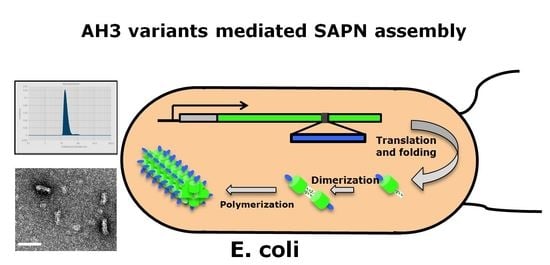
A Thermal-Stable Protein Nanoparticle That Stimulates Long Lasting Humoral Immune Response
Abstract
1. Introduction
2. Materials and Methods
2.1. Peptide Information, Expression and Purification of Recombinant Protein
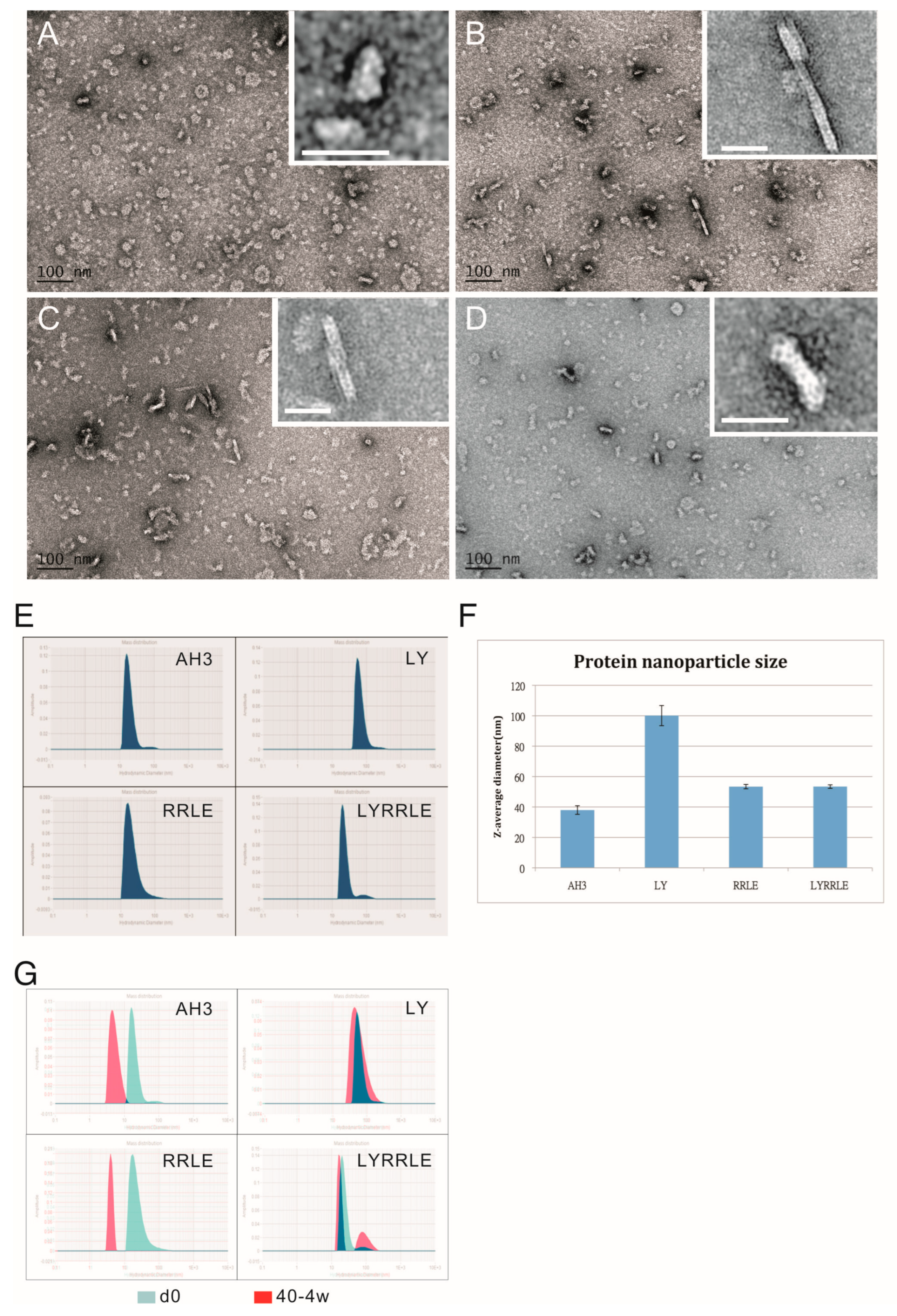
2.2. Biophysical Analysis of Protein Oligomerization and Hydrophobicity
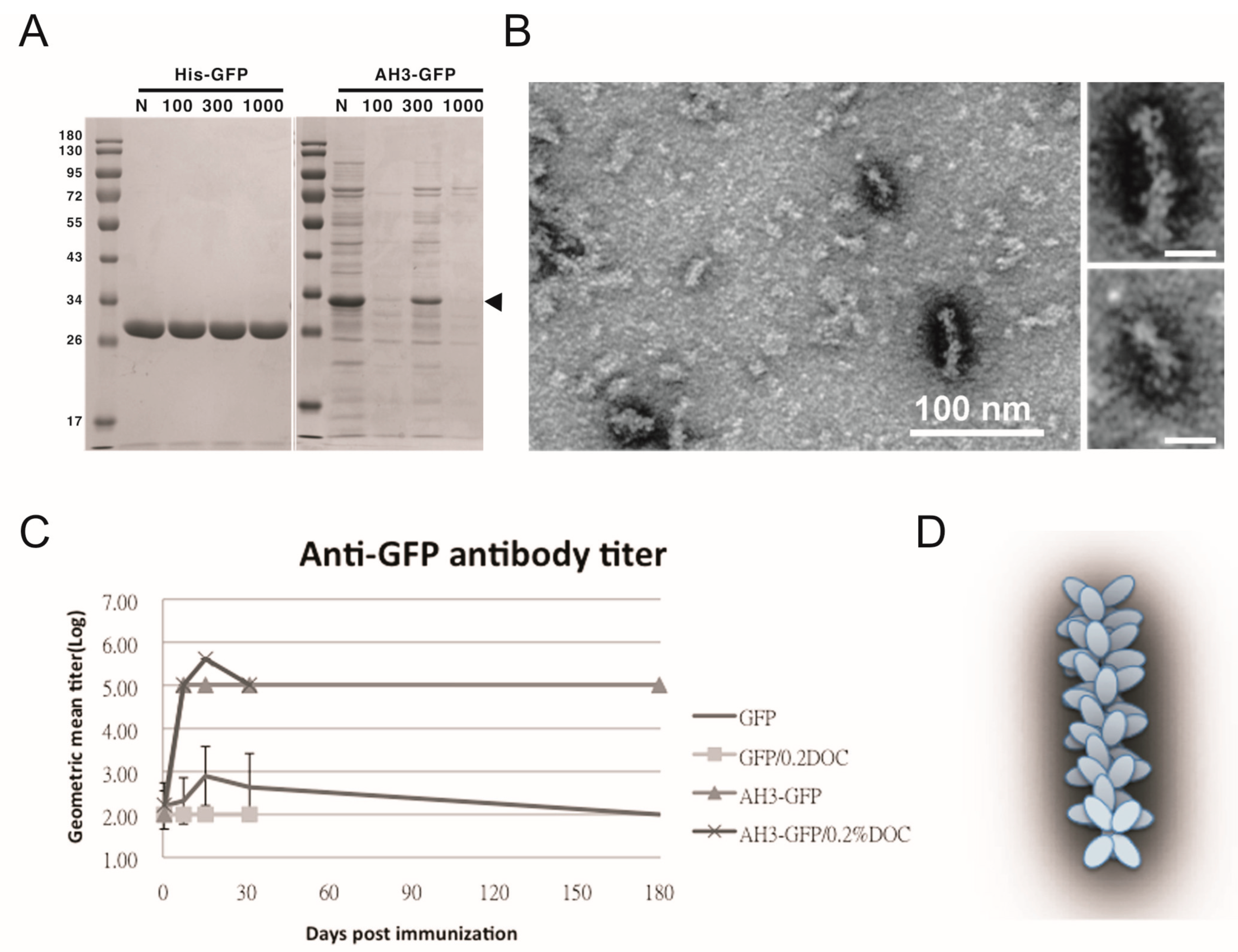
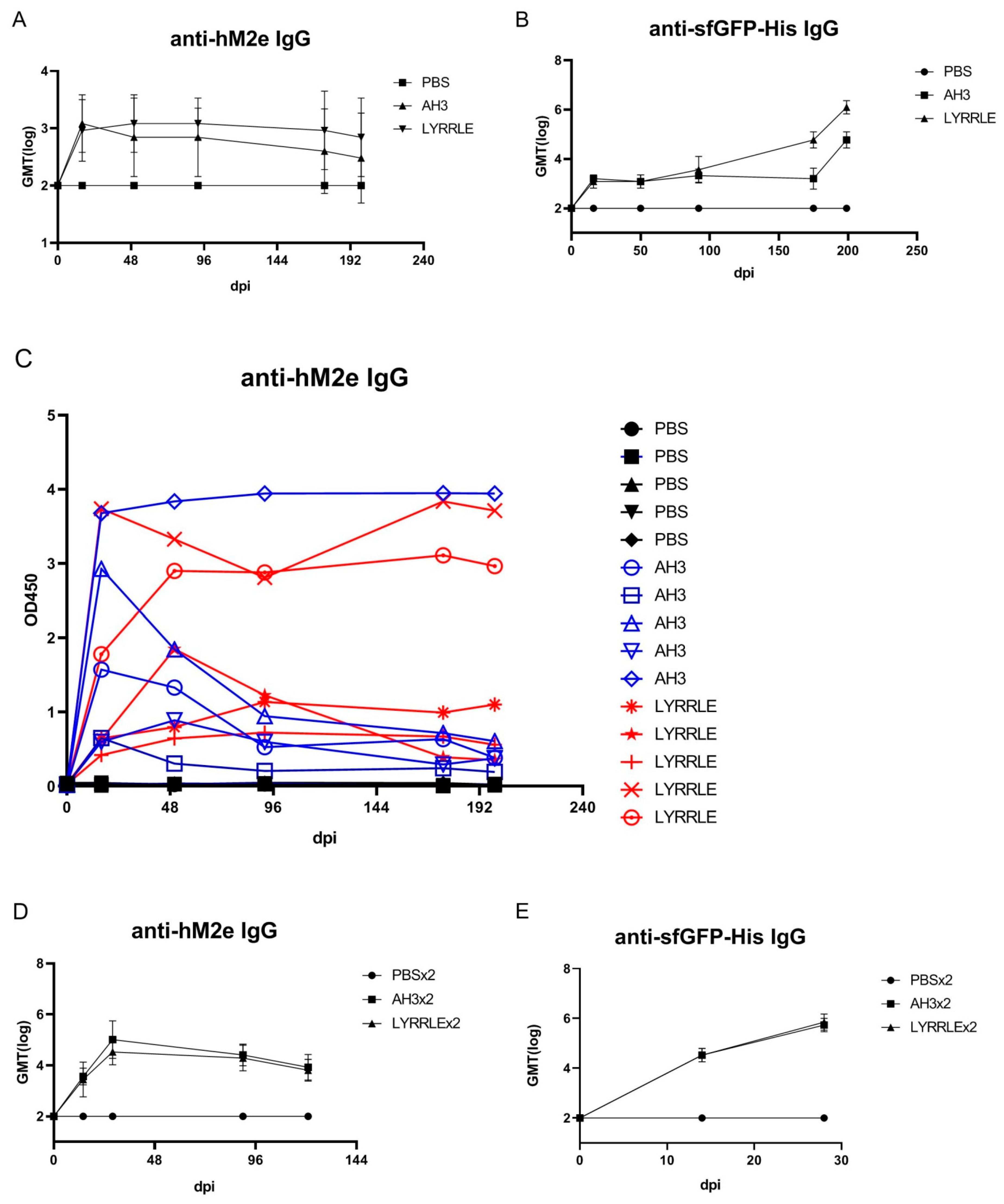
2.3. Animal Immunization and Antibody Titer Determination
2.4. Protein Structure Modeling and Intermolecular Force Calculation
2.5. Thermostability Determination
3. Results
3.1. Identification of a Protein Complex with High Antigenicity and Stability
3.2. Characterization of AH3–GFP Protein Complex
3.3. Modeling the AH3–GFP Protein Complex Structure
3.4. Designing a Vaccine Carrier That Enables Heterologous Antigen Insertion and High Stability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IEA. For the First Time in Decades, the Number of People without Access to Electricity is Set to Increase in 2022, IEA, Paris. Available online: https://www.iea.org/commentaries/for-the-first-time-in-decades-the-number-of-people-without-access-to-electricity-is-set-to-increase-in-2022 (accessed on 21 December 2022).
- Kumar, R.; Srivastava, V.; Baindara, P.; Ahmad, A. Thermostable vaccines: An innovative concept in vaccine development. Expert Rev. Vaccines 2022, 21, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Karp, C.L.; Lans, D.; Esparza, J.; Edson, E.B.; Owen, K.E.; Wilson, C.B.; Heaton, P.M.; Levine, O.S.; Rao, R. Evaluating the value proposition for improving vaccine thermostability to increase vaccine impact in low and middle-income countries. Vaccine 2015, 33, 3471–3479. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Sagaseta, J.; Malito, E.; Rappuoli, R.; Bottomley, M.J. Self-assembling protein nanoparticles in the design of vaccines. Comput. Struct. Biotechnol. J. 2016, 14, 58–68. [Google Scholar] [CrossRef]
- Ahsan, F.; Rivas, I.P.; Khan, M.A.; Torres Suárez, A.I. Targeting to macrophages: Role of physicochemical properties of particulate carriers-liposomes and microspheres-on the phagocytosis by macrophages. J. Control. Release 2002, 79, 29–40. [Google Scholar] [CrossRef]
- Bachmann, M.F.; Jennings, G.T. Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 2010, 10, 787–796. [Google Scholar] [CrossRef]
- Lu, Y.; Chan, W.; Ko, B.Y.; VanLang, C.C.; Swartz, J.R. Assessing sequence plasticity of a virus-like nanoparticle by evolution toward a versatile scaffold for vaccines and drug delivery. Proc. Natl. Acad. Sci. USA 2015, 112, 12360–12365. [Google Scholar] [CrossRef]
- Ravin, N.V.; Blokhina, E.A.; Kuprianov, V.V.; Stepanova, L.A.; Shaldjan, A.A.; Kovaleva, A.A.; Tsybalova, L.M.; Skryabin, K.G. Development of a candidate influenza vaccine based on virus-like particles displaying influenza M2e peptide into the immunodominant loop region of hepatitis B core antigen: Insertion of multiple copies of M2e increases immunogenicity and protective efficiency. Vaccine 2015, 33, 3392–3397. [Google Scholar] [PubMed]
- Manolova, V.; Flace, A.; Bauer, M.; Schwarz, K.; Saudan, P.; Bachmann, M.Ä. Nanoparticles target distinct dendritic cell populations according to their size. Eur. J. Immunol. 2008, 38, 1404–1413. [Google Scholar] [CrossRef]
- Gomes, A.C.; Roesti, E.S.; El-Turabi, A.; Bachmann, M.F. Type of RNA Packed in VLPs Impacts IgG Class Switching-Implications for an Influenza Vaccine Design. Vaccines 2019, 7, 47. [Google Scholar] [CrossRef]
- Johnston, S.C.; Ricks, K.M.; Lakhal-Naouar, I.; Jay, A.; Subra, C.; Raymond, J.L.; King, H.A.D.; Rossi, F.; Clements, T.L.; Fetterer, D.; et al. A SARS-CoV-2 Spike Ferritin Nanoparticle Vaccine Is Protective and Promotes a Strong Immunological Response in the Cynomolgus Macaque Coronavirus Disease 2019 (COVID-19) Model. Vaccines 2022, 10, 717. [Google Scholar] [CrossRef]
- Kim, S.A.; Kim, S.; Kim, G.B.; Goo, J.; Kim, N.; Lee, Y.; Nam, G.H.; Lim, S.; Kim, T.; Chang, K.H.; et al. A Multivalent Vaccine Based on Ferritin Nanocage Elicits Potent Protective Immune Responses against SARS-CoV-2 Mutations. Int. J. Mol. Sci. 2022, 23, 6123. [Google Scholar] [CrossRef] [PubMed]
- Bale, J.B.; Gonen, S.; Liu, Y.; Sheffler, W.; Ellis, D.; Thomas, C.; Cascio, D.; Yeates, T.O.; Gonen, T.; King, N.P.; et al. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science 2016, 353, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, G.L.; Lajoie, M.J.; Gustafson, H.H.; Sellers, D.L.; Nattermann, U.; Ellis, D.; Bale, J.B.; Ke, S.; Lenz, G.H.; Yehdego, A.; et al. Evolution of a designed protein assembly encapsulating its own RNA genome. Nature 2017, 552, 415–420. [Google Scholar] [CrossRef]
- Tsien, R.Y. THE GREEN FLUORESCENT PROTEIN. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef] [PubMed]
- Dove, S.G.; Hoegh-Guldberg, O.; Ranganathan, S. Major colour patterns of reef-building corals are due to a family of GFP-like proteins. Coral Reefs 2001, 19, 197–204. [Google Scholar] [CrossRef]
- Cody, C.W.; Prasher, D.C.; Westler, W.M.; Prendergast, F.G.; Ward, W.W. Chemical structure of the hexapeptide chromophore of the Aequorea green-fluorescent protein. Biochemistry 1993, 32, 1212–1218. [Google Scholar] [CrossRef]
- Close, D.W.; Paul, C.D.; Langan, P.S.; Wilce, M.C.; Traore, D.A.; Halfmann, R.; Rocha, R.C.; Waldo, G.S.; Payne, R.J.; Rucker, J.B.; et al. Thermal green protein, an extremely stable, nonaggregating fluorescent protein created by structure-guided surface engineering. Proteins 2014, 83, 1225–1237. [Google Scholar] [CrossRef] [PubMed]
- Kiss, C.; Temirov, J.; Chasteen, L.; Waldo, G.S.; Bradbury, A.R. Directed evolution of an extremely stable fluorescent protein. Protein Eng. Des. Sel. 2009, 22, 313–323. [Google Scholar] [CrossRef]
- Cormack, B.P.; Valdivia, R.H.; Falkow, S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 1996, 173, 33–38. [Google Scholar] [CrossRef]
- Crameri, A.; Whitehorn, E.A.; Tate, E.; Stemmer, W.P.C. Improved Green Fluorescent Protein by Molecular Evolution Using DNA Shuffling. Nat. Biotechnol. 1996, 14, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Pavoor, T.V.; Cho, Y.K.; Shusta, E.V. Development of GFP-based biosensors possessing the binding properties of antibodies. Proc. Natl. Acad. Sci. USA 2009, 106, 11895–11900. [Google Scholar] [CrossRef]
- Wang, R.; Xiang, S.; Zhang, Y.; Chen, Q.; Zhong, Y.; Wang, S. Development of a functional antibody by using a green fluorescent protein frame as the template. Appl. Environ. Microbiol. 2014, 80, 4126–4137. [Google Scholar] [CrossRef]
- Cabantous, S.p.; Terwilliger, T.C.; Waldo, G.S. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 2005, 23, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Romei, M.G.; Boxer, S.G. Split Green Fluorescent Proteins: Scope, Limitations, and Outlook. Annu. Rev. Biophys. 2019, 48, 19–44. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Weng, L.; Li, R.; Li, L.; Toyoda, T.; Zhong, J. The N-terminal helix alpha(0) of hepatitis C virus NS3 protein dictates the subcellular localization and stability of NS3/NS4A complex. Virology 2012, 422, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Horner, S.M.; Park, H.S.; Gale, M., Jr. Control of innate immune signaling and membrane targeting by the Hepatitis C virus NS3/4A protease are governed by the NS3 helix alpha0. J. Virol. 2012, 86, 3112–3120. [Google Scholar] [CrossRef]
- Tossi, A.; Sandri, L.; Giangaspero, A. Amphipathic, alpha-helical antimicrobial peptides. Pept. Sci. 2000, 55, 4–30. [Google Scholar] [CrossRef]
- Rossman, J.S.; Jing, X.; Leser, G.P.; Lamb, R.A. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 2010, 142, 902–913. [Google Scholar] [CrossRef]
- Roberts, K.L.; Leser, G.P.; Ma, C.; Lamb, R.A. The amphipathic helix of influenza A virus M2 protein is required for filamentous bud formation and scission of filamentous and spherical particles. J. Virol. 2013, 87, 9973–9982. [Google Scholar] [CrossRef]
- Kan, M.C. Novel Protein Structure Used for Efficient Antibody Production in Immunization. TWI683668B, 1 February 2018. [Google Scholar]
- Borisova, G.P.; Berzins, I.; Pushko, P.M.; Pumpen, P.; Gren, E.J.; Tsibinogin, V.V.; Loseva, V.; Ose, V.; Ulrich, R.; Siakkou, H.; et al. Recombinant core particles of hepatitis B virus exposing foreign antigenic determinants on their surface. FEBS Lett. 1989, 259, 121–124. [Google Scholar] [CrossRef]
- Kobayashi, T.; Morone, N.; Kashiyama, T.; Oyamada, H.; Kurebayashi, N.; Murayama, T. Engineering a novel multifunctional green fluorescent protein tag for a wide variety of protein research. PLoS ONE 2008, 3, e3822. [Google Scholar] [CrossRef] [PubMed]
- Pédelacq, J.-D.; Cabantous, S.; Tran, T.; Terwilliger, T.C.; Waldo, G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006, 24, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Jegerlehner, A.; Wiesel, M.; Dietmeier, K.; Zabel, F.; Gatto, D.; Saudan, P.; Bachmann, M.F. Carrier induced epitopic suppression of antibody responses induced by virus-like particles is a dynamic phenomenon caused by carrier-specific antibodies. Vaccine 2010, 28, 5503–5512. [Google Scholar] [CrossRef]
- Feldman, R.A.; Fuhr, R.; Smolenov, I.; Ribeiro, A.; Panther, L.; Watson, M.; Senn, J.J.; Smith, M.; Almarsson, R.; Pujar, H.S.; et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine 2019, 37, 3326–3334. [Google Scholar] [CrossRef] [PubMed]
- Li, J.X.; Hou, L.H.; Meng, F.Y.; Wu, S.P.; Hu, Y.M.; Liang, Q.; Chu, K.; Zhang, Z.; Xu, J.J.; Tang, R.; et al. Immunity duration of a recombinant adenovirus type-5 vector-based Ebola vaccine and a homologous prime-boost immunisation in healthy adults in China: Final report of a randomised, double-blind, placebo-controlled, phase 1 trial. Lancet Glob. Health 2016, 5, e324–e334. [Google Scholar] [CrossRef]
- Modjarrad, K.; Roberts, C.C.; Mills, K.T.; Castellano, A.R.; Paolino, K.; Muthumani, K.; Reuschel, E.L.; Robb, M.L.; Racine, T.; Oh, M.-D.; et al. Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine: A phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect. Dis. 2019, 19, 1013–1022. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.C.; Wurie, A.H.; Hou, L.H.; Liang, Q.; Li, Y.H.; Russell, J.B.; Wu, S.P.; Li, J.X.; Hu, Y.M.; Guo, Q.; et al. Safety and immunogenicity of a recombinant adenovirus type-5 vector-based Ebola vaccine in healthy adults in Sierra Leone: A single-centre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 2016, 389, 621–628. [Google Scholar] [CrossRef]
- Amanna, I.J.; Carlson, N.E.; Slifka, M.K. Duration of humoral immunity to common viral and vaccine antigens. N. Engl. J. Med. 2007, 357, 1903–1915. [Google Scholar] [CrossRef]
- Hu, B.; Siche, S.; Möller, L.; Veit, M. Amphipathic Helices of Cellular Proteins Can Replace the Helix in M2 of Influenza A Virus with Only Small Effects on Virus Replication. J. Virol. 2020, 94, e01605–e01619. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Liu, J.; Jin, S.; Wang, L. Development of tumour peptide vaccines: From universalization to personalization. Scand. J. Immunol. 2020, 91, e12875. [Google Scholar] [CrossRef]
- Li, W.; Joshi, M.D.; Singhania, S.; Ramsey, K.H.; Murthy, A.K. Peptide Vaccine: Progress and Challenges. Vaccines 2014, 2, 515–536. [Google Scholar] [CrossRef]
- Svendsen, O.; Edwards, C.N.; Lauritzen, B.; Rasmussen, A.D. Intramuscular injection of hypertonic saline: In vitro and in vivo muscle tissue toxicity and spinal neurone c-fos expression. Basic Clin. Pharmacol. Toxicol. 2005, 97, 52–57. [Google Scholar] [CrossRef]
- Luo, M.; Shao, B.; Yu, J.Y.; Liu, T.; Liang, X.; Lu, L.; Ye, T.H.; He, Z.Y.; Xiao, H.Y.; Wei, X.W. Simultaneous enhancement of cellular and humoral immunity by the high salt formulation of Al(OH)3 adjuvant. Cell Res. 2017, 27, 586–589. [Google Scholar] [CrossRef]
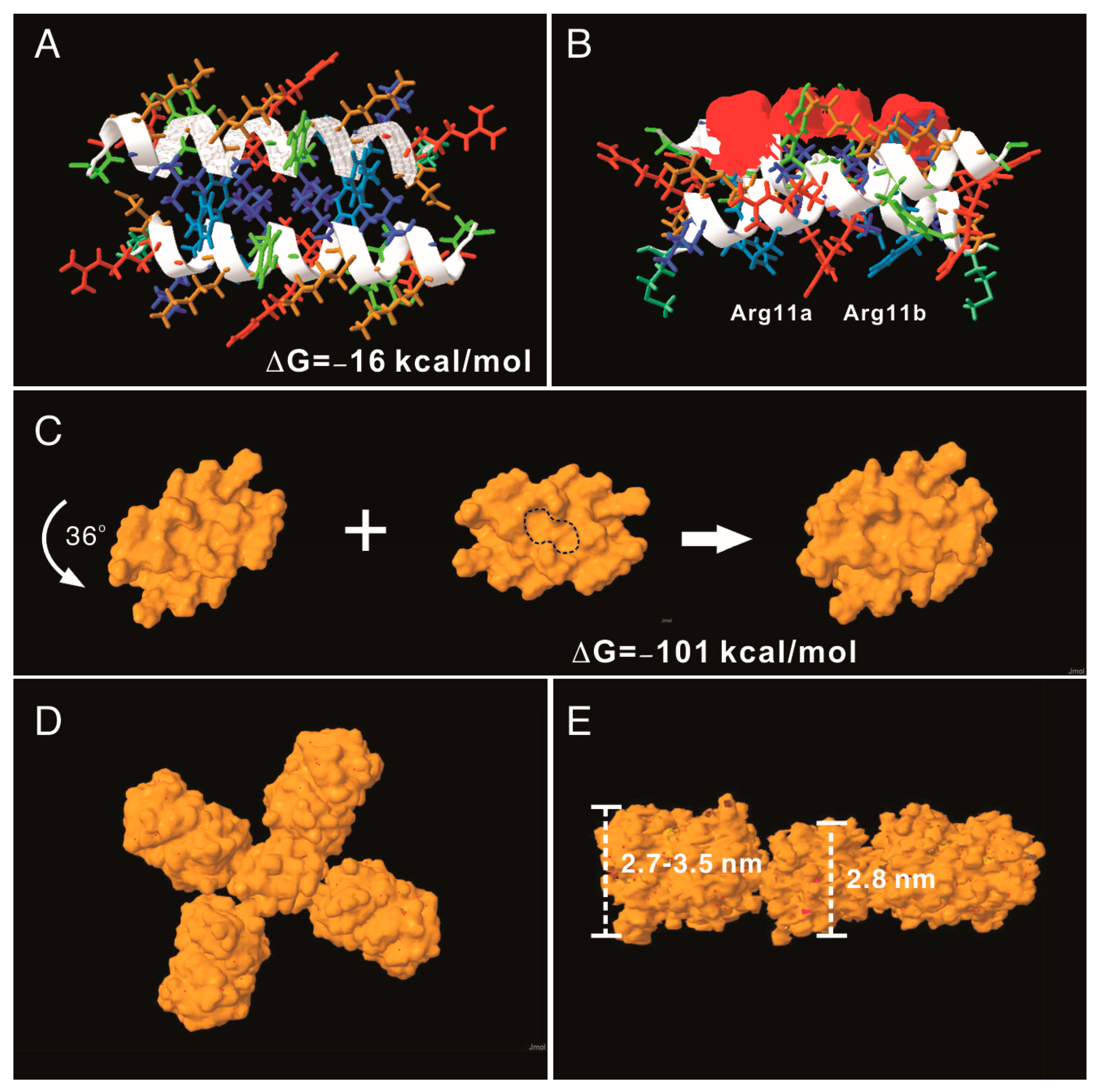
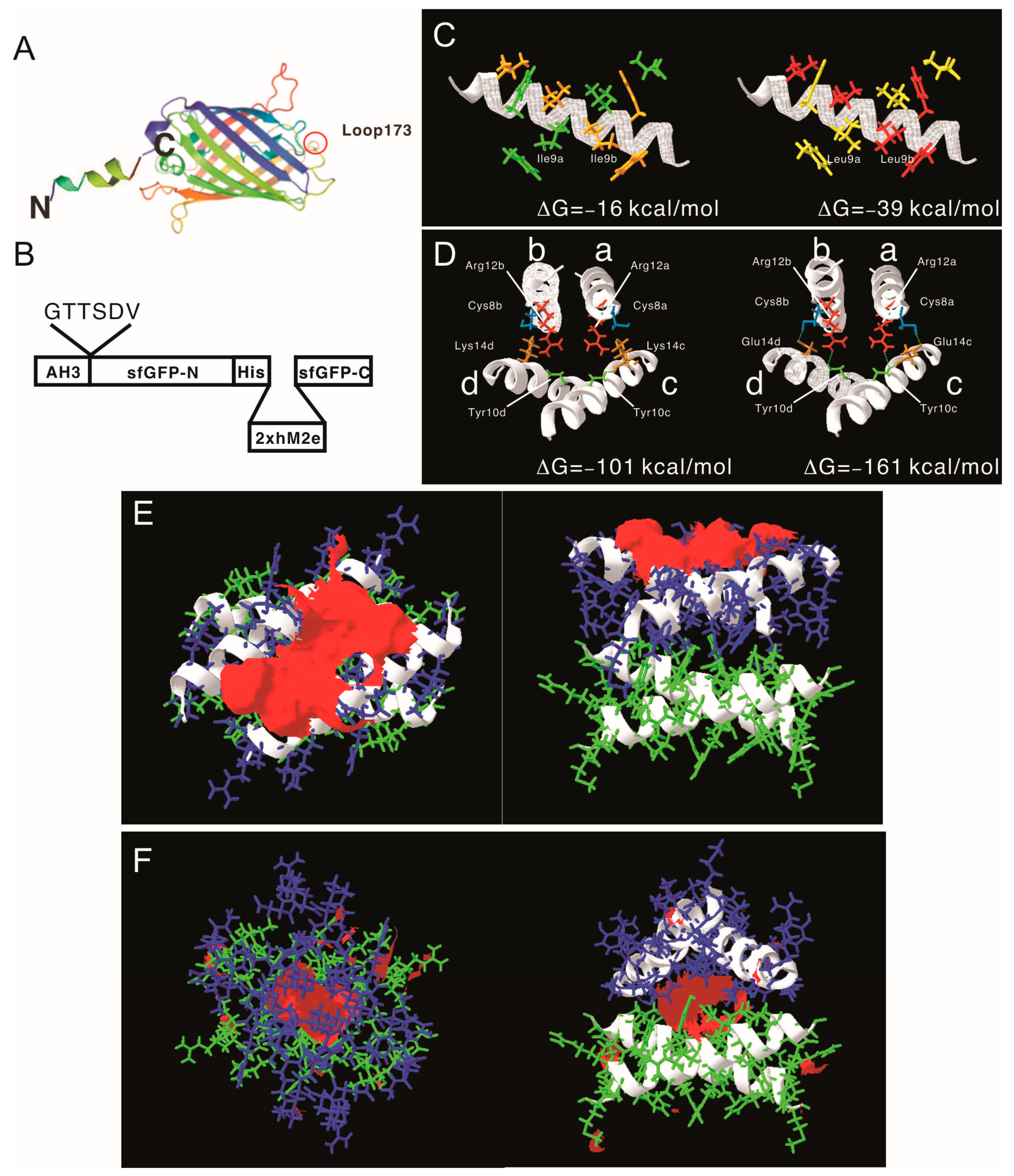


| Assembling Unit | Intermolecular Interactions (kcal/mol) | Hydrophobic Patch Area (A2) | Modelarchive Access Code | |
|---|---|---|---|---|
| AH3 | Monomer | na | na | ma-06g7k |
| Monomer–Monomer | −16 | 285 | ma-bhgiw | |
| Tandem dimer–dimer | −101 | 386 | ma-xhrzb | |
| Inverted dimer–dimer | na | na | na | |
| LY | Monomer | na | na | ma-0koys |
| Monomer–Monomer | −39 | 389 | ma-7x5gd | |
| Tandem dimer–dimer | na | na | na | |
| Inverted dimer–dimer | na | na | na | |
| RRLE | Monomer | na | na | ma-xkn28 |
| Monomer–Monomer | −23 | 429 | ma-ax78l | |
| Tandem dimer–dimer | −29 | 342 | ma-vyybf | |
| Inverted dimer–dimer | na | na | na | |
| LYRRLE | Monomer | na | na | ma-ivexo |
| Monomer–Monomer | −36 | 393 | ma-lptlj | |
| Tandem dimer–dimer | −161 | 433 | ma-izbbn | |
| Inverted dimer–dimer | −59 | 111 | ma-xomje |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wong, T.-T.; Liou, G.-G.; Kan, M.-C. A Thermal-Stable Protein Nanoparticle That Stimulates Long Lasting Humoral Immune Response. Vaccines 2023, 11, 426. https://doi.org/10.3390/vaccines11020426
Wong T-T, Liou G-G, Kan M-C. A Thermal-Stable Protein Nanoparticle That Stimulates Long Lasting Humoral Immune Response. Vaccines. 2023; 11(2):426. https://doi.org/10.3390/vaccines11020426
Chicago/Turabian StyleWong, Ten-Tsao, Gunn-Guang Liou, and Ming-Chung Kan. 2023. "A Thermal-Stable Protein Nanoparticle That Stimulates Long Lasting Humoral Immune Response" Vaccines 11, no. 2: 426. https://doi.org/10.3390/vaccines11020426
APA StyleWong, T.-T., Liou, G.-G., & Kan, M.-C. (2023). A Thermal-Stable Protein Nanoparticle That Stimulates Long Lasting Humoral Immune Response. Vaccines, 11(2), 426. https://doi.org/10.3390/vaccines11020426