Atypical Viral Infections in Gastroenterology
Abstract
1. Introduction
2. The Microbiome, Dysbiosis, and Viral Infections
3. Detection of Enteric Viruses
4. Atypical Viral Infections in Gastroenterology
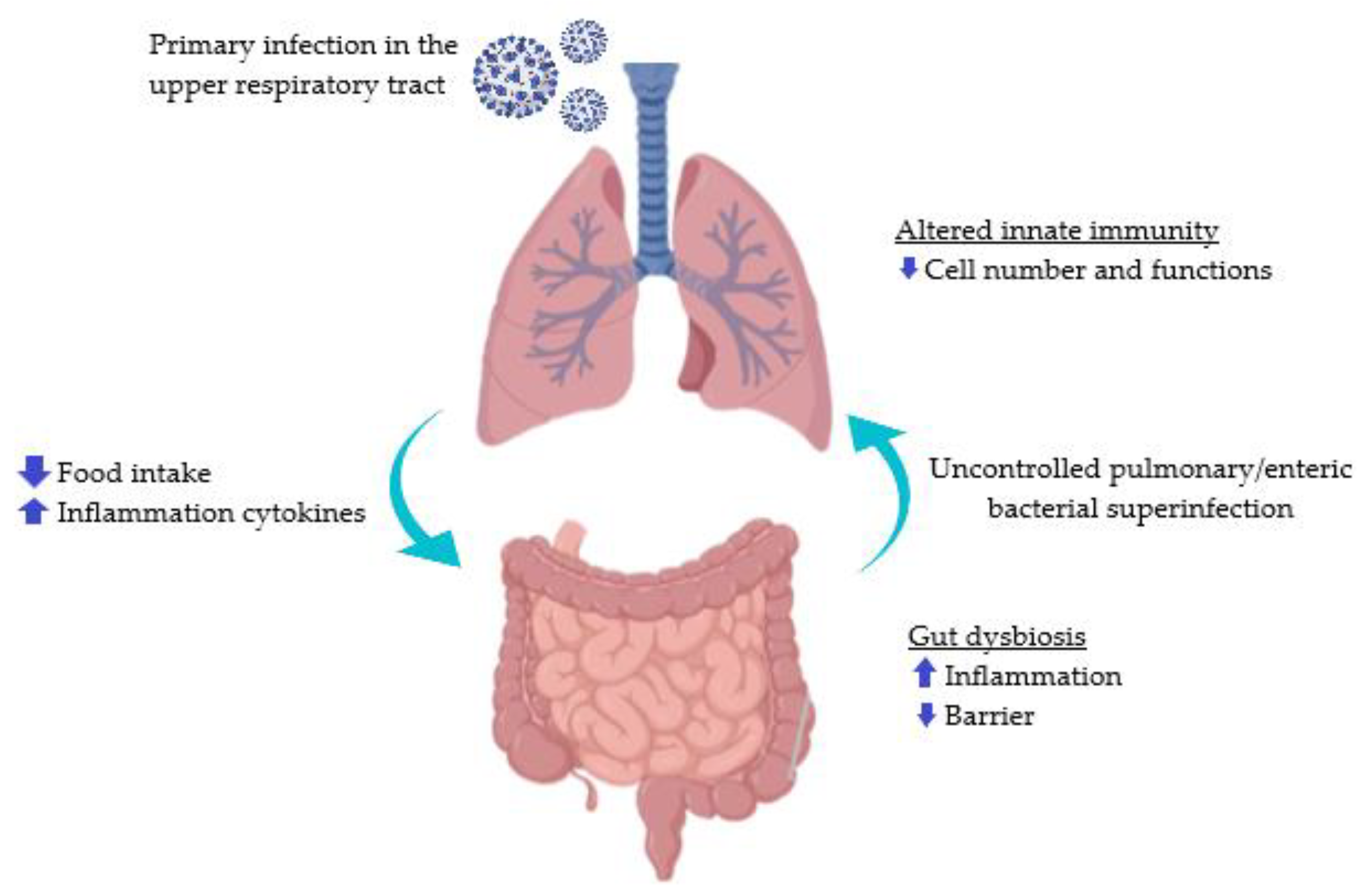
4.1. SARS-CoV-2
4.2. Hantavirus
4.3. Herpes Simplex Virus
4.4. Cytomegalovirus
4.5. Calicivirus
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Forstinus, N.; Ikechukwu, N.; Emenike, M.; Christiana, A. Water and Waterborne Diseases: A Review. Int. J. TROPICAL DISEASE Health 2016, 12, 1–14. [Google Scholar] [CrossRef]
- McClung, R.P.; Roth, D.M.; Vigar, M.; Roberts, V.A.; Kahler, A.M.; Cooley, L.A.; Hilborn, E.D.; Wade, T.J.; Fullerton, K.E.; Yoder, J.S.; et al. Waterborne Disease Outbreaks Associated with Environmental and Undetermined Exposures to Water—United States, 2013–2014. Morb. Mortal. Wkly. Rep. (MMWR) 2017, 66, 1222–1225. [Google Scholar] [CrossRef]
- World Health Organization. Guidelines for Drinking-Water Quality: Second Addendum. Vol 1, Recommendations [Internet]; World Health Organization: Geneva, Switzerland, 2008. [Google Scholar]
- Bosch, A. Human Enteric Viruses in the Water Environment: A Minireview. Int. Microbiol. Off. J. Span. Soc. Microbiol. 1998, 1, 191–196. [Google Scholar]
- Carter, M.J. Enterically Infecting Viruses: Pathogenicity, Transmission and Significance for Food and Waterborne Infection. J. Appl. Microbiol. 2005, 98, 1354–1380. [Google Scholar] [CrossRef]
- Svraka, S.; Duizer, E.; Vennema, H.; de Bruin, E.; van der Veer, B.; Dorresteijn, B.; Koopmans, M. Etiological Role of Viruses in Outbreaks of Acute Gastroenteritis in the Netherlands from 1994 through 2005. J. Clin. Microbiol. 2007, 45, 1389–1394. [Google Scholar] [CrossRef] [PubMed]
- Robinson, C.M. Enteric Viruses Exploit the Microbiota to Promote Infection. Curr. Opin. Virol. 2019, 37, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Bidawid, S.; Farber, J.M.; Sattar, S.A. Contamination of Foods by Food Handlers: Experiments on Hepatitis a Virus Transfer to Food and Its Interruption. Appl. Environ. Microbiol. 2000, 66, 2759–2763. [Google Scholar] [CrossRef]
- Meng, Q.S.; Gerba, C.P. Comparative Inactivation of Enteric Adenoviruses, Poliovirus and Coliphages by Ultraviolet Irradiation. Water Res. 1996, 30, 2665–2668. [Google Scholar] [CrossRef]
- Lippy, E.C.; Waltrip, S.C. Waterborne Disease Outbreaks—1946–1980: A Thirty-Five-Year Perspective. J. Am. Water Work. Assoc. 1984, 76, 60–67. [Google Scholar] [CrossRef]
- Jiménez-Clavero, M.A.; Fernández, C.; Ortiz, J.A.; Pro, J.; Carbonell, G.; Tarazona, J.V.; Roblas, N.; Ley, V. Teschoviruses as Indicators of Porcine Fecal Contamination of Surface Water. Appl. Environ. Microbiol. 2003, 69, 6311–6315. [Google Scholar] [CrossRef] [PubMed]
- Kaku, Y.; Sarai, A.; Murakami, Y. Genetic Reclassification of Porcine Enteroviruses. J. Gen. Virol. 2001, 82, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Ley, V.; Higgins, J.; Fayer, R. Bovine Enteroviruses as Indicators of Fecal Contamination. Appl. Environ. Microbiol. 2002, 68, 3455–3461. [Google Scholar] [CrossRef]
- Maluquer de Motes, C.; Clemente-Casares, P.; Hundesa, A.; Martín, M.; Girones, R. Detection of Bovine and Porcine Adenoviruses for Tracing the Source of Fecal Contamination. Appl. Environ. Microbiol. 2004, 70, 1448–1454. [Google Scholar] [CrossRef]
- Kuss, S.K.; Best, G.T.; Etheredge, C.A.; Pruijssers, A.J.; Frierson, J.M.; Hooper, L.V.; Dermody, T.S.; Pfeiffer, J.K. Intestinal Microbiota Promote Enteric Virus Replication and Systemic Pathogenesis. Science 2011, 334, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Kane, M.; Case, L.K.; Kopaskie, K.; Kozlova, A.; MacDearmid, C.; Chervonsky, A.V.; Golovkina, T.V. Successful Transmission of a Retrovirus Depends on the Commensal Microbiota. Science 2011, 334, 245–249. [Google Scholar] [CrossRef]
- Li, N.; Ma, W.-T.; Pang, M.; Fan, Q.-L.; Hua, J.-L. The Commensal Microbiota and Viral Infection: A Comprehensive Review. Front. Immunol. 2019, 10, 1551. [Google Scholar] [CrossRef]
- Schroeder, B.O. Fight Them or Feed Them: How the Intestinal Mucus Layer Manages the Gut Microbiota. Gastroenterol. Rep. 2019, 7, 3–12. [Google Scholar] [CrossRef]
- Lieleg, O.; Lieleg, C.; Bloom, J.; Buck, C.B.; Ribbeck, K. Mucin Biopolymers as Broad-Spectrum Antiviral Agents. Biomacromolecules 2012, 13, 1724–1732. [Google Scholar] [CrossRef]
- Nunn, K.L.; Wang, Y.-Y.; Harit, D.; Humphrys, M.S.; Ma, B.; Cone, R.; Ravel, J.; Lai, S.K. Enhanced Trapping of HIV-1 by Human Cervicovaginal Mucus Is Associated with Lactobacillus Crispatus-Dominant Microbiota. mBio 2015, 6, e01084-15. [Google Scholar] [CrossRef]
- Nagpal, R.; Yadav, H. Bacterial Translocation from the Gut to the Distant Organs: An Overview. Ann. Nutr. Metab. 2017, 71, 11–16. [Google Scholar] [CrossRef]
- Bron, P.A.; Kleerebezem, M.; Brummer, R.-J.; Cani, P.D.; Mercenier, A.; MacDonald, T.T.; Garcia-Ródenas, C.L.; Wells, J.M. Can Probiotics Modulate Human Disease by Impacting Intestinal Barrier Function? Br. J. Nutr. 2017, 117, 93–107. [Google Scholar] [CrossRef] [PubMed]
- Jenssen, H.; Hamill, P.; Hancock, R.E.W. Peptide Antimicrobial Agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [PubMed]
- Tabata, T.; Petitt, M.; Puerta-Guardo, H.; Michlmayr, D.; Wang, C.; Fang-Hoover, J.; Harris, E.; Pereira, L. Zika Virus Targets Different Primary Human Placental Cells, Suggesting Two Routes for Vertical Transmission. Cell Host Microbe 2016, 20, 155–166. [Google Scholar] [CrossRef]
- Serkedjieva, J.; Danova, S.; Ivanova, I. Antiinfluenza Virus Activity of a Bacteriocin Produced by Lactobacillus Delbrueckii. Appl. Biochem. Biotechnol. 2000, 88, 285–298. [Google Scholar] [CrossRef]
- Wachsman, M.B.; Castilla, V.; de Ruiz Holgado, A.P.; de Torres, R.A.; Sesma, F.; Coto, C.E. Enterocin CRL35 Inhibits Late Stages of HSV-1 and HSV-2 Replication in Vitro. Antivir. Res. 2003, 58, 17–24. [Google Scholar] [CrossRef]
- Petersen, C.; Round, J.L. Defining Dysbiosis and Its Influence on Host Immunity and Disease. Cell. Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef]
- McBurney, M.I.; Davis, C.; Fraser, C.M.; Schneeman, B.O.; Huttenhower, C.; Verbeke, K.; Walter, J.; Latulippe, M.E. Establishing What Constitutes a Healthy Human Gut Microbiome: State of the Science, Regulatory Considerations, and Future Directions. J. Nutr. 2019, 149, 1882–1895. [Google Scholar] [CrossRef]
- Borgdorff, H.; Gautam, R.; Armstrong, S.D.; Xia, D.; Ndayisaba, G.F.; van Teijlingen, N.H.; Geijtenbeek, T.B.H.; Wastling, J.M.; van de Wijgert, J.H.H.M. Cervicovaginal Microbiome Dysbiosis Is Associated with Proteome Changes Related to Alterations of the Cervicovaginal Mucosal Barrier. Mucosal Immunol. 2016, 9, 621–633. [Google Scholar] [CrossRef]
- Bosch, A.; Guix, S.; Sano, D.; Pintó, R.M. New Tools for the Study and Direct Surveillance of Viral Pathogens in Water. Curr. Opin. Biotechnol. 2008, 19, 295–301. [Google Scholar] [CrossRef]
- Cliver, D.O. Capsid and Infectivity in Virus Detection. Food Environ. Virol. 2009, 1, 123–128. [Google Scholar] [CrossRef]
- Rodriguez, R.A.; Pepper, I.L.; Gerba, C.P. Application of PCR-Based Methods to Assess the Infectivity of Enteric Viruses in Environmental Samples. Appl. Environ. Microbiol. 2008, 75, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Costafreda, M.I.; Bosch, A.; Pintó, R.M. Development, Evaluation, and Standardization of a Real-Time TaqMan Reverse Transcription-PCR Assay for Quantification of Hepatitis a Virus in Clinical and Shellfish Samples. Appl. Environ. Microbiol. 2006, 72, 3846–3855. [Google Scholar] [CrossRef] [PubMed]
- Wyn-Jones, A.P.; Sellwood, J. Enteric Viruses in the Aquatic Environment. J. Appl. Microbiol. 2001, 91, 945–962. [Google Scholar] [CrossRef] [PubMed]
- Mattison, K.; Bidawid, S. Analytical Methods for Food and Environmental Viruses. Food Environ. Virol. 2009, 1, 107–122. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Mez, J.; Daneshvar, D.H.; Kiernan, P.T.; Abdolmohammadi, B.; Alvarez, V.E.; Huber, B.R.; Alosco, M.L.; Solomon, T.M.; Nowinski, C.J.; McHale, L.; et al. Clinicopathological Evaluation of Chronic Traumatic Encephalopathy in Players of American Football. JAMA 2017, 318, 360–370. [Google Scholar] [CrossRef]
- Viana, S.D.; Nunes, S.; Reis, F. ACE2 Imbalance as a Key Player for the Poor Outcomes in COVID-19 Patients with Age-Related Comorbidities—Role of Gut Microbiota Dysbiosis. Ageing Res. Rev. 2020, 62, 101123. [Google Scholar] [CrossRef] [PubMed]
- Gheblawi, M.; Wang, K.; Viveiros, A.; Nguyen, Q.; Zhong, J.-C.; Turner, A.J.; Raizada, M.K.; Grant, M.B.; Oudit, G.Y. Angiotensin Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System. Circ. Res. 2020, 126, 1456–1474. [Google Scholar] [CrossRef] [PubMed]
- Saponaro, F.; Rutigliano, G.; Sestito, S.; Bandini, L.; Storti, B.; Bizzarri, R.; Zucchi, R. ACE2 in the Era of SARS-CoV-2: Controversies and Novel Perspectives. Front. Mol. Biosci. 2020, 7, 588618. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, T.; Hill, T.; Li, K.; Peters, C.J.; Tseng, C.-T.K. Severe Acute Respiratory Syndrome (SARS) Coronavirus-Induced Lung Epithelial Cytokines Exacerbate SARS Pathogenesis by Modulating Intrinsic Functions of Monocyte-Derived Macrophages and Dendritic Cells. J. Virol. 2008, 83, 3039–3048. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, L.-M.; Wan, L.; Xiang, T.-X.; Le, A.; Liu, J.-M.; Peiris, M.; Poon, L.L.M.; Zhang, W. Viral Dynamics in Mild and Severe Cases of COVID-19. Lancet Infect. Dis. 2020, 20, 656–657. [Google Scholar] [CrossRef]
- Jia, H.P.; Look, D.C.; Shi, L.; Hickey, M.; Pewe, L.; Netland, J.; Farzan, M.; Wohlford-Lenane, C.; Perlman, S.; McCray, P.B. ACE2 Receptor Expression and Severe Acute Respiratory Syndrome Coronavirus Infection Depend on Differentiation of Human Airway Epithelia. J. Virol. 2005, 79, 14614–14621. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.K.; Velkoska, E.; Burrell, L.M. Emerging Markers in Cardiovascular Disease: Where Does Angiotensin-Converting Enzyme 2 Fit in? Clin. Exp. Pharmacol. Physiol. 2013, 40, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Wenham, C.; Smith, J.; Morgan, R. COVID-19: The Gendered Impacts of the Outbreak. Lancet 2020, 395, 10227. [Google Scholar] [CrossRef]
- Jin, J.-M.; Bai, P.; He, W.; Wu, F.; Liu, X.-F.; Han, D.-M.; Liu, S.; Yang, J.-K. Gender Differences in Patients with COVID-19: Focus on Severity and Mortality. Front. Public Health 2020, 8, 152. [Google Scholar] [CrossRef]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833. [Google Scholar] [CrossRef]
- Sultan, S.; Altayar, O.; Siddique, S.M.; Davitkov, P.; Feuerstein, J.D.; Lim, J.K.; Falck-Ytter, Y.; El-Serag, H.B. AGA Institute Rapid Review of the GI and Liver Manifestations of COVID-19, Meta-Analysis of International Data, and Recommendations for the Consultative Management of Patients with COVID-19. Gastroenterology 2020, 159, 320–334. [Google Scholar] [CrossRef] [PubMed]
- Lovato, A.; de Filippis, C.; Marioni, G. Upper Airway Symptoms in Coronavirus Disease 2019 (COVID-19). Am. J. Otolaryngol. 2020, 41, 102474. [Google Scholar] [CrossRef] [PubMed]
- del Rio, C.; Malani, P.N. 2019 Novel Coronavirus—Important Information for Clinicians. JAMA 2020, 323, 1039–1040. [Google Scholar] [CrossRef]
- Cichoż-Lach, H.; Michalak, A. Liver Injury in the Era of COVID-19. World J. Gastroenterol. 2021, 27, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.; Ni, Z.; Hu, Y.; Liang, W.; Ou, C.; He, J.; Liu, L.; Shan, H.; Lei, C.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Ferm, S.; Fisher, C.; Pakala, T.; Tong, M.; Shah, D.; Schwarzbaum, D.; Cooley, V.; Hussain, S.; Kim, S.H. Analysis of Gastrointestinal and Hepatic Manifestations of SARS-CoV-2 Infection in 892 Patients in Queens, NY. Clin. Gastroenterol. Hepatol. 2020, 18, 2378–2379. [Google Scholar] [CrossRef] [PubMed]
- Bingula, R.; Filaire, M.; Radosevic-Robin, N.; Bey, M.; Berthon, J.-Y.; Bernalier-Donadille, A.; Vasson, M.-P.; Filaire, E. Desired Turbulence? Gut-Lung Axis, Immunity, and Lung Cancer. J. Oncol. 2017, 2017, 5035371. [Google Scholar] [CrossRef] [PubMed]
- Keely, S.; Talley, N.J.; Hansbro, P.M. Pulmonary-Intestinal Cross-Talk in Mucosal Inflammatory Disease. Mucosal Immunol. 2011, 5, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Dumas, A.; Bernard, L.; Poquet, Y.; Lugo-Villarino, G.; Neyrolles, O. The Role of the Lung Microbiota and the Gut-Lung Axis in Respiratory Infectious Diseases. Cell. Microbiol. 2018, 20, e12966. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Liu, Z.; Chen, Y.; Xiao, Y.; Huang, X.; Fan, X.-G. Bacterial and Fungal Infections in COVID-19 Patients: A Matter of Concern. Infect. Control Hosp. Epidemiol. 2020, 41, 1124–1125. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Xia, J.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; et al. Clinical Course and Outcomes of Critically Ill Patients with SARS-CoV-2 Pneumonia in Wuhan, China: A Single-Centered, Retrospective, Observational Study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients with COVID-19 during Time of Hospitalization. Gastroenterology 2020, 159, 944–955. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Gu, S.; Gong, Y.; Li, B.; Lu, H.; Li, Q.; Zhang, R.; Gao, X.; Wu, Z.; Zhang, J.; et al. Clinical Significance of the Correlation between Changes in the Major Intestinal Bacteria Species and COVID-19 Severity. Engineering 2020, 6, 1178–1184. [Google Scholar] [CrossRef]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the Gut Microbiota in Patients with Coronavirus Disease 2019 or H1N1 Influenza. Clin. Infect. Dis. 2020, 71, 2669–2678. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, X.; Zhu, B.; Liang, H.; Fang, C.; Gong, Y.; Guo, Q.; Sun, X.; Zhao, D.; Shen, J.; et al. Characteristics of Pediatric SARS-CoV-2 Infection and Potential Evidence for Persistent Fecal Viral Shedding. Nat. Med. 2020, 26, 502–505. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef]
- Gu, J.; Han, B.; Wang, J. COVID-19: Gastrointestinal Manifestations and Potential Fecal-Oral Transmission. Gastroenterology 2020, 158, 1518–1519. [Google Scholar] [CrossRef]
- D’Amico, F.; Baumgart, D.C.; Danese, S.; Peyrin-Biroulet, L. Diarrhea during COVID-19 Infection: Pathogenesis, Epidemiology, Prevention and Management. Clin. Gastroenterol. Hepatol. 2020, 18, 1663–1672. [Google Scholar] [CrossRef]
- Carvalho, A.; Alqusairi, R.; Adams, A.; Paul, M.; Kothari, N.; Peters, S.; DeBenedet, A.T. SARS-CoV-2 Gastrointestinal Infection Causing Hemorrhagic Colitis. Am. J. Gastroenterol. 2020, 115, 942–946. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Jiang, X.; Zhang, Z.; Huang, S.; Zhang, Z.; Fang, Z.; Gu, Z.; Gao, L.; Shi, H.; Mai, L.; et al. Gastrointestinal Symptoms of 95 Cases with SARS-CoV-2 Infection. Gut 2020, 69, 997–1001. [Google Scholar] [CrossRef]
- Seeliger, B.; Philouze, G.; Benotmane, I.; Mutter, D.; Pessaux, P. Is the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Present Intraperitoneally in Patients with Coronavirus Disease 2019 (COVID-19) Infection Undergoing Emergency Operations? Surgery 2020, 168, 220–221. [Google Scholar] [CrossRef] [PubMed]
- Walsh, K.A.; Jordan, K.; Clyne, B.; Rohde, D.; Drummond, L.; Byrne, P.; Ahern, S.; Carty, P.G.; O’Brien, K.K.; O’Murchu, E.; et al. SARS-CoV-2 Detection, Viral Load and Infectivity over the Course of an Infection. J. Infect. 2020, 81, 357–371. [Google Scholar] [CrossRef]
- Kopel, J.; Perisetti, A.; Gajendran, M.; Boregowda, U.; Goyal, H. Clinical Insights into the Gastrointestinal Manifestations of COVID-19. Dig. Dis. Sci. 2020, 65, 1932–1939. [Google Scholar] [CrossRef]
- Schmaljohn, C.S.; Dalrymple, J.M. Analysis of Hantaan Virus RNA: Evidence for a New Genus of Bunyaviridae. Virology 1983, 131, 482–491. [Google Scholar] [CrossRef]
- Plyusnin, A.; Vapalahti, O.; Vaheri, A. Hantaviruses: Genome Structure, Expression and Evolution. J. Gen. Virol. 1996, 77, 2677–2687. [Google Scholar] [CrossRef] [PubMed]
- Bridson, E. The English “Sweate” (Sudor Anglicus) and Hantavirus Pulmonary Syndrome. Br. J. Biomed. Sci. 2001, 58, 1–6. [Google Scholar] [PubMed]
- McCaughey, C.; Hart, C.A. Hantaviruses. J. Med. Microbiol. 2000, 49, 587–599. [Google Scholar] [CrossRef]
- Heyman, P.; Vaheri, A.; Lundkvist, A.; Avsic-Zupanc, T. Hantavirus Infections in Europe: From Virus Carriers to a Major Public-Health Problem. Expert Rev. Anti-Infect. Ther. 2009, 7, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, J.; Kramer, S.; Herrlinger, K.R.; Jeske, K.; Kuhns, M.; Weiss, S.; Ulrich, R.G.; Krüger, D.H. Tula Virus as Causative Agent of Hantavirus Disease in Immunocompetent Person, Germany. Emerg. Infect. Dis. 2021, 27, 1234–1237. [Google Scholar] [CrossRef]
- Avšič-Županc, T.; Saksida, A.; Korva, M. Hantavirus Infections. Clin. Microbiol. Infect. 2019, 21, e6–e16. [Google Scholar] [CrossRef] [PubMed]
- Deutz, A.; Fuchs, K.; Schuller, W.; Nowotny, N.; Auer, H.; Aspöck, H.; Stünzner, D.; Kerbl, U.; Klement, C.; Köfer, J. Seroepidemiological Studies of Zoonotic Infections in Hunters in Southeastern Austria--Prevalences, Risk Factors, and Preventive Methods. Berliner Und Munchener Tierarztliche Wochenschrift 2003, 116, 306–311. [Google Scholar]
- Zöller, L.; Faulde, M.; Meisel, H.; Ruh, B.; Kimmig, P.; Schelling, U.; Zeier, M.; Kulzer, P.; Becker, C.; Roggendorf, M. Seroprevalence of Hantavirus Antibodies in Germany as Determined by a New Recombinant Enzyme Immunoassay. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 1995, 14, 305–313. [Google Scholar] [CrossRef]
- Klempa, B. Hantaviruses and Climate Change. Clin. Microbiol. Infect. 2009, 15, 518–523. [Google Scholar] [CrossRef]
- Lednicky, J.A. Hantaviruses. A Short Review. Arch. Pathol. Lab. Med. 2003, 127, 30–35. [Google Scholar] [CrossRef]
- Chun, P.K.; Godfrey, L.J. Unique Selective Right Atrial Hemorrhage with Epidemic (Korean) Hemorrhagic Fever. Am. Heart J. 1984, 108, 410–412. [Google Scholar] [CrossRef]
- Alexeyev, O.A.; Morozov, V.G.; Efremov, A.G.; Settergren, B. A Case of Haemorrhagic Fever with Renal Syndrome Complicated by Spleen Haemorrhage. Scand. J. Infect. Dis. 1994, 26, 491–492. [Google Scholar] [CrossRef]
- Suh, D.C.; Park, J.S.; Park, S.K.; Lee, H.K.; Chang, K.H. Pituitary Hemorrhage as a Complication of Hantaviral Disease. AJNR Am. J. Neuroradiol. 1995, 16, 175–178; discussion 179–180. [Google Scholar] [PubMed]
- Roizman, B.; Sears, A.E. Herpes Simplex Viruses and Their Replication. Virology 1996, 2, 2231–2295. [Google Scholar]
- Becker, Y.; Dym, H.; Sarov, I. Herpes Simplex Virus DNA. Virology 1968, 36, 184–192. [Google Scholar] [CrossRef]
- Kieff, E.D.; Bachenheimer, S.L.; Roizman, B. Size, Composition, and Structure of the Deoxyribonucleic Acid of Herpes Simplex Virus Subtypes 1 and 2. J. Virol. 1971, 8, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Plummer, G.; Goodheart, C.R.; Henson, D.; Bowling, C.P. A Comparative Study of the DNA Density and Behavior in Tissue Cultures of Fourteen Different Herpesviruses. Virology 1969, 39, 134–137. [Google Scholar] [CrossRef]
- Furlong, D.; Swift, H.; Roizman, B. Arrangement of Herpesvirus Deoxyribonucleic Acid in the Core. J. Virol. 1972, 10, 1071–1074. [Google Scholar] [CrossRef] [PubMed]
- Cruz, A.T.; Freedman, S.B.; Kulik, D.M.; Okada, P.J.; Fleming, A.H.; Mistry, R.D.; Thomson, J.E.; Schnadower, D.; Arms, J.L.; Mahajan, P.; et al. Herpes Simplex Virus Infection in Infants Undergoing Meningitis Evaluation. Pediatrics 2018, 141, e20171688. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, H.; Wei, B. Immune Response of T Cells during Herpes Simplex Virus Type 1 (HSV-1) Infection. J. Zhejiang Univ.-Sci. B 2017, 18, 277–288. [Google Scholar] [CrossRef]
- Giraldo, D.; Wilcox, D.R.; Longnecker, R. The Type I Interferon Response and Age-Dependent Susceptibility to Herpes Simplex Virus Infection. DNA Cell Biol. 2017, 36, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Bin, L.; Li, X.; Richers, B.; Streib, J.E.; Hu, J.W.; Taylor, P.; Leung, D.Y.M. Ankyrin Repeat Domain 1 Regulates Innate Immune Responses against Herpes Simplex Virus 1: A Potential Role in Eczema Herpeticum. J. Allergy Clin. Immunol. 2018, 141, 2085–2093. [Google Scholar] [CrossRef] [PubMed]
- Rechenchoski, D.Z.; Faccin-Galhardi, L.C.; Linhares, R.E.C.; Nozawa, C. Herpesvirus: An Underestimated Virus. Folia Microbiol. 2016, 62, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Jawale, R.; Lai, K.K.; Lamps, L.W. Sexually Transmitted Infections of the Lower Gastrointestinal Tract. Virchows Arch. 2017, 472, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Yamamoto, R.; Yoshimitsu, S.; Shimazaki, K.; Ogawa, S.; Itoh, K.; Miura, S. Herpes Simplex Esophagitis in the Immunocompetent Host. Dis. Esophagus 2005, 18, 340–344. [Google Scholar] [CrossRef]
- Rattner, H.M.; Cooper, D.J.; Zaman, M.B. Severe Bleeding from Herpes Esophagitis. Am. J. Gastroenterol. 1985, 80, 523–525. [Google Scholar] [PubMed]
- Goodell, S.E.; Quinn, T.C.; Mkrtichian, E.; Schuffler, M.D.; Holmes, K.K.; Corey, L. Herpes Simplex Virus Proctitis in Homosexual Men. N. Engl. J. Med. 1983, 308, 868–871. [Google Scholar] [CrossRef] [PubMed]
- Greenson, J.K.; Beschorner, W.E.; Boitnott, J.K.; Yardley, J.H. Prominent Mononuclear Cell Infiltrate Is Characteristic of Herpes Esophagitis. Hum. Pathol. 1991, 22, 541–549. [Google Scholar] [CrossRef]
- Chetty, R.; Roskell, D.E. Cytomegalovirus Infection in the Gastrointestinal Tract. J. Clin. Pathol. 1994, 47, 968–972. [Google Scholar] [CrossRef] [PubMed]
- Kambham, N.; Vij, R.; Cartwright, C.A.; Longacre, T. Cytomegalovirus Infection in Steroid-Refractory Ulcerative Colitis. Am. J. Surg. Pathol. 2004, 28, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Kraus, M.D.; Feran-Doza, M.; Garcia-Moliner, M.L.; Antin, J.; Odze, R.D. Cytomegalovirus Infection in the Colon of Bone Marrow Transplantation Patients. Mod. Pathol. Off. J. United States Can. Acad. Pathol. 1998, 11, 29–36. [Google Scholar]
- Yan, Z.; Nguyen, S.; Poles, M.; Melamed, J.; Scholes, J.V. Adenovirus Colitis in Human Immunodeficiency Virus Infection: An Underdiagnosed Entity. Am. J. Surg. Pathol. 1998, 22, 1101–1106. [Google Scholar] [CrossRef]
- Green, K.Y.; Ando, T.; Balayan, M.S.; Berke, T.; Clarke, I.N.; Estes, M.K.; Matson, D.O.; Nakata, S.; Neill, J.D.; Studdert, M.J.; et al. Taxonomy of the Caliciviruses. J. Infect. Dis. 2000, 181 (Suppl. 2), S322–S330. [Google Scholar] [CrossRef]
- Hutson, A.M.; Atmar, R.L.; Estes, M.K. Norovirus Disease: Changing Epidemiology and Host Susceptibility Factors. Trends Microbiol. 2004, 12, 279–287. [Google Scholar] [CrossRef]
- Fankhauser, R.L.; Noel, J.S.; Monroe, S.S.; Ando, T.; Glass, R.I. Molecular Epidemiology of “Norwalk-like Viruses” in Outbreaks of Gastroenteritis in the United States. J. Infect. Dis. 1998, 178, 1571–1578. [Google Scholar] [CrossRef] [PubMed]
- Clarke, I.N.; Lambden, P.R. Viral Zoonoses and Food of Animal Origin: Caliciviruses and Human Disease. Viral Zoonoses Food Anim. Orig. 1997, 13, 141–152. [Google Scholar] [CrossRef]
- Dedman, D.; Laurichesse, H.; Caul, E.O.; Wall, P.G. Surveillance of Small Round Structured Virus (SRSV) Infection in England and Wales, 1990–1995. Epidemiol. Infect. 1998, 121, 139–149. [Google Scholar] [CrossRef]
- Vinje, J.; Koopmans, M.P.G. Molecular Detection and Epidemiology of Small Round-Structured Viruses in Outbreaks of Gastroenteritis in the Netherlands. J. Infect. Dis. 1996, 174, 610–615. [Google Scholar] [CrossRef] [PubMed]
- Atmar, R.L. Noroviruses—State of the Art. Food Environ. Virol. 2010, 2, 117–126. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority. The Community Summary Report on Food-Borne Outbreaks in the European Union in 2007. EFSA J. 2009, 7, 271r. [Google Scholar] [CrossRef]
- Götz, H.; Ekdahl, K.; Lindbäck, J.; de Jong, B.; Hedlund, K.; Giesecke, J. Clinical Spectrum and Transmission Characteristics of Infection with Norwalk-like Virus: Findings from a Large Community Outbreak in Sweden. Clin. Infect. Dis. 2001, 33, 622–628. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, S.S.; Chatterjee, N.K.; Fuschino, M.E.; Magid, M.S.; Gordon, R.E.; Morse, D.L.; Herold, B.C.; LeLeiko, N.S.; Tschernia, A.; Florman, S.S.; et al. Calicivirus Enteritis in an Intestinal Transplant Recipient. Am. J. Transplant. 2003, 3, 764–768. [Google Scholar] [CrossRef] [PubMed]
- Svraka, S.; Vennema, H.; van der Veer, B.; Hedlund, K.-O.; Thorhagen, M.; Siebenga, J.; Duizer, E.; Koopmans, M. Epidemiology and Genotype Analysis of Emerging Sapovirus-Associated Infections across Europe. J. Clin. Microbiol. 2010, 48, 2191–2198. [Google Scholar] [CrossRef]
- Lee, L.E.; Cebelinski, E.A.; Fuller, C.; Keene, W.E.; Smith, K.; Vinjé, J.; Besser, J.M. Sapovirus Outbreaks in Long-Term Care Facilities, Oregon and Minnesota, USA, 2002–2009. Emerg. Infect. Dis. 2012, 18, 873–876. [Google Scholar] [CrossRef] [PubMed]
- Medici, M.C.; Tummolo, F.; Albonetti, V.; Abelli, L.A.; Chezzi, C.; Calderaro, A. Molecular Detection and Epidemiology of Astrovirus, Bocavirus, and Sapovirus in Italian Children Admitted to Hospital with Acute Gastroenteritis, 2008–2009. J. Med. Virol. 2012, 84, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Sala, M.R.; Broner, S.; Moreno, A.; Arias, C.; Godoy, P.; Minguell, S.; Martínez, A.; Torner, N.; Bartolomé, R.; de Simón, M.; et al. Cases of Acute Gastroenteritis due to Calicivirus in Outbreaks: Clinical Differences by Age and Aetiological Agent. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2014, 20, 793–798. [Google Scholar] [CrossRef] [PubMed][Green Version]
| Atypical Viral Infection | Genetic Material | Symptoms | Risk Groups | |
|---|---|---|---|---|
| Common | Rare | |||
| SARS-CoV-2 | RNA | Shortness of breath [50] Fever [50] Dry cough [50] | Diarrhea [51] Abdominal pain [51] Nausea [51] Vomiting [51] | Older males with cardiovascular diseases, hypertension, or diabetes [38] |
| Hantavirus | RNA | Abdominal pain [81] Myalgia [81] Neurological [81] | Mucosal bleeding [82] Epistaxis [82] Spleen hemorrhage [83] Panhypopituitarism [84] | Immunocompromised patients [76] |
| HSV | DNA | Herpes [94] Ulcer [98] Chest pain [97] Fever [97] | Multinucleated large cells [99] Distinctive viral inclusions [99] | Immunocompromised patients [96] |
| CMV | DNA | Weight loss [100] Fever [100] Stomach pain [100] Diarrhea [100] | Hypertrophic gastropathy [100] Protein-losing enteropathy [100] | Immunocompromised patients [100] |
| Calicivirus | RNA | Diarrhea [114] Vomiting [114] Gastroenteritis [106,107,108,109] Low-grade fever [114] | / | Infants [114] Immunocompromised patients [114] Elderly patients [114] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crnčević, N.; Rifatbegović, Z.; Hukić, M.; Deumić, S.; Pramenković, E.; Selimagić, A.; Gavrankapetanović, I.; Avdić, M. Atypical Viral Infections in Gastroenterology. Diseases 2022, 10, 87. https://doi.org/10.3390/diseases10040087
Crnčević N, Rifatbegović Z, Hukić M, Deumić S, Pramenković E, Selimagić A, Gavrankapetanović I, Avdić M. Atypical Viral Infections in Gastroenterology. Diseases. 2022; 10(4):87. https://doi.org/10.3390/diseases10040087
Chicago/Turabian StyleCrnčević, Neira, Zijah Rifatbegović, Mirsada Hukić, Sara Deumić, Emina Pramenković, Amir Selimagić, Ismet Gavrankapetanović, and Monia Avdić. 2022. "Atypical Viral Infections in Gastroenterology" Diseases 10, no. 4: 87. https://doi.org/10.3390/diseases10040087
APA StyleCrnčević, N., Rifatbegović, Z., Hukić, M., Deumić, S., Pramenković, E., Selimagić, A., Gavrankapetanović, I., & Avdić, M. (2022). Atypical Viral Infections in Gastroenterology. Diseases, 10(4), 87. https://doi.org/10.3390/diseases10040087