Mapping Protein Targets of Carnosol, a Molecule Identified in Rosmarinus officinalis: In Silico Docking Studies and Network Pharmacology
Abstract
1. Introduction
2. Materials and Methods
2.1. Analysis of the Hydro-Alcoholic Extract of Rosemary by HPLC-QTOF-MS/MS
2.1.1. Plant Material and Extraction
2.1.2. Sample Preparation and HPLC-QTOF-MS/MS Conditions
2.2. Mapping Targets of Carnosol by Virtual Screening
2.2.1. Preparation of Crystallographic Structures of Human Proteins for Molecular Docking
2.2.2. Preparation of Carnosol Structures
2.2.3. Docking Calculations on Human Proteins
2.2.4. Molecular Docking Validation
2.3. Mapping Targets of Carnosol by Comparative Toxicogenomics Database (CTD)
Docking Calculations on CTD Targets
2.4. Docking Visualization
2.5. Network Pharmacology
2.5.1. Genetic Ontology and Functional Interaction Pathway Enrichment
2.5.2. Pharmacological Network Analysis
2.6. Molecular Dynamics Simulation
3. Results
3.1. Analysis of the Hydro-Alcoholic Extract of Rosemary by HPLC-QTOF-MS/MS
3.2. Mapping Targets for Carnosol by Molecular Docking
3.3. Mapping Targets of Carnosol by Comparative Toxicogenomics Database (CTD)
3.4. Molecular Docking Validation
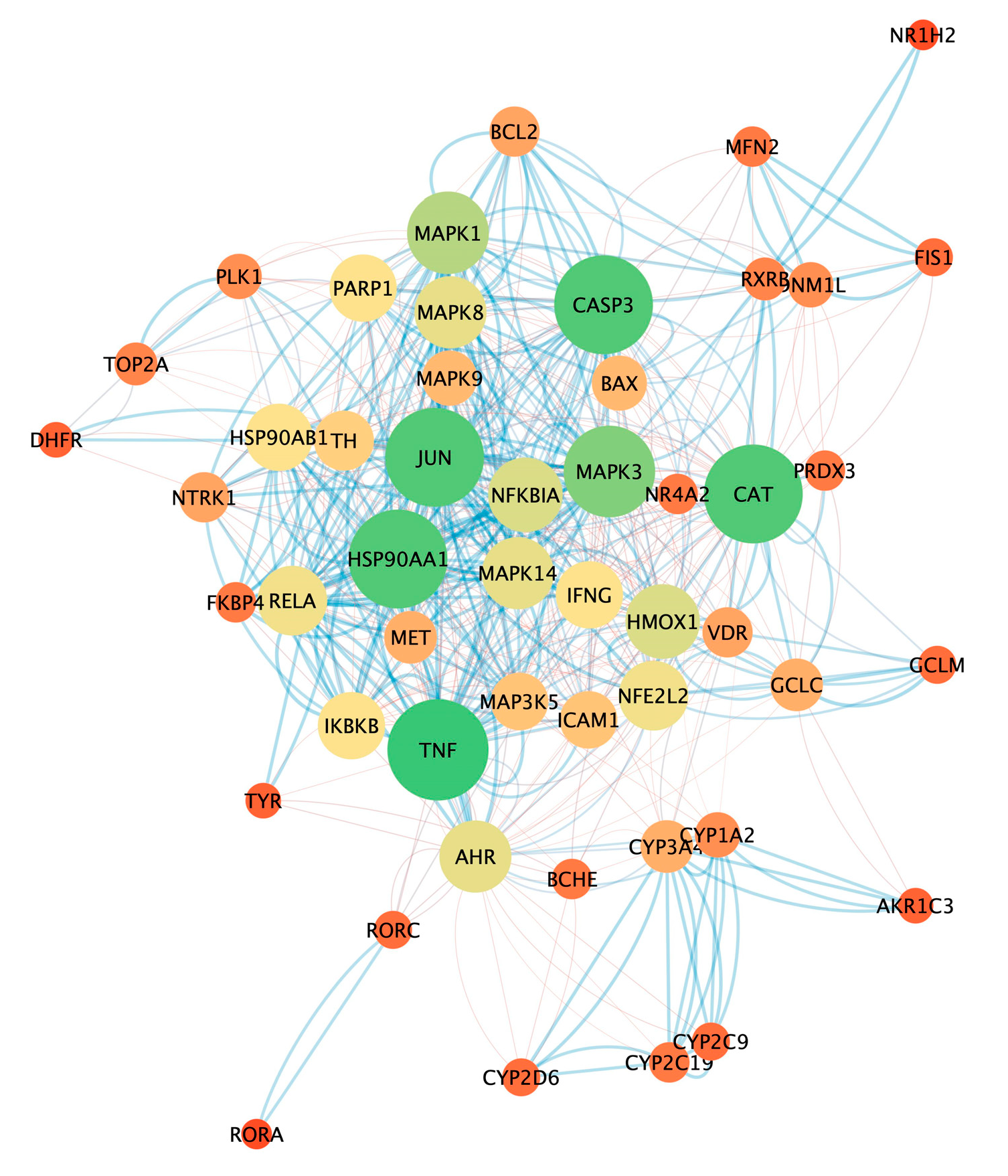
3.5. Analysis of Pharmacological Network
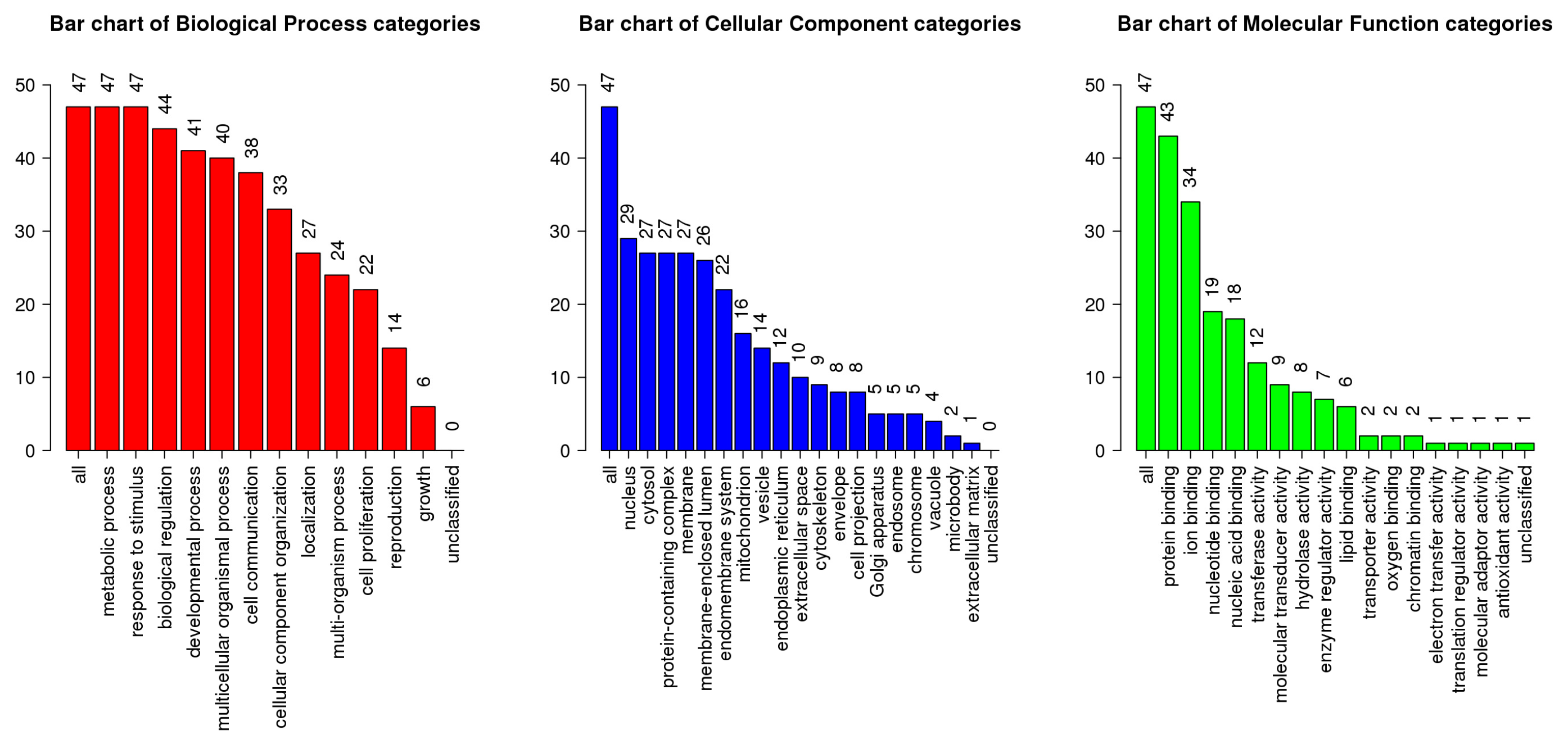
3.6. GO and KEGG Pathway Enrichment Analysis
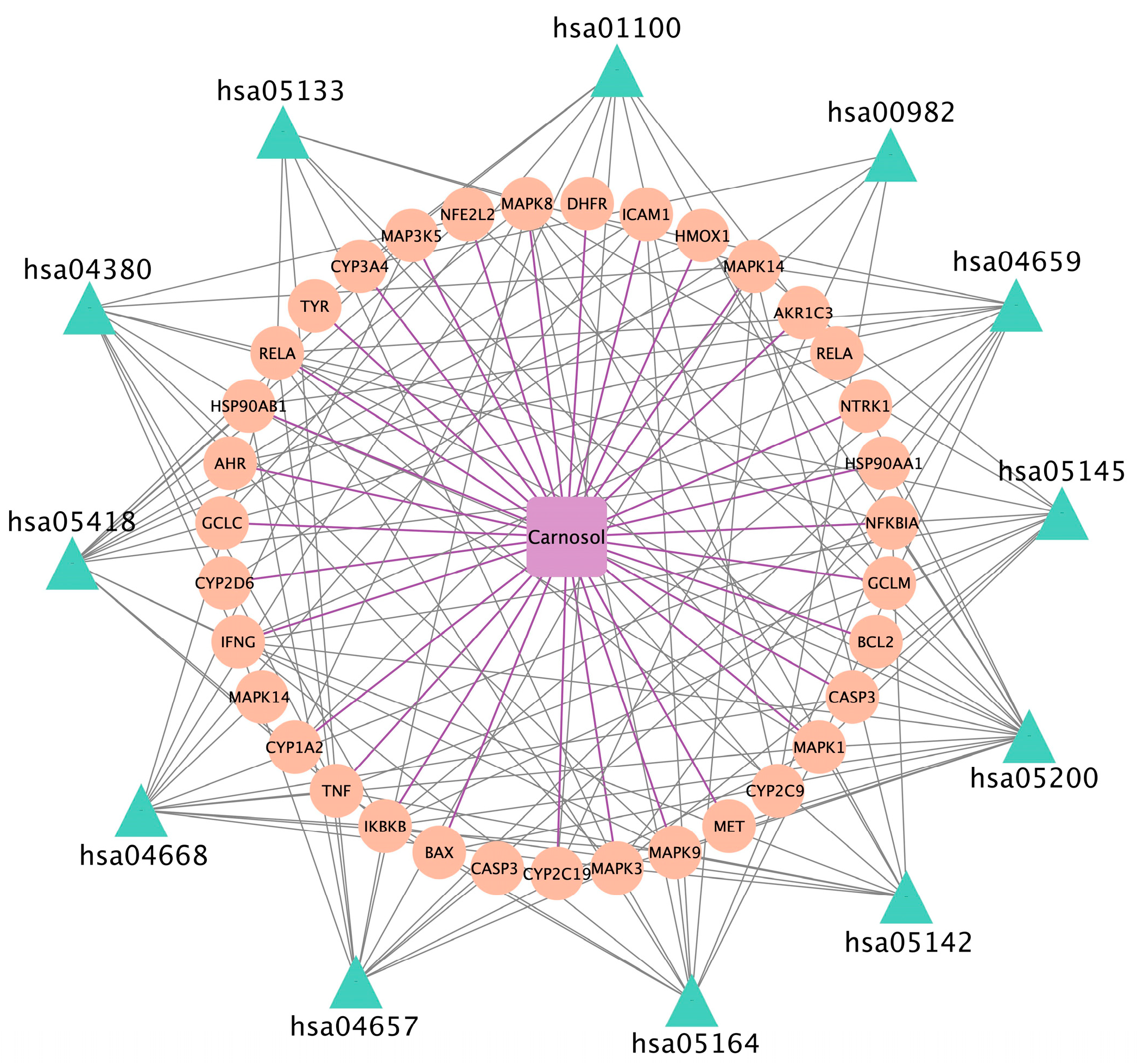
3.7. Chemical Compound–Target–Pathway
3.8. Molecular Docking Results
3.9. Molecular Dynamic (MD) Simulation
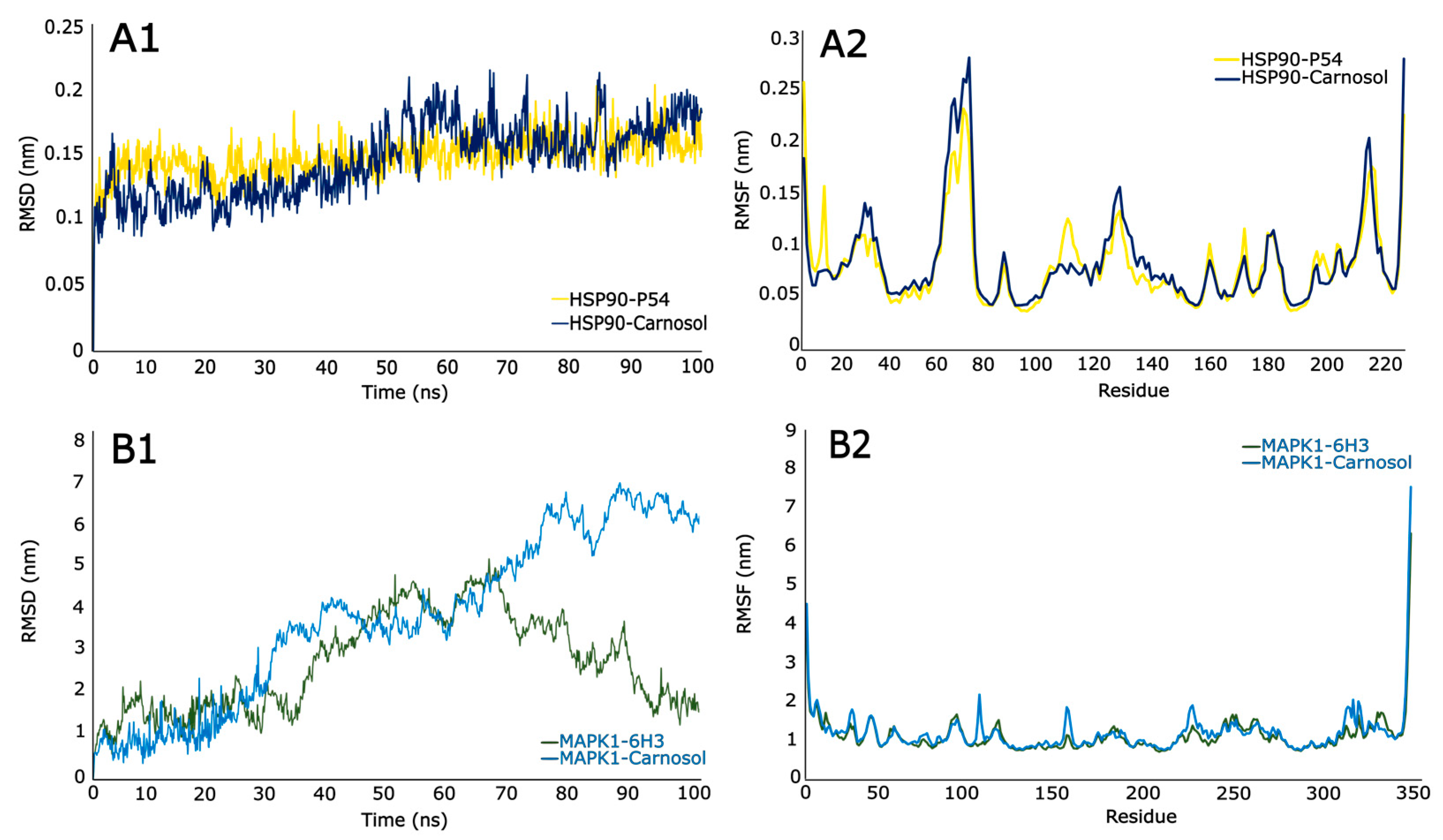
3.9.1. Root Mean Square Deviations (RSMD)
3.9.2. Root Mean Square Fluctuations (RSMF)
3.9.3. Molecular Mechanics Energies Combined with Surface Area Continuum Solvation (MMGBSA)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharifi-Rad, J.; Ezzat, S.M.; El Bishbishy, M.H.; Mnayer, D.; Sharopov, F.; Kılıç, C.S.; Neagu, M.; Constantin, C.; Sharifi-Rad, M.; Atanassova, M.; et al. Rosmarinus plants: Key farm concepts towards food applications. Phytother. Res. 2020, 34, 1474–1518. [Google Scholar] [CrossRef]
- Napoli, E.; Siracusa, L.; Ruberto, G. New Tricks for Old Guys: Recent Developments in the Chemistry, Biochemistry, Applications and Exploitation of Selected Species from the Lamiaceae Family. Chem. Biodivers. 2020, 17, e1900677. [Google Scholar] [CrossRef]
- Bower, A.; Marquez, S.; de Mejia, E.G. The health benefits of selected culinary herbs and spices found in the traditional Mediterranean diet. Crit. Rev. Food Sci. Nutr. 2016, 56, 2728–2746. [Google Scholar] [CrossRef]
- Moore, J.; Yousef, M.; Tsiani, E. Anticancer Effects of Rosemary (Rosmarinus officinalis L.) Extract and Rosemary Extract Polyphenols. Nutrients 2016, 8, 731. [Google Scholar] [PubMed]
- Lu, S.; Li, Y.; Shen, Q.; Zhang, W.; Gu, X.; Ma, M.; Li, Y.; Zhang, L.; Liu, X.; Zhang, X. Carnosol and its analogues attenuate muscle atrophy and fat lipolysis induced by cancer cachexia. J. Cachexia Sarcopenia Muscle 2021, 12, 779–795. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, T.; Li, K.; Xu, H.; Liang, R.; Wang, W.; Li, H.; Shao, A.; Yang, B. Screening active ingredients of rosemary based on spectrum-effect relationships between UPLC fingerprint and vasorelaxant activity using three chemometrics. J. Chromatogr. B 2019, 1134, 121854. [Google Scholar] [CrossRef] [PubMed]
- Nieto, G.; Ros, G.; Castillo, J. Antioxidant and antimicrobial properties of rosemary (Rosmarinus officinalis L.): A review. Medicines 2018, 5, 98. [Google Scholar] [CrossRef]
- Colica, C.; Di Renzo, L.; Aiello, V.; De Lorenzo, A.; Abenavoli, L. Rosmarinic Acid as Potential Anti-Inflammatory Agent. Rev. Recent Clin. Trials 2018, 13, 240–242. [Google Scholar] [CrossRef]
- Naimi, M.; Vlavcheski, F.; Shamshoum, H.; Tsiani, E. Rosemary Extract as a Potential Anti-Hyperglycemic Agent: Current Evidence and Future Perspectives. Nutrients 2017, 9, 968. [Google Scholar] [CrossRef]
- Zhao, Y.; Sedighi, R.; Wang, P.; Chen, H.; Zhu, Y.; Sang, S. Carnosic acid as a major bioactive component in rosemary extract ameliorates high-fat-diet-induced obesity and metabolic syndrome in mice. J. Agric. Food Chem. 2015, 63, 4843–4852. [Google Scholar] [CrossRef]
- Pérez-Sánchez, A.; Barrajón-Catalán, E.; Ruiz-Torres, V.; Agulló-Chazarra, L.; Herranz-López, M.; Valdés, A.; Cifuentes, A.; Micol, V. Rosemary (Rosmarinus officinalis) extract causes ROS-induced necrotic cell death and inhibits tumor growth in vivo. Sci. Rep. 2019, 9, 808. [Google Scholar] [CrossRef] [PubMed]
- Erkan, N.; Ayranci, G.; Ayranci, E. Antioxidant activities of rosemary (Rosmarinus officinalis L.) extract, blackseed (Nigella sativa L.) essential oil, carnosic acid, rosmarinic acid and sesamol. Food Chem. 2008, 110, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.J. Carnosol: A promising anti-cancer and anti-inflammatory agent. Cancer Lett. 2011, 305, 1–7. [Google Scholar] [CrossRef]
- Park, S.Y.; Song, H.; Sung, M.K.; Kang, Y.H.; Lee, K.W.; Park, J.H.Y. Carnosic acid inhibits the epithelial-mesenchymal transition in B16F10 melanoma cells: A possible mechanism for the inhibition of cell migration. Int. J. Mol. Sci. 2014, 15, 12698–12713. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Dai, F.; Li, Y.; Jia, L.; Luan, Y.; Zhao, Y. The research on the mechanism of Tsoong inhibiting for colon cancer. Saudi J. Biol. Sci. 2019, 26, 605–613. [Google Scholar] [CrossRef]
- Jakovljević, M.; Jokić, S.; Molnar, M.; Jašić, M.; Babić, J.; Jukić, H.; Banjari, I. Bioactive Profile of Various Salvia officinalis L. Preparations. Plants 2019, 8, 55. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Liang, X.; Yang, L.; Su, M.; Lai, K.P. Network Pharmacology and bioinformatics analyses identify intersection genes of niacin and COVID-19 as potential therapeutic targets. Brief. Bioinform. 2020, 22, 1279–1290. [Google Scholar] [CrossRef]
- Gogoi, B.; Gogoi, D.; Silla, Y.; Kakoti, B.B.; Bhau, B.S. Network pharmacology-based virtual screening of natural products from Clerodendrum species for identification of novel anti-cancer therapeutics. Mol. Biosyst. 2017, 13, 406–416. [Google Scholar] [CrossRef]
- Duran-Izquierdo, M.; Taboada-Alquerque, M.; Sierra-Marquez, L.; Alvarez-Ortega, N.; Stashenko, E.; Olivero-Verbel, J. Hydroalcoholic extract of Haematoxylum brasiletto protects Caenorhabditis elegans from cadmium-induced toxicity. BMC Complement. Med. Ther. 2022, 22, 184. [Google Scholar] [CrossRef]
- Cabarcas-Montalvo, M.; Maldonado-Rojas, W.; Montes-Grajales, D.; Bertel-Sevilla, A.; Wagner-Dobler, I.; Sztajer, H.; Reck, M.; Flechas-Alarcon, M.; Ocazionez, R.; Olivero-Verbel, J. Discovery of antiviral molecules for dengue: In silico search and biolog-ical evaluation. Eur. J. Med. Chem. 2016, 110, 87–97. [Google Scholar] [CrossRef]
- Coronado-Posada, N.; Mercado-Camargo, J.; Olivero-Verbel, J. In Silico Analysis to Identify Molecular Targets for Chemicals of Concern: The Case Study of Flocoumafen, an Anticoagulant Pesticide. Environ. Toxicol. Chem. 2021, 40, 2032–2041. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 442009, Carnosol. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/442009#section=3D-Conformer (accessed on 9 February 2023).
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Fedorov, A.A.; Martí-Arbona, R.; Nemmara, V.V.; Hitchcock, D.; Fedorov, E.V.; Almo, S.C.; Raushel, F.M. Structure of N-formimino-L-glutamate iminohydrolase from Pseudomonas aeruginosa. Biochemistry 2015, 54, 890–897. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2. 0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Lohning, A.E.; Levonis, S.M.; Williams-Noonan, B.; Schweiker, S.S. A Practical Guide to Molecular Docking and Homology Modelling for Medicinal Chemists. Curr. Top. Med. Chem. 2017, 17, 2023–2040. [Google Scholar] [CrossRef]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; McMorran, R.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. The Comparative Toxicogenomics Database: Update 2019. Nucleic Acids Res. 2019, 47, D948–D954. [Google Scholar] [CrossRef]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the scope of the protein-ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System; Delano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Tavirani, M.R.; Mansouri, V.; Tavirani, S.R.; Tackallou, S.H.; Rostami-Nejad, M. Gliosarcoma protein-protein interaction network analysis and gene ontology. Int. J. Cancer Manag. 2018, 11, e65701. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- MacKerell, A.D., Jr.; Bashford, D.; Bellott, M.; Dunbrack Jr, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef] [PubMed]
- Vanommeslaeghe, K.; Hatcher, E.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; Mackerell, A.D., Jr. CHARMM general force field: A force field for druglike molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef] [PubMed]
- Mark, P.; Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Miller III, B.R.; McGee Jr, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA. py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Zou, D.; Wang, J.; Zhang, B.; Xie, S.; Wang, Q.; Xu, K.; Lin, R. Analysis of chemical constituents in Wuzi-Yanzong-Wan by UPLC-ESI-LTQ-orbitrap-MS. Molecules 2015, 20, 21373–21404. [Google Scholar] [CrossRef]
- Mena, P.; Cirlini, M.; Tassotti, M.; Herrlinger, K.A.; Dall’Asta, C.; Del Rio, D. Phytochemical profiling of flavonoids, phenolic acids, terpenoids, and volatile fraction of a rosemary (Rosmarinus officinalis L.) extract. Molecules 2016, 21, 1576. [Google Scholar] [CrossRef]
- Wang, L.; Gan, C.; Wang, Z.; Liu, L.; Gao, M.; Li, Q.; Yang, C. Determination and pharmacokinetic study of three diterpenes in rat plasma by UHPLC-ESI-MS/MS after oral administration of Rosmarinus officinalis L. extract. Molecules 2017, 22, 934. [Google Scholar] [CrossRef]
- Zhang, Y.; Smuts, J.P.; Dodbiba, E.; Rangarajan, R.; Lang, J.C.; Armstrong, D.W. Degradation study of carnosic acid, carnosol, rosmarinic acid, and rosemary extract (Rosmarinus officinalis L.) assessed using HPLC. J. Agric. Food Chem. 2012, 60, 9305–9314. [Google Scholar] [CrossRef] [PubMed]
- Birtić, S.; Dussort, P.; Pierre, F.-X.; Bily, A.C.; Roller, M. Carnosic acid. Phytochemistry 2015, 115, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Loussouarn, M.; Krieger-Liszkay, A.; Svilar, L.; Bily, A.; Birtić, S.; Havaux, M. Carnosic acid and carnosol, two major antioxidants of rosemary, act through different mechanisms. Plant Physiol. 2017, 175, 1381–1394. [Google Scholar] [CrossRef] [PubMed]
- Labbé, C.; Faini, F.; Calderón, D.; Molina, J.; Arredondo, S. Variations of Carnosic Acid and Carnosol Concentrations in Ethanol Extracts of Wild Lepechinia salviae in Spring (2008–2011). Nat. Prod. Commun. 2014, 9, 1413–1416. [Google Scholar] [CrossRef]
- Luis, J.C.; Johnson, C.B. Seasonal variations of rosmarinic and carnosic acids in rosemary extracts. Analysis of their in vitro antiradical activity. Span. J. Agric. Res. 2005, 3, 106–112. [Google Scholar] [CrossRef]
- Macías Alonso, M.; Sancén, S.; Gonzalez-Marrero, J. Carnosic Acid and its Derivatives: Diterpenes of Biological Interest. Biomed. J. Sci. Tech. Res. 2019, 16, 12172–12174. [Google Scholar] [CrossRef]
- Patel, P.D.; Yan, P.; Seidler, P.M.; Patel, H.J.; Sun, W.; Yang, C.; Que, N.; Taldone, T.; Finotti, P.; Stephani, R.A.; et al. Paralog-selective Hsp90 inhibitors define tumor-specific regulation of HER2. Nat. Chem. Biol. 2013, 9, 677–684. [Google Scholar] [CrossRef]
- Kim, C.Y.; Chang, J.S.; Doyon, J.B.; Baird, T.T.; Fierke, C.A.; Jain, A.; Christianson, D.W. Contribution of fluorine to protein− ligand affinity in the binding of fluoroaromatic inhibitors to carbonic anhydrase II. J. Am. Chem. Soc. 2000, 122, 12125–12134. [Google Scholar] [CrossRef]
- Mittl, P.R.E.; Berry, A.; Scrutton, N.S.; Perham, R.N.; Schulz, G.E. Anatomy of an engineered NAD-binding site. Protein Sci. 1994, 3, 1504–1514. [Google Scholar] [CrossRef]
- Putnam, C.D.; Arvai, A.S.; Bourne, Y.; Tainer, J.A. Active and inhibited human catalase structures: Ligand and NADPH binding and catalytic mechanism. J. Mol. Biol. 2000, 296, 295–309. [Google Scholar] [CrossRef]
- Chen, L.; Glover, J.N.M.; Hogan, P.G.; Rao, A.; Harrison, S.C. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature 1998, 392, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Schulte, K.W.; Green, E.; Wilz, A.; Platten, M.; Daumke, O. Structural basis for aryl hydrocarbon receptor-mediated gene activation. Structure 2017, 25, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Bresinsky, M.; Strasser, J.M.; Vallaster, B.; Liu, P.; McCue, W.M.; Fuller, J.; Hubmann, A.; Singh, G.; Nelson, K.M.; Cuellar, M.E.; et al. Structure-based design and biological evaluation of novel caspase-2 inhibitors based on the peptide AcVDVAD-CHO and the caspase-2-mediated tau cleavage sequence YKPVD314. ACS Pharmacol. Transl. Sci. 2022, 5, 20–40. [Google Scholar] [CrossRef] [PubMed]
- Banner, D.W.; D’Arcy, A.; Janes, W.; Gentz, R.; Schoenfeld, H.J.; Broger, C.; Loetscher, H.; Lesslauer, W. Crystal structure of the soluble human 55 kd TNF receptor-human TNFβ complex: Implications for TNF receptor activation. Cell 1993, 73, 431–445. [Google Scholar] [CrossRef]
- Lin, W.W.; Karin, M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J. Clin. Investig. 2007, 117, 1175–1183. [Google Scholar] [CrossRef]
- Pacheco de Souza, A.; Costa, V.L.; da Costa Silva, M.; de Oliveira Araújo, I.B.; Castro Trindade, S.; Moura-Costa, L.F.; Costa Rodrigues, G.; Santana Sales, T.; Alves do Santos, H.; de Carvalho-Filho, P.C.; et al. MAPK involvement in cytokine production in response to Corynebacterium pseudotuberculosis infection. BMC Microbiol. 2014, 14, 230. [Google Scholar]
- Shi, J.; Zhao, Y.; Wang, Y.; Gao, W.; Ding, J.; Li, P.; Hu, L.; Shao, F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 2014, 514, 187–192. [Google Scholar] [CrossRef]
- Neavin, D.R.; Liu, D.; Ray, B.; Weinshilboum, R.M. The role of the aryl hydrocarbon receptor (AHR) in immune and inflammatory diseases. Int. J. Mol. Sci. 2018, 19, 3851. [Google Scholar] [CrossRef]
- Shi, W.; Xu, G.; Zhan, X.; Gao, Y.; Wang, Z.; Fu, S.; Qin, N.; Hou, X.; Ai, Y.; Wang, C.; et al. Carnosol inhibits inflammasome activation by directly targeting HSP90 to treat inflammasome-mediated diseases. Cell Death Dis. 2020, 11, 252. [Google Scholar] [CrossRef]
- Mohebati, A.; Guttenplan, J.B.; Kochhar, A.; Zhao, Z.L.; Kosinska, W.; Subbaramaiah, K.; Dannenberg, A.J. Carnosol, a Constituent of Zyflamend, Inhibits Aryl Hydrocarbon Receptor–Mediated Activation of CYP1A1 and CYP1B1 Transcription and MutagenesisBMI and Invasive Breast Cancer Risk in NSABP P-1 and STAR Trials. Cancer Prev. Res. 2012, 5, 593–602. [Google Scholar] [CrossRef]
- Cecchi, F.; Rabe, D.C.; Bottaro, D.P. The hepatocyte growth factor receptor: Structure, function, and pharmacological targeting in cancer. Curr. Signal Transduct. Ther. 2011, 6, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Baldanzi, G.; Graziani, A. Physiological signaling and structure of the HGF receptor MET. Biomedicines 2014, 3, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Altomare, D.A.; Testa, J.R. Perturbations of the AKT signaling pathway in human cancer. Oncogene 2005, 24, 7455–7464. [Google Scholar] [CrossRef] [PubMed]
- García-Gómez, R.; Bustelo, X.R.; Crespo, P. Protein–protein interactions: Emerging oncotargets in the RAS-ERK pathway. Trends Cancer 2018, 4, 616–633. [Google Scholar] [CrossRef]
- Aliebrahimi, S.; Kouhsari, S.M.; Arab, S.S.; Shadboorestan, A.; Ostad, S.N. Phytochemicals, withaferin A and carnosol, overcome pancreatic cancer stem cells as c-Met inhibitors. Biomed. Pharmacother. 2018, 106, 1527–1536. [Google Scholar] [CrossRef] [PubMed]
- Valdés, A.; García-Cañas, V.; Artemenko, K.A.; Simó, C.; Bergquist, J.; Cifuentes, A. Nano-liquid chromatography-orbitrap MS-based quantitative proteomics reveals differences between the mechanisms of action of carnosic acid and carnosol in colon cancer cells. Mol. Cell Proteom. 2017, 16, 8–22. [Google Scholar] [CrossRef]
- Penning, T.M. Aldo-Keto Reductase (AKR) 1C3 inhibitors: A patent review. Expert Opin. Ther. Pat. 2017, 27, 1329–1340. [Google Scholar] [CrossRef]
- Adeniji, A.O.; Chen, M.; Penning, T.M. AKR1C3 as a target in castrate resistant prostate cancer. J. Steroid Biochem. Mol. Biol. 2013, 137, 136–149. [Google Scholar] [CrossRef]
- Jaglanian, A.; Termini, D.; Tsiani, E. Rosemary (Rosmarinus officinalis L.) extract inhibits prostate cancer cell proliferation and survival by targeting Akt and mTOR. Biomed. Pharmacother. 2020, 131, 110717. [Google Scholar] [CrossRef]
- Dickmann, L.J.; VandenBrink, B.M.; Lin, Y.S. In vitro hepatotoxicity and cytochrome P450 induction and inhibition characteristics of carnosic acid, a dietary supplement with antiadipogenic properties. Drug Metab. Dispos. 2012, 40, 1263–1267. [Google Scholar] [CrossRef]
- Li, X.; Zhao, L.; Han, J.; Zhang, F.; Liu, S.; Zhu, L.; Wang, Z.; Zhang, G.; Zhang, Y. Carnosol Modulates Th17 Cell Differentiation and Microglial Switch in Experimental Autoimmune Encephalomyelitis. Front. Immunol. 2018, 9, 1807. [Google Scholar] [CrossRef]
- De Albuquerque, U.P.; de Medeiros, P.M.; de Almeida, A.L.S.; Monteiro, J.M.; Neto, E.M.F.L.; de Melo, J.G.; dos Santos, J.P. Medicinal plants of the caatinga (semi-arid) vegetation of NE Brazil: A quantitative approach. J. Ethnopharmacol. 2007, 114, 325–354. [Google Scholar] [CrossRef]
- Shiravi, A.; Akbari, A.; Mohammadi, Z.; Khalilian, M.S.; Zeinalian, A.; Zeinalian, M. Rosemary and its protective potencies against COVID-19 and other cytokine storm associated infections: A molecular review. Mediterr. J. Nutr. Metab. 2021, 14, 401–416. [Google Scholar] [CrossRef]
- Aslani, A.; Zolfaghari, B.; Fereidani, Y. Design, formulation, and evaluation of a herbal gel contains melissa, sumac, licorice, rosemary, and geranium for treatment of recurrent labial herpes infections. Dent. Res. J. 2018, 15, 191. [Google Scholar] [CrossRef] [PubMed]
- Nejati, H.; Farahpour, M.R.; Nagadehi, M.N. Topical Rosemary officinalis essential oil improves wound healing against disseminated Candida albicans infection in rat model. Comp. Clin. Pathol. 2015, 24, 1377–1383. [Google Scholar] [CrossRef]
- Kalantar, H.; Sadeghi, E.; Abolnezhadian, F.; Goudarzi, M.; Hemmati, A.A.; Basir, Z.; Kalantar, M. Carnosol attenuates bleomycin-induced lung damage via suppressing fibrosis, oxidative stress and inflammation in rats. Life Sci. 2021, 287, 120059. [Google Scholar] [CrossRef] [PubMed]
- Weckesser, S.; Engel, K.; Simon-Haarhaus, B.; Wittmer, A.; Pelz, K.; Schempp, C.Á. Screening of plant extracts for antimicrobial activity against bacteria and yeasts with dermatological relevance. Phytomedicine 2007, 14, 508–516. [Google Scholar] [CrossRef]

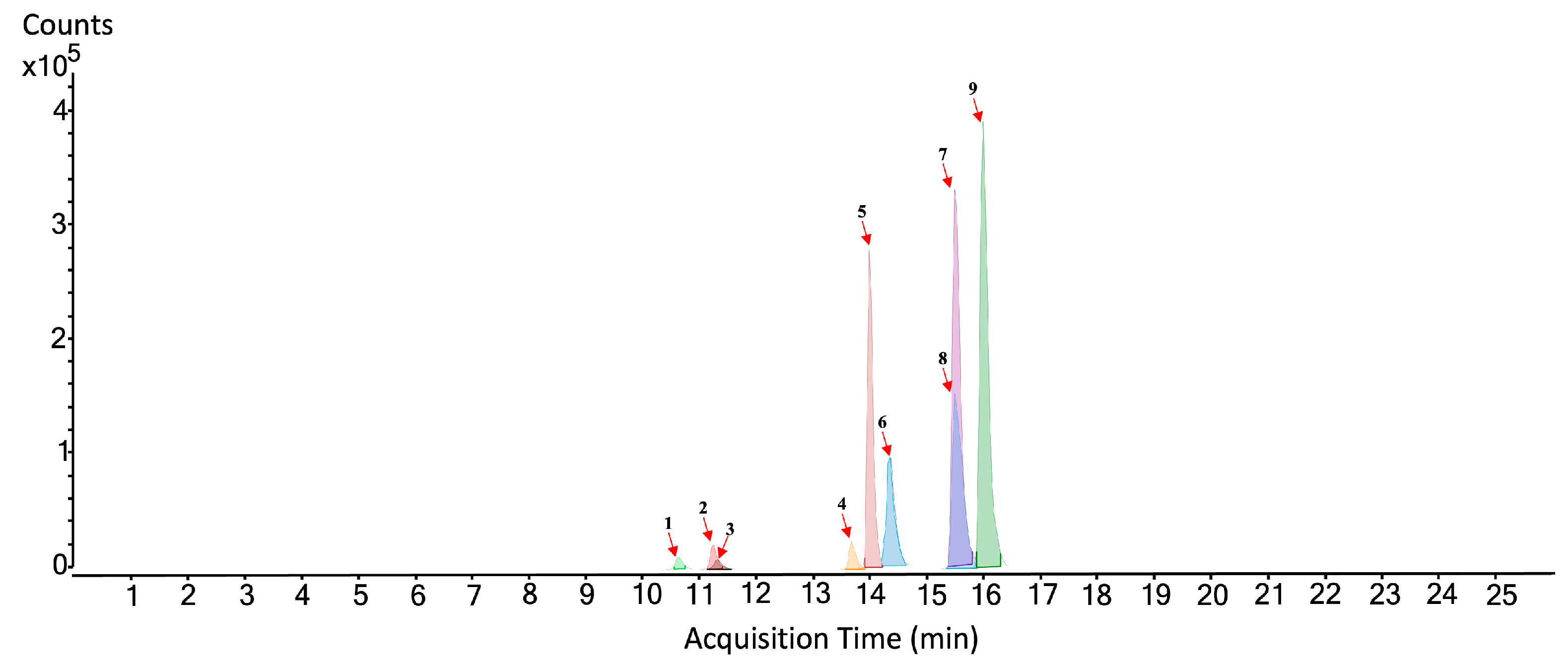

| No. Figure 2 | RT (min) | Tentative Annotation | Structure | Formula | Ion | Experimental Mass | Calculated Mass | Δ ppm |
|---|---|---|---|---|---|---|---|---|
| 1 | 10.639 | Flavonoid-glycosylated type | C22H22O12 | [M-H]− | 478.11044 | 478.111675 | −2.58 | |
| 2 | 11.256 | Rosmarinic acid |  | C18H16O8 | [M-H]− | 360.08439 | 360.085066 | −1.88 |
| 3 | 11.35 | Isorhamnetin-rutinoside | 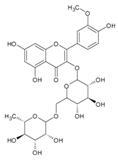 | C31H28O14 | [M-H]− | 624.14747 | 624.148454 | −1.58 |
| 4 | 13.726 | Dihydroxy-dimethoxyflavone | 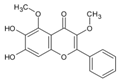 | C17H14O6 | [M-H]− | 314.07821 | 314.079587 | −4.38 |
| 5 | 14.012 | Diterpene type | C38H48O8 | [M+CH3COO]− | 632.33497 | 632.335467 | −0.78 | |
| 6 | 14.478 | Rosmanol | 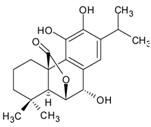 | C20H26O5 | [M-H]− | 346.17808 | 346.178573 | −1.42 |
| 7 | 15.558 | Rosmadial | 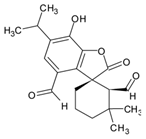 | C20H24O5 | [M-H]− | 344.16241 | 344.162922 | −1.49 |
| 8 | 15.597 | Diterpene type | C24H26O9 | [M-H]− | 458.15621 | 458.158231 | −4.41 | |
| 9 | 16.077 | Carnosol | 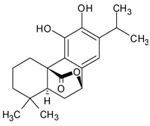 | C20H26O4 | [M-H]− | 330.18354 | 330.183658 | −0.36 |
| No. | PDB ID | Gene | Uniprot ID | Description | AV Binding Energy (kcal/mol) Exhaustiveness: 15 | AV Binding Energy (kcal/mol) Exhaustiveness: 32 |
|---|---|---|---|---|---|---|
| 1 | 3O0I | HSP90AA1 | P07900 | Heat shock protein HSP 90-α | −10.9 | −10.8 |
| 2 | 3NMQ | HSP90AB1 | P08238 | Heat shock protein HSP 90-beta | −10.4 | −10.4 |
| 3 | 1RY8 | AKR1C3 | P42330 | Aldo-keto reductase family 1 member C3 | −10.4 | −10.3 |
| 4 | 2WKM | MET | P08581 | Hepatocyte growth factor receptor | −10.3 | −10.2 |
| 5 | 1N83 | RORA | P35398 | Nuclear receptor ROR-alpha | −10.0 | −10.3 |
| 6 | 1HFQ | DHFR | P00374 | Dihydrofolate reductase (DHFR) | −10.0 | −10.0 |
| 7 | 3WHW | NUDT1 | P36639 | 7,8-dihydro-8-oxoguanine triphosphatase | −9.9 | −9.8 |
| 8 | 3L0L | RORC | P51449 | Nuclear receptor ROR-gamma | −9.9 | −9.9 |
| 9 | 2XVT | RAMP2 | O60895 | Receptor activity-modifying protein 2 | −9.9 | −9.9 |
| 10 | 1TQN | CYP3A4 | P08684 | Cytochrome P450 3A4 | −9.8 | −9.8 |
| 11 | 1DGB | CAT | P04040 | Catalase | −9.8 | −9.6 |
| 12 | 4XII | BCHE | P06276 | Cholinesterase | −9.8 | −9.5 |
| 13 | 1P8D | NR1H2 | P55055 | Oxysterols receptor LXR-beta | −9.7 | −10.3 |
| 14 | 4DRJ | FKBP4 | Q02790 | Peptidyl-prolyl cis-trans isomerase FKBP4 | −9.7 | −9.6 |
| 15 | 4AOJ | NTRK1 | P04629 | High-affinity nerve growth factor receptor | −9.6 | −9.5 |
| 16 | 1UHL | RXRB | P28702 | Retinoic acid receptor RXR-beta | −9.6 | −9.6 |
| 17 | 1ZXM | TOP2A | P11388 | DNA topoisomerase 2-alpha | −9.6 | −9.8 |
| 18 | 4FA2 | MAPK14 | Q16539 | Mitogen-activated protein kinase 14 | −9.5 | −9.3 |
| 19 | 4J52 | PLK1 | P53350 | Serine/threonine-protein kinase PLK1 | −9.5 | −9.5 |
| 20 | 3X36 | VDR | P11473 | Vitamin D3 receptor | −9.2 | −9.2 |
| No. | PDB ID | Gene | Uniprot ID | Description |
|---|---|---|---|---|
| 1 | 1TNR | TNF | P19438 | Tumor necrosis factor |
| 2 | 5UCX | PRDX3 | P30048 | Peroxiredoxin 3 |
| 3 | 2VGE | RELA | Q8WUF5 | RELA proto-oncogene, NF-kB subunit |
| 4 | 6EHA | HMOX1 | P09601 | Heme oxygenase 1 |
| 5 | 7O7B | NFE2L2 | Q16236 | NFE2-like bZIP transcription factor 2 |
| 6 | 3E2M | ICAM1 | P20701 | Intercellular adhesion molecule 1 |
| 7 | 4Q7H | IFNG | Interferon gamma | |
| 8 | 6ZZU | TH | P07101 | Tyrosine hydroxylase |
| 9 | 4S0O | BAX | Q07812 | BCL2-associated X, apoptosis regulator |
| 10 | 5UUK | BCL2 | Q16548 | BCL2 apoptosis regulator |
| 11 | 4H1V | DNM1L | O00429 | Dynamin 1-like |
| 12 | 1NZN | FIS1 | Q9Y3D6 | Fission, mitochondrial 1 |
| 13 | 6OYW | MAP3K5 | Q99683 | Mitogen-activated protein kinase kinase kinase 5 |
| 14 | 4G1W | MAPK8 | P45983 | Mitogen-activated protein kinase 8 |
| 15 | 7CML | MAPK9 | P45984 | Mitogen-activated protein kinase 9 |
| 16 | 6JFL | MFN2 | O95140 | Mitofusin 2 |
| 17 | 7KKM | PARP1 | O95271 | Poly(ADP-ribose) polymerase 1 |
| 18 | 7RNF | CASP3 | P42574 | Caspase 3 |
| 19 | 5M8R | TYR | P17643 | Tyrosinase |
| 20 | 5NJ8 | AHR | P35869 | Aryl hydrocarbon receptor |
| 21 | 2HI4 | CYP1A2 | P05177 | Cytochrome P450 family 1 subfamily A member 2 |
| 22 | 4GQS | CYP2C19 | P33261 | Cytochrome P450 family 2 subfamily C member 19 |
| 23 | 6VLT | CYP2C9 | P11712 | Cytochrome P450 family 2 subfamily C member 9 |
| 24 | 4XRY | CYP2D6 | P10635 | Cytochrome P450 family 2 subfamily D member 6 |
| 25 | 4D7D | CYP3A4 | P08684 | Cytochrome P450 family 3 subfamily A member 4 |
| 26 | - | GCLC | Glutamate–cysteine ligase catalytic subunit | |
| 27 | - | GCLM | Glutamate–cysteine ligase modifier subunit | |
| 28 | 4KIK | IKBKB | O14920 | Inhibitor of nuclear factor kappa B kinase subunit beta |
| 29 | 1A02 | JUN | P05412 | Jun proto-oncogene, AP-1 transcription factor subunit |
| 30 | 6G54 | MAPK1 | P28482 | Mitogen-activated protein kinase 1 |
| 31 | 6GES | MAPK3 | P27361 | Mitogen-activated protein kinase 3 |
| 32 | 6Y1J | NFKBIA | P25963 | NFKB inhibitor alpha |
| 33 | 5YD6 | NR4A2 | P43354 | Nuclear receptor subfamily 4 group A member 2 |
| Name | Degree | Betweenness Centrality | Closeness Centrality |
|---|---|---|---|
| TNF | 30 | 3.4008 | 0.7101 |
| CAT | 29 | 3.0840 | 0.6901 |
| JUN | 29 | 1.4134 | 0.6901 |
| HSP90AA1 | 29 | 2.2865 | 0.6901 |
| CASP3 | 29 | 1.5676 | 0.6622 |
| MAPK3 | 26 | 1.4197 | 0.6364 |
| MAPK1 | 22 | 1.0351 | 0.6050 |
| AHR | 18 | 3.2344 | 0.6050 |
| No. | PDB ID | Gene | Ref | Compound | AV Binding Energy (kcal/mol) | Interactions |
|---|---|---|---|---|---|---|
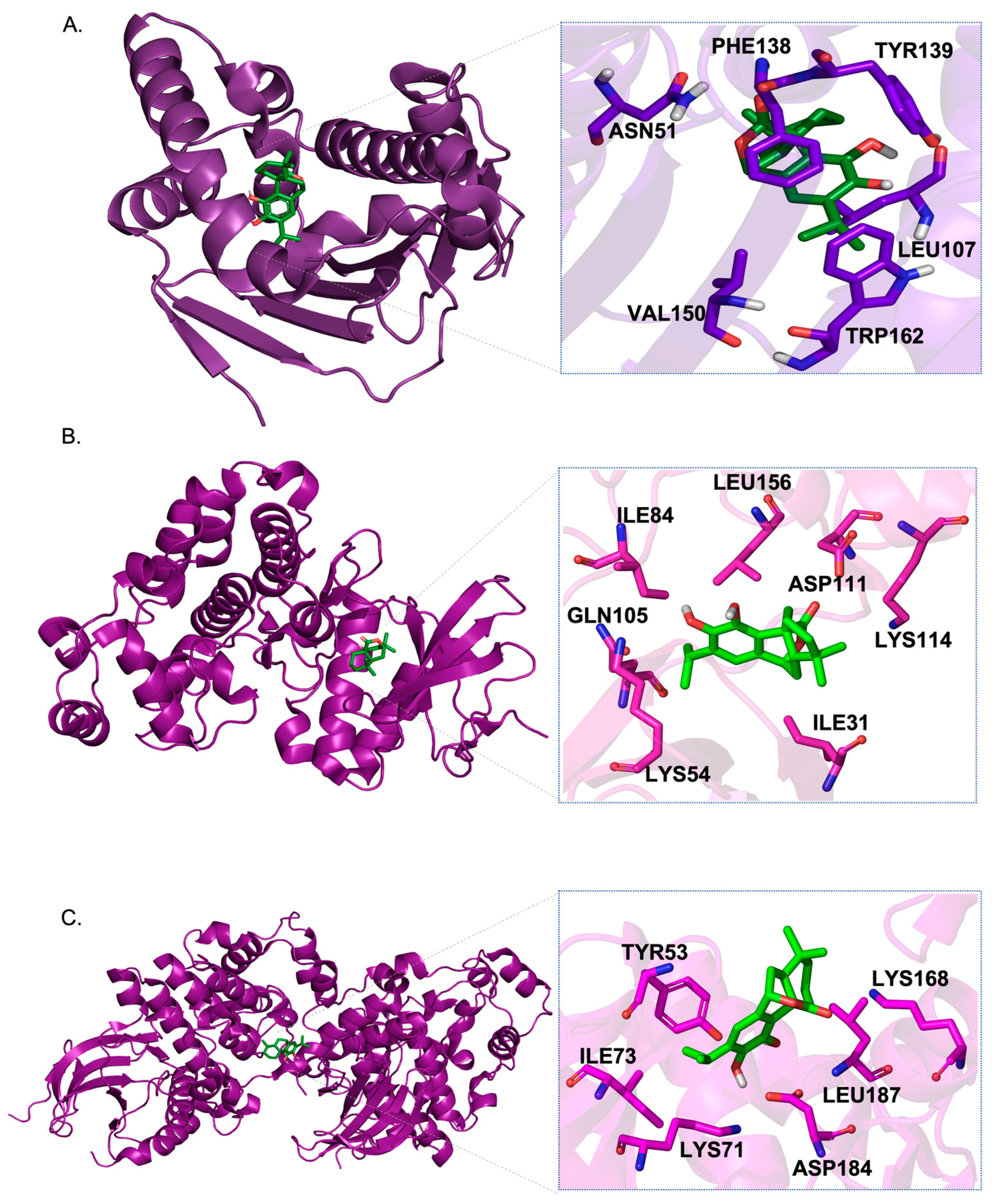
| 1 | 3O0I | HSP90AA1 | [49] | Carnosol | −10.8 | LEU107A, PHE138A, VAL150A, TRP162A, ASN51A, TYR139A |
| 2 | 1G54 | MAPK1 | [50] | −8.5 | ILE31A, LYS54A, ILE84A, GLN105A, LYS114A, LEU156A, ASP111A | |
| 3 | 6GES | MAPK3 | [51] | −8.3 | TYR53A, LYS71A, ILE73A, LEU187A, ASP184A, LYS168A | |
| 4 | 1DGB | CAT | [52] | −7.9 | PHE198A, VAL302A ALA445A, PHE446A, VAL450A, HIS305A | |
| 5 | 1A02 | JUN | [53] | −7.6 | ARG541N, PRO566N, GLN669N ASP464N | |
| 6 | 5NJ8 | AHR | [54] | −7.5 | TYR76A, TYR137A, LYS80A | |
| 7 | 7RNF | CASP3 | [55] | −7.3 | PHE247D, PHE250D, ASN208D, GLU246D, PHE247D | |
| 8 | 1TNR | TNF | [56] | −6.3 | ALA30A, PHE53A, PHE169A, LEU171A, ALA170A |
| Complex | Total Binding Free Energy (kcal/mol) | Standard Deviation |
|---|---|---|
| HSP90-P54 | −6.1978 | 2.8193 |
| HSP90-Carnosol | −28.4237 | 2.9445 |
| MAPK1-6H3 | −27.6204 | 2.7037 |
| MAPK1-Carnosol | −27.7457 | 3.7537 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taboada-Alquerque, M.; Pajaro-Valenzuela, D.; Caballero-Gallardo, K.; Cifuentes, A.; Ibáñez, E.; Ahumedo-Monterrosa, M.; Stashenko, E.E.; Olivero-Verbel, J. Mapping Protein Targets of Carnosol, a Molecule Identified in Rosmarinus officinalis: In Silico Docking Studies and Network Pharmacology. Sci. Pharm. 2023, 91, 19. https://doi.org/10.3390/scipharm91020019
Taboada-Alquerque M, Pajaro-Valenzuela D, Caballero-Gallardo K, Cifuentes A, Ibáñez E, Ahumedo-Monterrosa M, Stashenko EE, Olivero-Verbel J. Mapping Protein Targets of Carnosol, a Molecule Identified in Rosmarinus officinalis: In Silico Docking Studies and Network Pharmacology. Scientia Pharmaceutica. 2023; 91(2):19. https://doi.org/10.3390/scipharm91020019
Chicago/Turabian StyleTaboada-Alquerque, María, Danilo Pajaro-Valenzuela, Karina Caballero-Gallardo, Alejandro Cifuentes, Elena Ibáñez, Maicol Ahumedo-Monterrosa, Elena E. Stashenko, and Jesus Olivero-Verbel. 2023. "Mapping Protein Targets of Carnosol, a Molecule Identified in Rosmarinus officinalis: In Silico Docking Studies and Network Pharmacology" Scientia Pharmaceutica 91, no. 2: 19. https://doi.org/10.3390/scipharm91020019
APA StyleTaboada-Alquerque, M., Pajaro-Valenzuela, D., Caballero-Gallardo, K., Cifuentes, A., Ibáñez, E., Ahumedo-Monterrosa, M., Stashenko, E. E., & Olivero-Verbel, J. (2023). Mapping Protein Targets of Carnosol, a Molecule Identified in Rosmarinus officinalis: In Silico Docking Studies and Network Pharmacology. Scientia Pharmaceutica, 91(2), 19. https://doi.org/10.3390/scipharm91020019










