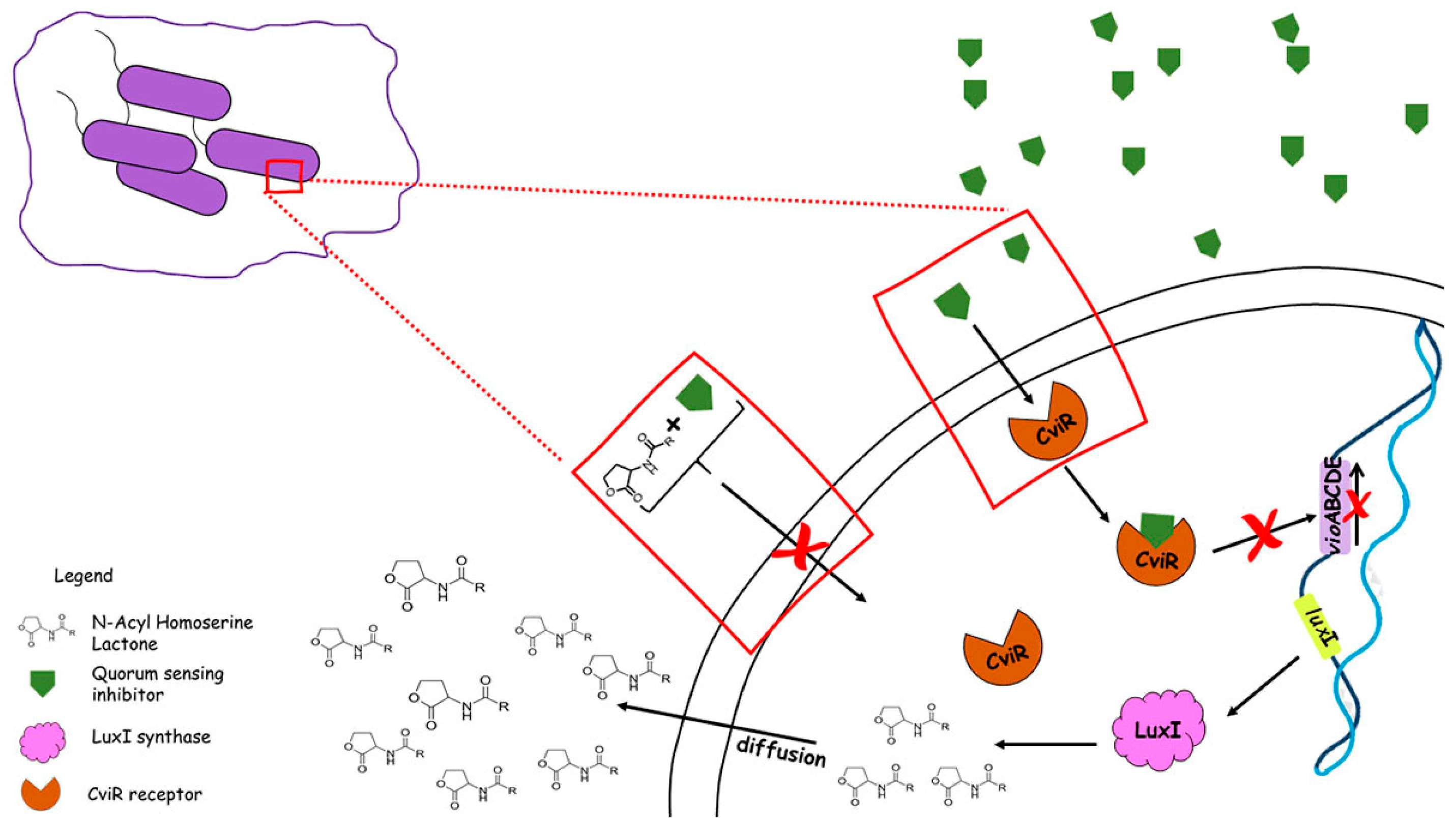
4. Plant Inhibitors: A New Way to Control Bacterial Communication
Quorum-Sensing Inhibitory Potential of Plants
| Plant | Compound | Strain | Mechanism of Action or Effect | Ref. |
|---|---|---|---|---|
| Combretum albiflorum | Catechin | C. violaceum CV026 | Inhibition of violacein production | [121] |
| Rosa rugosa | Epigallocatechin gallate Epicatechin | C. violaceum CV026 | Reduction in violacein production | [110] |
| Vernonia blumeoides | Sesquiterpene lactone | C. violaceum CV026 C. violaceum VIR07 C. violaceum ATCC 12472 C. violaceum ATCC 31532 | Antagonist effect against CviR | [2] |
| Drimys winteri | Cinnamolide Valdiviolide | C. violaceum ATCC 12472 | Inhibition of QS and violacein reduction | [122] |
| Polydora serratuloides | Sesquiterpene lactone (13-acetoxy 1(4β),5(6)βdiepoxy-8α-(senecioyloxy) 3-oxo-1,7(11)-germacradiene-12,6-olide 1) | C. violaceum ATCC 12472 | Inhibition of QS mediators | [123] |
| Allium sativum | P-Coumaric acid | C. violaceum 5999 and wt 494 | Inhibition of biofilm formation and the expression of bacterial virulence factor; antagonizes the activity of LuxR, ahyR, and TraR receptors | [2] |
| Caffeine (1,3,7-trimethylxanthine) | C. violaceum CV026 | Inhibition of violacein production; inhibition of CviI synthase | [25] | |
| Isothiocyanates | C. violaceum CV12472 | Modulation of AHL activity and synthesis | [25] | |
| N, N-disubstituted biguanides | C. violaceum ATCC 12472 | Reduces the synthesis of violacein; inhibition of the transcription factor CviR | [25] | |
| Psidium guajava L. | Quercetin (2-(3,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-one) | C. violaceum ATCC 31532 | Inhibition of violacein synthesis; binds to transcription factor CviR | [53] |
| Gnaphalium hypoleucum DC | Apigenin and luteolin | C. violaceum ATCC 12472 | Effects on violacein pigment biosynthesis, biofilm formation, and motility; downregulation of the vioB, vioC, and vioD genes | [119] |
| Quercetin 4′-O-β-D-glucopyranoside | C. violaceum ATCC 12472 C. violaceum CV026 | Reduction in violacein synthesis, biofilm formation, EPS production, motility, and alginate production; inhibition of the C6-AHL communication molecule | [120] | |
| Myristica Cinnamomea | Malabaricone C | C. violaceum CV026 | Inhibition of violacein production | [54] |
| Bitter orange | Naringin | C. violaceum (CECT 494) | Inhibition of the production of violacein | [2] |
| Vanilla planifolia Andrews | Vanillin (4-hydroxy-3 methoxybenzaldehyde) | C. violaceum CV026 | Reduced violacein production | [20] |
| Amphypterygium adstringens | Anacardic acids mixture | C. violaceum ATCC 12472 | Inhibition of violacein production | [20] |
| Syzygium aromaticum | Eugenol | C. violaceum CV026 | Dose-dependent inhibitory effect on violacein synthesis | [20] |
| Syzygium cumini | Malvidin | C. violaceum CV026 (CECT 5999) C. violaceum MTCC2656 | Inhibition of violacein production; reduction in biofilm biomass | [82] |
| Origanum vulgare | Carvacrol | C. violaceum ATCC 12472 | Reductions in biofilm formation, violacein production, and chitinase activity; reduces the expression of CviI | [82] |
| Coumarin (2H-chromen-2-one) | C. violaceum ATCC 12472 C. violaceum CV026 | Inhibition of violacein biosynthesis | [53] | |
| Cinnamic acid derivatives | C. violaceum ATCC 12472 | Reduces the production of virulence factors—violacein, hemolysin, chitinase, and biofilm formation; downregulation of some QS-related metabolites (ethanolamine and L-methionine); decreases the expression of cviI and cviR genes; inhibition of the C10-HSL synthesis | [124] | |
| Methyl gallate | C. violaceum ATCC 12472 C. violaceum ATCC 31532 C. Violaceum CV026 | Suppression of the synthesis and activity of AHL | [125] | |
| Phytol | C. violaceum ATCC 12472 C. Violaceum ATCC 31532 | Reducing QS-regulated traits—biofilm formation, cell aggregation, and alkaline protease activity; binds to CviR | [47] | |
| Thymol | C. violaceum ATCC 12472 | Inhibition of violacein synthesis, biofilm formation, and EPS production; binds to CviR | [126] |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| QS | Quorum sensing |
| QSIs | Quorum-sensing inhibitors |
| AI | Autoinducer |
| AHLs | Acyl-homoserine lactones |
References
- Kumar, P.; Sharma, S.; Tripathi, V.N. Quorum sensing in bacteria of rice rhizospheres from Chhattisgarh, India. Bioinformation 2023, 19, 199–205. [Google Scholar] [CrossRef]
- Bouyahya, A.; Dakka, N.; Et-Touys, A.; Abrini, J.; Bakri, Y. Medicinal plant products targeting quorum sensing for combating bacterial infections. Asian Pac. J. Trop. Med. 2017, 10, 729–743. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Hu, W.; Tian, Z.; Yuan, D.; Yi, G.; Zhou, Y.; Cheng, Q.; Zhu, J.; Li, M. Developing natural products as potential anti-biofilm agents. Chin. Med. 2019, 14, 11. [Google Scholar] [CrossRef]
- Rasmussen, T.B.; Manefield, M.; Andersen, J.B.; Eberl, L.; Anthoni, U.; Christophersen, C.; Steinberg, P.; Kjelleberg, S.; Givskov, M. How Delisea pulchra furanones affect quorum sensing and swarming motility in Serratia liquefaciens MG1. Microbiology 2000, 146, 3237–3244. [Google Scholar] [CrossRef] [PubMed]
- Parker, C.T.; Sperandio, V. Cell-to-cell signalling during pathogenesis. Cell. Microbiol. 2009, 11, 363–369. [Google Scholar] [CrossRef]
- Rutherford, S.T.; Bassler, B.L. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- Kalia, V.C. Quorum sensing inhibitors: An overview. Biotechnol. Adv. 2013, 31, 224–245. [Google Scholar] [CrossRef]
- Yi, L.; Dong, X.; Grenier, D.; Wang, K.; Wang, Y. Research progress of bacterial quorum sensing receptors: Classification, structure, function and characteristics. Sci. Total Environ. 2021, 763, 143031. [Google Scholar] [CrossRef]
- Stoitsova, S.R.; Paunova-Krasteva, T.S.; Borisova, D.B. Modulation of Biofilm Growth by Sub-Inhibitory Amounts of Antibacterial Substances. In Microbial Biofilms—Importance and Applications; Dhanasekaran, D., Thajuddin, N., Eds.; InTech: Rijeka, Croatia, 2016; ISBN 978-953-51-2436-8. [Google Scholar] [CrossRef]
- Trendafilova, A.; Ivanova, V.; Rangelov, M.; Todorova, M.; Ozek, G.; Yur, S.; Ozek, T.; Aneva, I.; Veleva, R.; Moskova-Doumanova, V.; et al. Caffeoylquinic Acids, Cytotoxic, Antioxidant, Acetylcholinesterase and Tyrosinase Enzyme Inhibitory Activities of Six Inula Species from Bulgaria. Chem. Biodivers. 2020, 17, e2000051. [Google Scholar] [CrossRef]
- Stoitsova, S.; Paunova-Krasteva, T.; Dimitrova, P.D.; Damyanova, T. The concept for the antivirulence therapeutics approach as alternative to antibiotics: Hope or still a fiction? Biotechnol. Biotechnol. Equip. 2022, 36, 697–705. [Google Scholar] [CrossRef]
- Hemdan, B.A.; Mostafa, A.; Elbatanony, M.M.; El-Feky, A.M.; Paunova-Krasteva, T.; Stoitsova, S.; El-Liethy, M.A.; El-Taweel, G.E.; Abu Mraheil, M. Bioactive Azadirachta indica and Melia azedarach leaves extracts with anti-SARS-CoV-2 and antibacterial activities. PLoS ONE 2023, 18, e0282729. [Google Scholar] [CrossRef]
- Paunova-Krasteva, T.; Hemdan, B.A.; Dimitrova, P.D.; Damyanova, T.; El-Feky, A.M.; Elbatanony, M.M.; Stoitsova, S.; El-Liethy, M.A.; El-Taweel, G.E.; El Nahrawy, A.M. Hybrid Chitosan/CaO-Based Nanocomposites Doped with Plant Extracts from Azadirachta indica and Melia azedarach: Evaluation of Antibacterial and Antibiofilm Activities. BioNanoScience 2023, 13, 88–102. [Google Scholar] [CrossRef]
- Nealson, K.H.; Platt, T.; Hastings, J.W. Cellular Control of the Synthesis and Activity of the Bacterial Luminescent System. J. Bacteriol. 1970, 104, 313–322. [Google Scholar] [CrossRef]
- Nealson, K.H.; Hastings, J.W. Bacterial Bioluminescence: Its Control and Ecological Significance. Microbiol. Rev. 1979, 43, 496–518. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Bassler, B.L. Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef]
- Abisado, R.G.; Benomar, S.; Klaus, J.R.; Dandekar, A.A.; Chandler, J.R. Bacterial Quorum Sensing and Microbial Community Interactions. MBio 2018, 9, e02331-17. [Google Scholar] [CrossRef]
- Chugani, S.; Greenberg, E.P. An evolving perspective on the Pseudomonas aeruginosa orphan quorum sensing regulator QscR. Front. Cell. Infect. Microbiol. 2014, 4, 152. [Google Scholar] [CrossRef]
- Durán, N.; Justo, G.Z.; Durán, M.; Brocchi, M.; Cordi, L.; Tasic, L.; Castro, G.R.; Nakazato, G. Advances in Chromobacterium violaceum and properties of violacein-Its main secondary metabolite: A review. Biotechnol. Adv. 2016, 34, 1030–1045. [Google Scholar] [CrossRef]
- Rajamanikandan, S.; Srinivasan, P. Exploring the selectivity of auto-inducer complex with LuxR using molecular docking, mutational studies and molecular dynamics simulations. J. Mol. Struct. 2017, 1131, 281–293. [Google Scholar] [CrossRef]
- Vadakkan, K.; Choudhury, A.A.; Gunasekaran, R.; Hemapriya, J.; Vijayanand, S. Quorum sensing intervened bacterial signaling: Pursuit of its cognizance and repression. J. Genet. Eng. Biotechnol. 2018, 16, 239–252. [Google Scholar] [CrossRef]
- Pena, R.T.; Blasco, L.; Ambroa, A.; González-Pedrajo, B.; Fernández-García, L.; López, M.; Bleriot, I.; Bou, G.; García-Contreras, R.; Wood, T.K.; et al. Relationship Between Quorum Sensing and Secretion Systems. Front. Microbiol. 2019, 10, 1100. [Google Scholar] [CrossRef]
- Saeki, E.K.; Kobayashi, R.K.T.; Nakazato, G. Quorum sensing system: Target to control the spread of bacterial infections. Microb. Pathog. 2020, 142, 104068. [Google Scholar] [CrossRef] [PubMed]
- Haque, S.; Ahmad, F.; Dar, S.A.; Jawed, A.; Mandal, R.K.; Wahid, M.; Lohani, M.; Khan, S.; Singh, V.; Akhter, N. Developments in strategies for Quorum Sensing virulence factor inhibition to combat bacterial drug resistance. Microb. Pathog. 2018, 121, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Majdura, J.; Jankiewicz, U.; Gałązka, A.; Orzechowski, S. The Role of Quorum Sensing Molecules in Bacterial–Plant Interactions. Metabolites 2023, 13, 114. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-Cell Communication in Bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [PubMed]
- De Keersmaecker, S.C.J.; Sonck, K.; Vanderleyden, J. Let LuxS speak up in AI-2 signaling. Trends Microbiol. 2006, 14, 114–119. [Google Scholar] [CrossRef]
- Stephens, K.; Bentley, W.E. Quorum Sensing from Two Engineers’ Perspectives. Isr. J. Chem. 2023, 63, e202200083. [Google Scholar] [CrossRef]
- Ha, C.; Park, S.J.; Im, S.J.; Park, S.J.; Lee, J.H. Interspecies signaling through QscR, a quorum receptor of Pseudomonas aeruginosa. Mol. Cells 2012, 33, 53–59. [Google Scholar] [CrossRef]
- Stauff, D.L.; Bassler, B.L. Quorum Sensing in Chromobacterium violaceum: DNA Recognition and Gene Regulation by the CviR Receptor. J. Bacteriol. 2011, 193, 3871–3878. [Google Scholar] [CrossRef]
- Kothari, V.; Sharma, S.; Padia, D. Recent research advances on Chromobacterium violaceum. Asian Pac. J. Trop. Med. 2017, 10, 744–752. [Google Scholar] [CrossRef] [PubMed]
- Venkatramanan, M.; Sankar Ganesh, P.; Senthil, R.; Akshay, J.; Veera Ravi, A.; Langeswaran, K.; Vadivelu, J.; Nagarajan, S.; Rajendran, K.; Shankar, E.M. Inhibition of Quorum Sensing and Biofilm Formation in Chromobacterium violaceum by Fruit Extracts of Passiflora edulis. ACS Omega 2020, 5, 25605–25616. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, N.; Shin, J.Y.; Jung, K. Activity, Abundance, and Localization of Quorum Sensing Receptors in Vibrio harveyi. Front. Microbiol. 2017, 8, 634. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lu, Y.; Ye, X.; Emam, M.; Zhang, H.; Wang, H. Current advances in Vibrio harveyi quorum sensing as drug discovery targets. Eur. J. Med. Chem. 2020, 207, 112741. [Google Scholar] [CrossRef] [PubMed]
- Watve, S.; Barrasso, K.; Jung, S.A.; Davis, K.J.; Hawver, L.A.; Khataokar, A.; Palaganas, R.G.; Neiditch, M.B.; Perez, L.J.; Ng, W.-L. Parallel quorum-sensing system in Vibrio cholerae prevents signal interference inside the host. PLoS Pathog. 2020, 16, e1008313. [Google Scholar] [CrossRef] [PubMed]
- Brazilian National Genome Project Consortium. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc. Natl. Acad. Sci. USA 2003, 100, 11660–11665. [Google Scholar] [CrossRef] [PubMed]
- Kamaeva, A.A.; Vasilchenko, A.S.; Deryabin, D.G. Atomic Force Microscopy Reveals a Morphological Differentiation of Chromobacterium violaceum Cells Associated with Biofilm Development and Directed by N-Hexanoyl-L-Homoserine Lactone. PLoS ONE 2014, 9, e103741. [Google Scholar] [CrossRef]
- Kaniyarakkal, V.; Orvankundil, S.; Lalitha, S.K.; Thazhethekandi, R.; Thottathil, J. Chromobacterium violaceum Septicaemia and Urinary Tract Infection: Case Reports from a Tertiary Care Hospital in South India. Case Rep. Infect. Dis. 2016, 2016, 6795743. [Google Scholar] [CrossRef]
- de Oca-Mejía, M.M.; Castillo-Juárez, I.; Martínez-Vázquez, M.; Soto-Hernandez, M.; García-Contreras, R. Influence of quorum sensing in multiple phenotypes of the bacterial pathogen Chromobacterium violaceum. Pathog. Dis. 2015, 73, 1–4. [Google Scholar] [CrossRef]
- Durán, M.; Faljoni-Alario, A.; Durán, N. Chromobacterium violaceum and its important metabolites—Review. Folia Microbiol. 2010, 55, 535–547. [Google Scholar] [CrossRef]
- Batista, J.H.; da Silva Neto, J.F. Chromobacterium violaceum Pathogenicity: Updates and Insights from Genome Sequencing of Novel Chromobacterium Species. Front. Microbiol. 2017, 10, 2213. [Google Scholar] [CrossRef]
- Evans, K.C.; Benomar, S.; Camuy-Vélez, L.A.; Nasseri, E.B.; Wang, X.; Neuenswander, B.; Chandler, J.R. Quorum-sensing control of antibiotic resistance stabilizes cooperation in Chromobacterium violaceum. ISME J. 2018, 12, 1263–1272. [Google Scholar] [CrossRef]
- Cheng, W.J.; Zhou, J.W.; Zhang, P.P.; Luo, H.Z.; Tang, S.; Li, J.J.; Deng, S.M.; Jia, A.Q. Quorum sensing inhibition and tobramycin acceleration in Chromobacterium violaceum by two natural cinnamic acid derivatives. Appl. Microbiol. Biotechnol. 2020, 104, 5025–5037. [Google Scholar] [CrossRef] [PubMed]
- Gohil, N.; Bhattacharjee, G.; Gayke, M.; Narode, H.; Alzahrani, K.J.; Singh, V. Enhanced production of violacein by Chromobacterium violaceum using agro-industrial waste soybean meal. J. Appl. Microbiol. 2022, 132, 1121–1133. [Google Scholar] [CrossRef] [PubMed]
- Grandclément, C.; Tannières, M.; Moréra, S.; Dessaux, Y.; Faure, D. Quorum quenching: Role in nature and applied developments. FEMS Microbiol. Rev. 2016, 40, 86–116. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Chen, W.J.; Bhatt, K.; Zhou, Z.; Huang, Y.; Zhang, L.H.; Chen, S.; Wang, J. Innovative microbial disease biocontrol strategies mediated by quorum quenching and their multifaceted applications: A review. Front. Plant Sci. 2023, 13, 1063393. [Google Scholar] [CrossRef]
- Devescovi, G.; Kojic, M.; Covaceuszach, S.; Cámara, M.; Williams, P.; Bertani, I.; Subramoni, S.; Venturi, V. Negative Regulation of Violacein Biosynthesis in Chromobacterium violaceum. Front. Microbiol. 2017, 8, 349. [Google Scholar] [CrossRef]
- Mookherjee, A.; Singh, S.; Maiti, M.K. Quorum sensing inhibitors: Can endophytes be prospective sources? Arch. Microbiol. 2018, 200, 355–369. [Google Scholar] [CrossRef]
- Snoussi, M.; Noumi, E.; Punchappady-Devasya, R.; Trabelsi, N.; Kanekar, S.; Nazzaro, F.; Fratianni, F.; Flamini, G.; De Feo, V.; Al-Sieni, A. Antioxidant properties and anti-quorum sensing potential of Carum copticum essential oil and phenolics against Chromobacterium violaceum. J. Food Sci. Technol. 2018, 55, 2824–2832. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.P.; Zeng, H.; Wan, C.X.; Zhou, Z.B. Amicoumacins from a desert bacterium: Quorum sensing inhibitor against Chromobacterium violaceum. Nat. Prod. Res. 2021, 35, 5508–5512. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.L.; Sam, C.K.; Yin, W.F.; Tan, L.; Krishnan, T.; Chong, Y.M.; Chan, K.G. Plant-Derived Natural Products as Sources of Anti-Quorum Sensing Compounds. Sensors 2013, 13, 6217–6228. [Google Scholar] [CrossRef] [PubMed]
- Deryabin, D.; Galadzhieva, A.; Kosyan, D.; Duskaev, G. Plant-Derived Inhibitors of AHL-Mediated Quorum Sensing in Bacteria: Modes of Action. Int. J. Mol. Sci. 2019, 20, 5588. [Google Scholar] [CrossRef] [PubMed]
- Truchado, P.; Larrosa, M.; Castro-Ibáñez, I.; Allende, A. Plant food extracts and phytochemicals: Their role as Quorum Sensing Inhibitors. Trends Food Sci. Technol. 2015, 43, 189–204. [Google Scholar] [CrossRef]
- Joo, H.S.; Deyrup, S.T.; Shim, S.H. Endophyte-produced antimicrobials: A review of potential lead compounds with a focus on quorum-sensing disruptors. Phytochem. Rev. 2021, 20, 543–568. [Google Scholar] [CrossRef]
- Bassler, B.L.; Losick, R. Bacterially Speaking. Cell 2006, 125, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Bassler, B.L. How bacteria talk to each other: Regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 1999, 2, 582–587. [Google Scholar] [CrossRef]
- Yang, M.; Meng, F.; Gu, W.; Li, F.; Tao, Y.; Zhang, Z.; Zhang, F.; Yang, X.; Li, J.; Yu, J. Effects of Natural Products on Bacterial Communication and Network-Quorum Sensing. BioMed Res. Int. 2020, 2020, 8638103. [Google Scholar] [CrossRef]
- Rasmussen, T.B.; Givskov, M. Quorum sensing inhibitors: A bargain of effects. Microbiology 2006, 152, 895–904. [Google Scholar] [CrossRef]
- Fleitas Martínez, O.; Rigueiras, P.O.; da Silva Pires, Á.; Porto, W.F.; Silva, O.N.; de la Fuente-Nunez, C.; Franco, O.L. Interference with Quorum-Sensing Signal Biosynthesis as a Promising Therapeutic Strategy Against Multidrug-Resistant Pathogens. Front. Cell Infect. Microbiol. 2019, 8, 444. [Google Scholar] [CrossRef]
- Rasko, D.A.; Sperandio, V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 2010, 9, 117–128. [Google Scholar] [CrossRef]
- Gao, M.; Teplitski, M.; Robinson, J.B.; Bauer, W.D. Production of Substances by Medicago truncatula that Affect Bacterial Quorum Sensing. Mol. Plant-Microbe Interact. 2003, 16, 827–834. [Google Scholar] [CrossRef]
- Hentzer, M.; Givskov, M. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J. Clin. Investig. 2003, 112, 1300–1307. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.C.; Andersen, B.; Thrane, U. The use of secondary metabolite profiling in chemotaxonomy of filamentous fungi. Mycol. Res. 2008, 112, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Dobretsov, S.; Teplitski, M.; Bayer, M.; Gunasekera, S.; Proksch, P.; Paul, V.J. Inhibition of marine biofouling by bacterial quorum sensing inhibitors. Biofouling 2011, 27, 893–905. [Google Scholar] [CrossRef]
- Wahjudi, M.; Papaioannou, E.; Hendrawati, O.; van Assen, A.H.G.; van Merkerk, R.; Cool, R.H.; Poelarends, G.J.; Quax, W.J. PA0305 of Pseudomonas aeruginosa is a quorum quenching acylhomoserine lactone acylase belonging to the Ntn hydrolase superfamily. Microbiology 2011, 157, 2042–2055. [Google Scholar] [CrossRef]
- Terwagne, M.; Mirabella, A.; Lemaire, J.; Deschamps, C.; De Bolle, X.; Letesson, J.J. Quorum Sensing and Self-Quorum Quenching in the Intracellular Pathogen Brucellamelitensis. PLoS ONE 2013, 8, e82514. [Google Scholar] [CrossRef] [PubMed]
- Teiber, J.F.; Horke, S.; Haines, D.C.; Chowdhary, P.K.; Xiao, J.; Kramer, G.L.; Haley, R.W.; Draganov, D.I. Dominant Role of Paraoxonases in Inactivation of the Pseudomonas aeruginosa Quorum-Sensing Signal N-(3-Oxododecanoyl)-l-Homoserine Lactone. Infect. Immun. 2008, 76, 2512–2519. [Google Scholar] [CrossRef]
- Martinelli, D.; Grossmann, G.; Séquin, U.; Brandl, H.; Bachofen, R. Effects of natural and chemically synthesized furanones on quorum sensing in Chromobacterium violaceum. BMC Microbiol. 2004, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Amaya, S.; Pereira, J.A.; Borkosky, S.A.; Valdez, J.C.; Bardón, A.; Arena, M.E. Inhibition of quorum sensing in Pseudomonas aeruginosa by sesquiterpene lactones. Phytomedicine. 2012, 19, 1173–1177. [Google Scholar] [CrossRef]
- Martín-Rodríguez, A.J.; Ticona, J.C.; Jiménez, I.A.; Flores, N.; Fernández, J.J.; Bazzocchi, I.L. Flavonoids from Piper delineatum modulate quorum-sensing-regulated phenotypes in Vibrio harveyi. Phytochemistry 2015, 117, 98–106. [Google Scholar] [CrossRef]
- Majik, M.S.; Gawas, U.B.; Mandrekar, V.K. Next generation quorum sensing inhibitors: Accounts on structure activity relationship studies and biological activities. Bioorg. Med. Chem. 2020, 28, 115728. [Google Scholar] [CrossRef]
- Ahmed, S.O.; Zedan, H.H.; Ibrahim, Y.M. Quorum sensing inhibitory effect of bergamot oil and aspidosperma extract against Chromobacterium violaceum and Pseudomonas aeruginosa. Arch. Microbiol. 2021, 203, 4663–4675. [Google Scholar] [CrossRef]
- Kazemian, H.; Ghafourian, S.; Heidari, H.; Amiri, P.; Yamchi, J.K.; Shavalipour, A.; Houri, H.; Maleki, A.; Sadeghifard, N. Antibacterial, anti-swarming and anti-biofilm formation activities of Chamaemelum nobile against Pseudomonas aeruginosa. Rev. Soc. Bras. Med. Trop. 2015, 48, 432–436. [Google Scholar] [CrossRef]
- Bacha, K.; Tariku, Y.; Gebreyesus, F.; Zerihun, S.; Mohammed, A.; Weiland-Bräuer, N.; Schmitz, R.A.; Mulat, M. Antimicrobial and anti-Quorum Sensing activities of selected medicinal plants of Ethiopia: Implication for development of potent antimicrobial agents. BMC Microbiol. 2016, 16, 139. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, I.; Paunova-Krasteva, T.; Petrova, Z.; Grozdanov, P.; Nikolova, N.; Tsonev, G.; Triantafyllidis, A.; Andreev, S.; Trepechova, M.; Milkova, V.; et al. Bulgarian Medicinal Extracts as Natural Inhibitors with Antiviral and Antibacterial Activity. Plants 2022, 11, 1666. [Google Scholar] [CrossRef] [PubMed]
- Olivero-Verbel, J.; Barreto-Maya, A.; Bertel-Sevilla, A.; Stashenko, E.E. Composition, anti-quorum sensing and antimicrobial activity of essential oils from Lippia alba. Braz. J. Microbiol. 2014, 45, 759–767. [Google Scholar] [CrossRef]
- Choo, J.H.; Rukayadi, Y.; Hwang, J.K. Inhibition of bacterial quorum sensing by vanilla extract. Lett. Appl. Microbiol. 2006, 42, 637–641. [Google Scholar] [CrossRef]
- Al-Hussaini, R.; Mahasneh, A. Microbial Growth and Quorum Sensing Antagonist Activities of Herbal Plants Extracts. Molecules 2009, 14, 3425–3435. [Google Scholar] [CrossRef]
- Singh, B.N.; Singh, B.R.; Singh, R.L.; Prakash, D.; Sarma, B.K.; Singh, H.B. Antioxidant and anti-quorum sensing activities of green pod of Acacia nilotica L. Food Chem. Toxicol. 2009, 47, 778–786. [Google Scholar] [CrossRef]
- Singh, B.N.; Singh, B.R.; Singh, R.L.; Prakash, D.; Dhakarey, R.; Upadhyay, G.; Singh, H.B. Oxidative DNA damage protective activity, antioxidant and anti-quorum sensing potentials of Moringa oleifera. Food Chem. Toxicol. 2009, 47, 1109–1116. [Google Scholar] [CrossRef] [PubMed]
- Asfour, H. Anti-quorum sensing natural compounds. J. Microsc. Ultrastruct. 2018, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Swem, L.R.; Swem, D.L.; Stauff, D.L.; O’Loughlin, C.T.; Jeffrey, P.D.; Bassler, B.L.; Hughson, F.M. A Strategy for Antagonizing Quorum Sensing. Mol. Cell 2011, 42, 199–209. [Google Scholar] [CrossRef]
- Aliyu, A.B.; Koorbanally, N.A.; Moodley, B.; Singh, P.; Chenia, H.Y. Quorum sensing inhibitory potential and molecular docking studies of sesquiterpene lactones from Vernonia blumeoides. Phytochemistry 2016, 126, 23–33. [Google Scholar] [CrossRef]
- Peña-González, M.C.; Muñoz-Cázares, N.; Peña-Rodríguez, L.M. Natural Inhibitors of Quorum-Sensing Factors: A Novel Strategy to Control Pathogenic Bacteria. Rev. Bras. Farmacogn. 2020, 30, 743–755. [Google Scholar] [CrossRef]
- Takó, M.; Kerekes, E.B.; Zambrano, C.; Kotogán, A.; Papp, T.; Krisch, J.; Vágvölgyi, C. Plant Phenolics and Phenolic-Enriched Extracts as Antimicrobial Agents against Food-Contaminating Microorganisms. Antioxidants 2020, 9, 165. [Google Scholar] [CrossRef] [PubMed]
- Koh, K.H.; Tham, F.Y. Screening of traditional Chinese medicinal plants for quorum-sensing inhibitors activity. J. Microbiol. Immunol. Infect. 2011, 44, 144–148. [Google Scholar] [CrossRef]
- Fatima, Q.; Zahin, M.; Khan, M.S.A.; Ahmad, I. Modulation of quorum sensing controlled behaviour of bacteria by growing seedling, seed and seedling extracts of leguminous plants. Indian. J. Microbiol. 2010, 50, 238–242. [Google Scholar] [CrossRef]
- Song, C.; Ma, H.; Zhao, Q.; Song, S.; Jia, Z. Inhibition of Quorum Sensing Activity by Ethanol Extract of Scutellaria baicalensis Georgi. J. Plant Pathol. Microbiol. 2012, 7, 1–4. [Google Scholar] [CrossRef]
- Chong, Y.M.; Yin, W.F.; Ho, C.Y.; Mustafa, M.R.; Hadi, A.H.A.; Awang, K.; Narrima, P.; Koh, C.L.; Appleton, D.R.; Chan, K.G. Malabaricone C from Myristica cinnamomea Exhibits Anti-Quorum Sensing Activity. J. Nat. Prod. 2011, 74, 2261–2264. [Google Scholar] [CrossRef]
- Musthafa, K.S.; Ravi, A.V.; Annapoorani, A.; Packiavathy, I.S.V.; Pandian, S.K. Evaluation of Anti-Quorum-Sensing Activity of Edible Plants and Fruits through Inhibition of the N-Acyl-Homoserine Lactone System in Chromobacterium violaceum and Pseudomonas aeruginosa. Chemotherapy 2010, 56, 333–339. [Google Scholar] [CrossRef]
- Chenia, H. Anti-Quorum Sensing Potential of Crude Kigelia africana Fruit Extracts. Sensors 2013, 13, 2802–2817. [Google Scholar] [CrossRef]
- Olivero, V.; Jesús, T.; Pájaro, C.; Nerlis, P.; Stashenko, E. Antiquorum sensing activity of essential oils isolated from different species of the genus Piper. Vitae 2011, 18, 77–82. [Google Scholar] [CrossRef]
- Zhou, L.; Zheng, H.; Tang, Y.; Yu, W.; Gong, Q. Eugenol inhibits quorum sensing at sub-inhibitory concentrations. Biotechnol. Lett. 2013, 35, 631–637. [Google Scholar] [CrossRef]
- Musthafa, K.S.; Sahu, S.K.; Ravi, A.V.; Kathiresan, K. Anti-quorum sensing potential of the mangrove Rhizophora annamalayana. World J. Microbiol. Biotechnol. 2013, 29, 1851–1858. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.A. Assessment of Anti-Quorum Sensing Activity for Some Ornamental and Medicinal Plants Native to Egypt. Sci. Pharm. 2013, 81, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Vasavi, H.S.; Arun, A.B.; Rekha, P.D. Inhibition of quorum sensing in Chromobacterium violaceum by Syzygium cumini L. and Pimenta dioica L. Asian Pac. J. Trop. Biomed. 2013, 3, 954–959. [Google Scholar] [CrossRef]
- Ceylan, O.; Sahin, M.; Akdamar, G. Antioxidant and Anti-quorum Sensing Potential of Acer monspessulanum subsp. monspessulanum Extracts. Planta Med. 2016, 82, 1335–1340. [Google Scholar] [CrossRef] [PubMed]
- Tamfu, A.N.; Kucukaydin, S.; Ceylan, O.; Sarac, N.; Duru, M.E. Phenolic Composition, Enzyme Inhibitory and Anti-quorum Sensing Activities of Cinnamon (Cinnamomum zeylanicum Blume) and Basil (Ocimum basilicum Linn). Chem. Afr. 2021, 4, 759–767. [Google Scholar] [CrossRef]
- Moradi, F.; Hadi, N.; Bazargani, A. Evaluation of quorum-sensing inhibitory effects of extracts of three traditional medicine plants with known antibacterial properties. New Microbes New Infect. 2020, 38, 100769. [Google Scholar] [CrossRef]
- Alibi, S.; Ben Selma, W.; Ramos-Vivas, J.; Smach, M.A.; Touati, R.; Boukadida, J.; Navas, J.; Ben Mansour, H. Anti-oxidant, antibacterial, anti-biofilm, and anti-quorum sensing activities of four essential oils against multidrug-resistant bacterial clinical isolates. Curr. Res. Transl. Med. 2020, 68, 59–66. [Google Scholar] [CrossRef]
- Li, Y.; Xiao, P.; Wang, Y.; Hao, Y. Mechanisms and Control Measures of Mature Biofilm Resistance to Antimicrobial Agents in the Clinical Context. ACS Omega 2020, 5, 22684–22690. [Google Scholar] [CrossRef] [PubMed]
- Qais, F.A.; Khan, M.S.; Ahmad, I. Broad-spectrum quorum sensing and biofilm inhibition by green tea against gram-negative pathogenic bacteria: Deciphering the role of phytocompounds through molecular modelling. Microb. Pathog. 2019, 126, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.R.M.; Asfour, H.Z.; Elshali, K.Z.; Shaaban, M.I.A.; Al-Attas, A.A.M.; Mohamed, G.A.A. Antimicrobial, antiquorum sensing, and antiproliferative activities of sesquiterpenes from Costus speciosus rhizomes. Pak. J. Pharm. Sci. 2019, 32, 109–115. [Google Scholar]
- Rahman, M.R.T.; Lou, Z.; Yu, F.; Wang, P.; Wang, H. Anti-quorum sensing and anti-biofilm activity of Amomum tsaoko (Amommum tsao-ko Crevost et Lemarie) on foodborne pathogens. Saudi J. Biol. Sci. 2017, 24, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wang, L.; Gao, J.; Liu, X.; Feng, Y.; Wu, Q.; Baloch, A.B.; Cui, L.; Xia, X. Tannin-Rich Fraction from Pomegranate Rind Inhibits Quorum Sensing in Chromobacterium violaceum and Biofilm Formation in Escherichia coli. Foodborne Pathog. Dis. 2016, 13, 28–35. [Google Scholar] [CrossRef]
- Poli, J.P.; Guinoiseau, E.; de Rocca Serra, D.; Sutour, S.; Paoli, M.; Tomi, F.; Quilichini, Y.; Berti, L.; Lorenzi, V. Anti-Quorum Sensing Activity of 12 Essential Oils on Chromobacterium violaceum and Specific Action of cis-cis-p-Menthenolide from Corsican Mentha suaveolens ssp. Insularis. Molecules 2018, 23, 2125. [Google Scholar] [CrossRef]
- Noumi, E.; Merghni, A.; Alreshidi, M.M.; Haddad, O.; Akmadar, G.; De Martino, L.; Mastouri, M.; Ceylan, O.; Snoussi, M.; Al-sieni, A.; et al. Chromobacterium violaceum and Pseudomonas aeruginosa PAO1: Models for Evaluating Anti-Quorum Sensing Activity of Melaleuca alternifolia Essential Oil and Its Main Component Terpinen-4-ol. Molecules 2018, 23, 2672. [Google Scholar] [CrossRef]
- Shukla, V.; Bhathena, Z. Broad Spectrum Anti-Quorum Sensing Activity of Tannin-Rich Crude Extracts of Indian Medicinal Plants. Scientifica. 2016, 2016, 5823013. [Google Scholar] [CrossRef]
- Zhang, J.; Rui, X.; Wang, L.; Guan, Y.; Sun, X.; Dong, M. Polyphenolic extract from Rosa rugosa tea inhibits bacterial quorum sensing and biofilm formation. Food Control 2014, 42, 125–131. [Google Scholar] [CrossRef]
- Li, G.; Yan, C.; Xu, Y.; Feng, Y.; Wu, Q.; Lv, X.; Yang, B.; Wang, X.; Xia, X. Punicalagin Inhibits Salmonella Virulence Factors and Has Anti-Quorum-Sensing Potential. Appl. Environ. Microbiol. 2014, 80, 6204–6211. [Google Scholar] [CrossRef]
- Deryabin, D.; Tolmacheva, A. Antibacterial and Anti-Quorum Sensing Molecular Composition Derived from Quercus cortex (Oak bark) Extract. Molecules. 2015, 20, 17093–17108. [Google Scholar] [CrossRef]
- Paliya, B.S.; Mathew, J.; Singh, B.N. Evaluation of Anti-quorum Sensing Potential of Saraca asoca (Family Caesalpiniaceae) against Chromobacterium violaceum and Pseudomonas aeruginosa PA01. J. Pharm. Res. Int. 2021, 33, 71–82. [Google Scholar] [CrossRef]
- Ma, L.; Yao, L. Antiviral Effects of Plant-Derived Essential Oils and Their Components: An Updated Review. Molecules 2020, 25, 2627. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Jurado, F.; Navarro-Cruz, A.R.; Ochoa-Velasco, C.E.; Palou, E.; López-Malo, A.; Ávila-Sosa, R. Essential oils in vapor phase as alternative antimicrobials: A review. Crit. Rev. Food Sci. Nutr. 2020, 60, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhao, X. Effects of quorum sensing on the biofilm formation and viable but non-culturable state. Food Res. Int. 2020, 137, 109742. [Google Scholar] [CrossRef]
- Bhattacharya, S.P.; Karmakar, S.; Acharya, K.; Bhattacharya, A. Quorum sensing inhibition and antibiofilm action of triterpenoids: An updated insight. Fitoterapia 2023, 167, 105508. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Lahiri, D.; Nag, M.; Dey, A.; Pandit, S.; Sarkar, T.; Pati, S.; Abdul Kari, Z.; Ishak, A.R.; Edinur, H.A.; et al. Phytocompound Mediated Blockage of Quorum Sensing Cascade in ESKAPE Pathogens. Antibiotics 2022, 11, 61. [Google Scholar] [CrossRef]
- Li, Y.L.; Chu, Z.Y.; Liu, G.M.; Yang, S.Q.; Zeng, H. The Derived Components of Gnaphalium hypoleucum DC. Reduce Quorum Sensing of Chromobacterium violaceum. Molecules 2022, 27, 4881. [Google Scholar] [CrossRef]
- Bouyahya, A.; Chamkhi, I.; Balahbib, A.; Rebezov, M.; Shariati, M.A.; Wilairatana, P.; Mubarak, M.S.; Benali, T.; El Omari, N. Mechanisms, Anti-Quorum-Sensing Actions, and Clinical Trials of Medicinal Plant Bioactive Compounds against Bacteria: A Comprehensive Review. Molecules 2022, 27, 1484. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, O.M.; Kiendrebeogo, M.; Rajaonson, S.; Diallo, B.; Mol, A.; El Jaziri, M.; Baucher, M. Identification of Catechin as One of the Flavonoids from Combretum albiflorum Bark Extract That Reduces the Production of Quorum-Sensing-Controlled Virulence Factors in Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 2010, 76, 243–253. [Google Scholar] [CrossRef]
- Cárcamo, G.; Silva, M.; Becerra, J.; Urrutia, H.; Sossa, K.; Paz, C. Inhibition of quorum sensing by drimane lactones from chilean flora. J. Chil. Chem. Soc. 2014, 59, 2622–2624. [Google Scholar] [CrossRef]
- Aliyu, A.B.; Koorbanally, N.A.; Moodley, B.; Chenia, H.Y. Sesquiterpene lactones from Polydora serratuloides and their quorum sensing inhibitory activity. Nat. Prod. Res. 2021, 35, 4517–4523. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chu, W.; Ye, C.; Gaeta, B.; Tao, H.; Wang, M.; Qiu, Z. Chlorogenic acid attenuates virulence factors and pathogenicity of Pseudomonas aeruginosa by regulating quorum sensing. Appl. Microbiol. Biotechnol. 2019, 103, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Rayan, M.; Abu Lafi, S.; Falah, M.; Kacergius, T.; Kirkliauskiene, A.; Gabe, V.; Rayan, A. Alkyl Gallates as Potential Antibiofilm Agents: A Review. Molecules 2023, 28, 1751. [Google Scholar] [CrossRef]
- Saptami, K.L.; Arokia, D.; Chandrasekaran, J.; Rekha, P.D. Competitive interaction of thymol with cviR inhibits quorum sensing and associated biofilm formation in Chromobacterium violaceum. Int. Microbiol. 2022, 25, 629–638. [Google Scholar] [CrossRef]
| Sources of QSIs | Active Component | Bacteria | Inhibition Characteristics and Mode of Action | Ref.: |
|---|---|---|---|---|
| Prunella vulgaris (whole plant) Imperata cylindrica (underground stem) Nelumbo nucifera (leaf) Panax notoginseng (flower) Punica granatum (bark) Areca catechu (seed) | Acetone/water extracts | C. violaceum CV026 | QS and antimicrobial activities | [87] |
| Pisum sativum L. (seedling) Trigonella foenum graecum (seed) | Methanol and ethanol seed extracts | C. violaceum CV026, C. violaceum ATCC 12472 | Violacein production | [88] |
| Acacia nilotica (L.) (green pod) | Phenol and polyphenol compounds | C. violaceum ATCC 12472 | Violacein production | [80] |
| Scutellaria baicalensis Georgi | Ethanol extract | C. violaceum CV026 | Violacein production | [89] |
| Myristica cinnamomea King (bark) | Methanol extract and Malabaricone C | C. violaceum CV026 | Violacein | [90] |
| Ananas comosus Musa paradiciaca Manilkara zapota Ocimum sanctum | Fruit aqueous extracts | C. violaceum CV026, C. violaceum ATCC 12472 | Violacein production | [91] |
| Kigelia africana (Lam.) Benth. | Fruit ethyl acetate, dichloromethane, hexane, and methanol extracts | C. violaceum ATCC 12472, C. violaceum CV026, C. violaceum ATCC 31532 | Competitive binding to AHL-receptor, antimicrobial activity, and violacein production | [92] |
| Laurus nobilis L. Populus alba L. Populus nigra L. Lavandula angustifolia Rosmarinus officinalis L. Sonchus oleraceus L. Tecoma capensis Thunb. Lindl. Jasminum sambac Ait. | Ethanolic extracts | C. violaceum | Antimicrobial activities | [79] |
| Piper bredemeyer Piper bogotense Piper brachypodon (Benth.) | Essential oils | C. violaceum CV026 | Competitive binding to AHL-receptor, violacein production, and cell growth | [93] |
| Syzygium aromaticum (L.) Merrill, Perry (clove) | Extracts | C. violaceum CV026 | Violacein production | [94] |
| Rhizophora annamalayana Kathiresan (bark) | Bark extracts | C. violaceum ATCC 12472 | Antagonistic/allosteric inhibitors causing conformational changes in the receptor; violacein production | [95] |
| Adhatoda vasica L. (leaves) Bauhinia purpurea L. (leaves) Myoporum laetum G. Forst. (leaves) Lantana camara L. (leaves) Piper longum L. (fruits) Taraxacum officinale F.H. Wigg. (aerial parts) | Ethanol fractions | C. violaceum ATCC 12472 | Antimicrobial activities | [96] |
| Syzygium cumini (L.) Skeels. Pimenta dioica (L.) Merr. | Ethyl acetate fractions | C. violaceum ATCC 12472, C. violaceum ATCC 31532, C. violaceum CV026 | Inhibition of AHL activity; violacein production | [97] |
| Acer monspessulanum subsp. monspessulanum | Ethanol and ethyl acetate extracts | C. violaceum CV026, C. violaceum ATCC 12472 | Violacein production; antimicrobial activities | [98] |
| Cinnamomum zeylanicum, Ocimum basilicum | Ethanol extracts | C. violaceum CV026, C. violaceum ATCC 12472 | Anti-QS activities; violacein production | [99] |
| Rubus rosaefolius | Phenolic extracts | C. violaceum ATCC 12472 | Cluster movement, biofilm formation, and violacein production | [58] |
| Astilbe rivularis, Fragaria nubicola, Osbeckia nepalensis | Extracts | C. violaceum MTCC 2656 | Violacein | |
| Melicope lunuankenda (Gaertn.) T. G. Hartley | Hexane, chloroform, and methanol extracts | C. violaceum CV026 | Violacein production | |
| Nymphaea tetragona | Water extracts | C. violaceum | Violacein production | |
| Camellia sinensis L. | Water extracts | C. violaceum ATCC 12472 | Violacein production | |
| Allium cepa Lineu | Phenolic compounds | C. violaceum | Violacein production; swarming motility | |
| Elletaria cardamomum | Essential oils | C. violaceum | Violacein production | [24] |
| Eucalyptus radiate | ||||
| Origanum vulgare | ||||
| Rubus rosaefolius | Phenolic extracts | |||
| Syzygium aromaticum | Extracts | C. violaceum CV026 | QS inhibition assay; violacein production | [100] |
| Dionysia revoluta Boiss. | ||||
| Eucalyptus camaldulensis Dehnh. | ||||
| Cinnamomum verum | Essential oils | C. violaceum CV026 | Violacein production | [101] |
| Origanum majorana | ||||
| Thymus vulgaris | ||||
| Eugenia caryophyllata | ||||
| Lemon | Essential oils | C. violaceum SZMC 6269 | Biofilm formation | [102] |
| Juniper | ||||
| Cuminum cyminum | Methanol extract | C. violaceum ATCC 12472 | Violacein production | |
| Green tea | Extracts | C. violaceum ATCC 12472 | Ability to bind to CviR; violacein production | [103] |
| Costus speciosus | Methanol extract | C. violaceum | Violacein production | [104] |
| Amomum tsaoko | Crude extract | C. violaceum ATCC 12472 | Violacein production | [105] |
| Punica granatum | Tannin-rich fraction | C. violaceum ATCC 12472 | Violacein production | [106] |
| Mentha suaveolens ssp. insularis | Essential oils | C. violaceum wild-type strain—103350T | Violacein production; biofilm formation | [107] |
| Melaleuca alternifolia | Essential oils | C. violaceum ATCC 12472 | Violacein production | [108] |
| Syzygium cumini | Tannin-rich extracts | C. violaceum ATCC 12472 | Affect luxI; violacein production | [109] |
| Embelia ribes | C. violaceum ATCC 12472 | Violacein production | ||
| Phyllanthus emblica | C. violaceum CV026 | Affect cviR; violacein synthesis | ||
| Terminalia bellirica | C. violaceum ATCC 31532 | Affect the production of C6-HSL; violacein synthesis | ||
| Terminalia chebula | ||||
| Punica granatum | Pericarp | C. violaceum ATCC 12472 | Affect both cviI and cviR; violacein synthesis | |
| Mangifera indica | Flowers and seed kernel | C. violaceum ATCC 31532 | Affect both cviI and cviR; violacein synthesis | |
| Acacia arabica, | Barks | C. violaceum ATCC 12472 | Violacein production | |
| Terminalia arjuna | ||||
| Thespesia populnea | ||||
| Casuarina equisetifolia | ||||
| Rosa rugosa tea | Polyphenol (RTP) extract | C. violaceum CV026 | Violacein production | [110] |
| Punica granatum L. | Punicalagin | C. violaceum ATCC 12472 | Violacein production; growth | [111] |
| Quercus cortex (Oak bark) | Phytochemicals | C. violaceum CV026 | Violacein production; growth | [112] |
| Saraca asoca barks (stem) | Extracts | C. violaceum ATCC 12472 | Violacein production; anti-QS activities | [113] |
| Raspberry and cloudberry | Phenol extracts | C. violaceum | AHL inhibitors | [86] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dimitrova, P.D.; Damyanova, T.; Paunova-Krasteva, T. Chromobacterium Violaceum: A Model for Evaluating the Anti-Quorum Sensing Activities of Plant Substances. Sci. Pharm. 2023, 91, 33. https://doi.org/10.3390/scipharm91030033
Dimitrova PD, Damyanova T, Paunova-Krasteva T. Chromobacterium Violaceum: A Model for Evaluating the Anti-Quorum Sensing Activities of Plant Substances. Scientia Pharmaceutica. 2023; 91(3):33. https://doi.org/10.3390/scipharm91030033
Chicago/Turabian StyleDimitrova, Petya D., Tsvetozara Damyanova, and Tsvetelina Paunova-Krasteva. 2023. "Chromobacterium Violaceum: A Model for Evaluating the Anti-Quorum Sensing Activities of Plant Substances" Scientia Pharmaceutica 91, no. 3: 33. https://doi.org/10.3390/scipharm91030033
APA StyleDimitrova, P. D., Damyanova, T., & Paunova-Krasteva, T. (2023). Chromobacterium Violaceum: A Model for Evaluating the Anti-Quorum Sensing Activities of Plant Substances. Scientia Pharmaceutica, 91(3), 33. https://doi.org/10.3390/scipharm91030033








