MALDI-TOF MS for Rapid Analysis of Bacterial Pathogens Causing Urinary Tract Infections in the Riyadh Region
Abstract
1. Introduction
2. Materials and Methods
2.1. Urine Collection
2.2. Bacterial Culture
2.3. Standard Plate Count (SPC)
2.4. Molecular Analysis by MALDI-TOF MS
2.5. Antimicrobial Susceptibility Tests
2.6. DNA Isolation
2.7. PCR & MLST Analysis
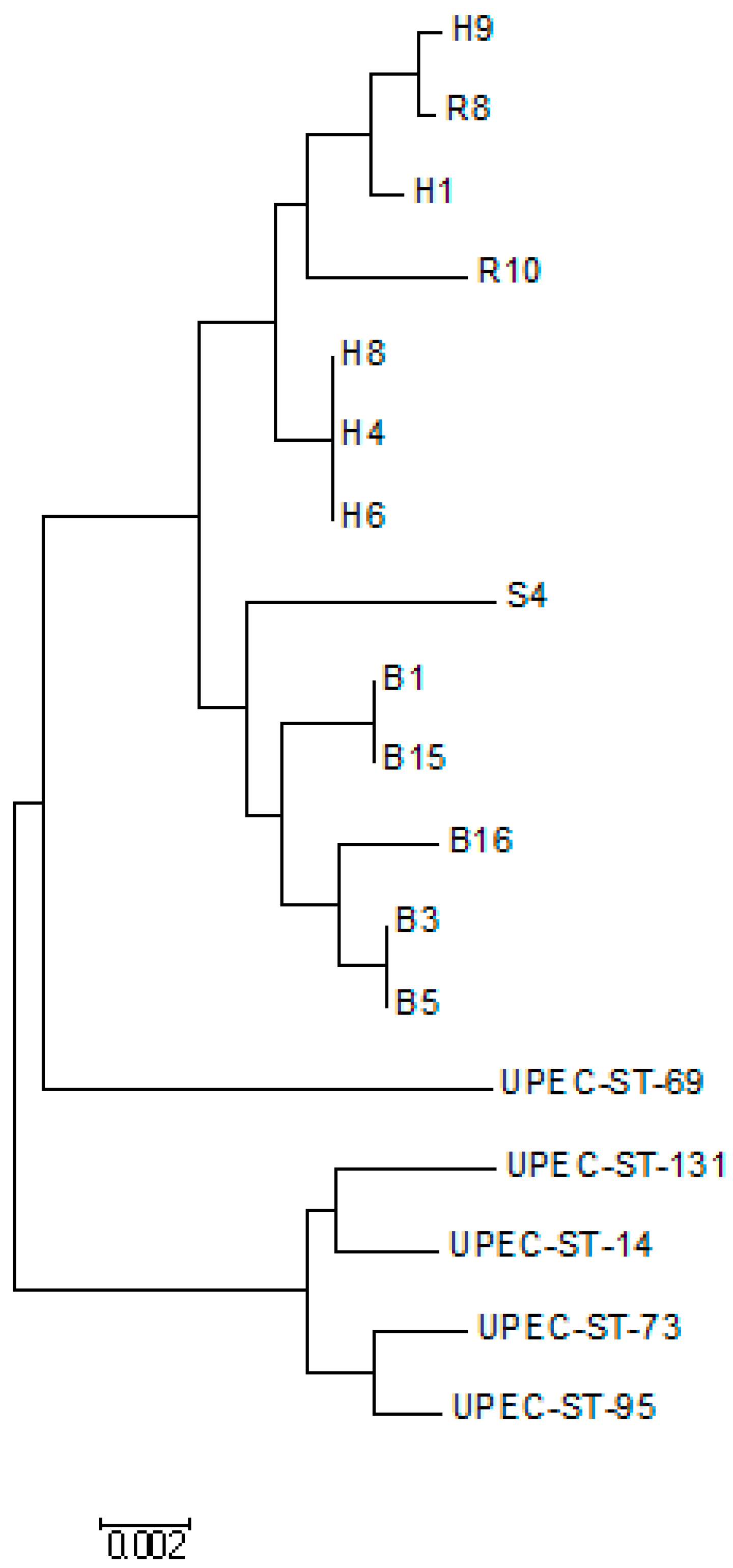
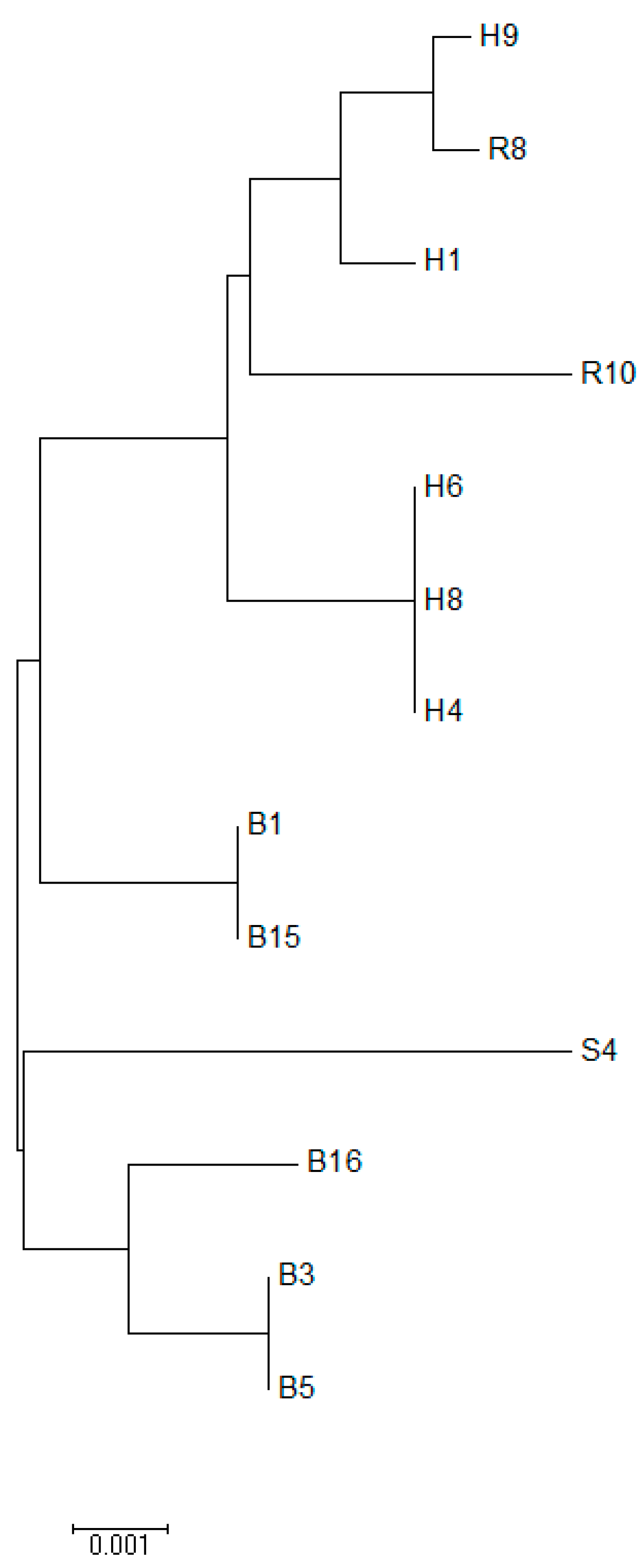
2.8. Phylogenetic Tree Analysis
2.9. Data Analysis
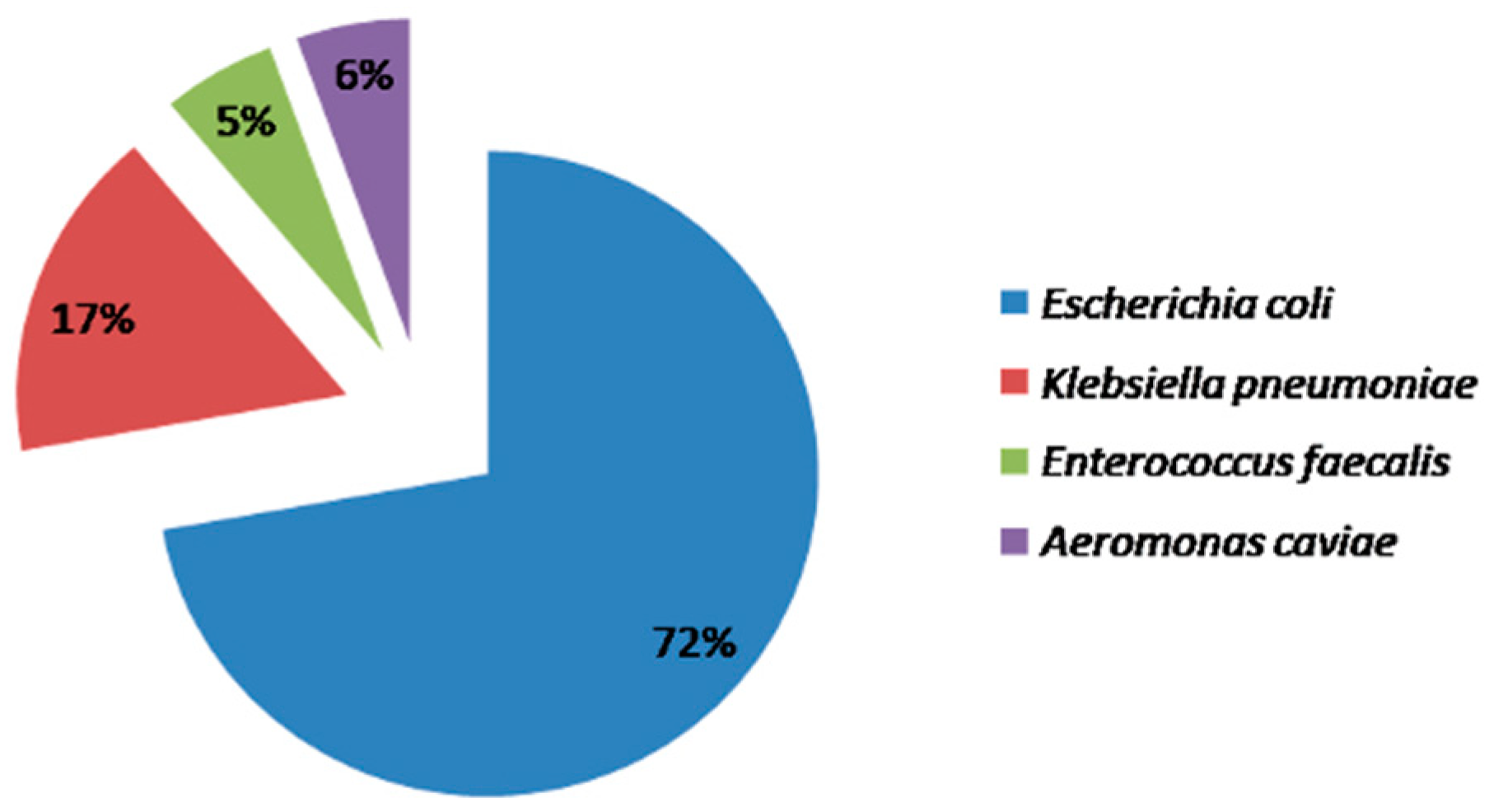
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Clark, A.E.; Kaleta, E.J.; Arora, A.; Wolk, D.M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: A fundamental shift in the routine practice of clinical microbiology. Clin. Microbiol. Rev. 2013, 26, 547–603. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.; Sánchez-Juanes, F.; García-Fraile, P.; Rivas, R.; Mateos, P.F.; Martínez-Molina, E.; González-Buitrago, J.M.; Velázquez, E. MALDI-TOF mass spectrometry is a fast and reliable platform for identification and ecological studies of species from family Rhizobiaceae. PLoS ONE 2011, 6, e20223. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.; Vega Castaño, S.; Sánchez-Juanes, F.; González-Cabrero, S.; Menegotto, F.; Orduña-Domingo, A.; González-Buitrago, J.M.; Muñoz-Bellido, J.L. Identification of Brucella by MALDI-TOF mass spectrometry. Fast and reliable identification from agar plates and blood cultures. PLoS ONE 2010, 5, e14235. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, Z.; Guo, Y. Chemical and biochemical applications of MALDI TOF-MS based on analyzing the small organic compounds. Top Curr. Chem. 2013, 331, 165–192. [Google Scholar] [CrossRef]
- Burillo, A.; Rodríguez-Sánchez, B.; Ramiro, A.; Cercenado, E.; Rodríguez-Créixems, M.; Bouza, E. Gram-stain plus MALDI-TOF MS (Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry) for a rapid diagnosis of urinary tract infection. PLoS ONE 2014, 9, e86915. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, M.; Zhu, M.; Wang, M.; Sun, Y.; Gu, H.; Cao, J.; Li, X.; Zhang, S.; Wang, J.; et al. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry systems for the identification of clinical filamentous fungi. World J. Microbiol. Biotechnol. 2017, 33, 142. [Google Scholar] [CrossRef]
- Li, W.; Sun, E.; Wang, Y.; Pan, H.; Zhang, Y.; Li, Y.; Zhang, X.; Li, C.; Du, L.; Wang, C. Rapid Identification and Antimicrobial Susceptibility Testing for Urinary Tract Pathogens by Direct Analysis of Urine Samples Using a MALDI-TOF MS-Based Combined Protocol. Front. Microbiol. 2019, 10, 1182. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef]
- Foxman, B. Epidemiology of urinary tract infections: Incidence, morbidity, and economic costs. Am. J. Med. 2002, 113 (Suppl. S1A), 5S–13S. [Google Scholar] [CrossRef]
- Terlizzi, M.E.; Gribaudo, G.; Maffei, M.E. UroPathogenic Escherichia coli (UPEC) Infections: Virulence Factors, Bladder Responses, Antibiotic, and Non-antibiotic Antimicrobial Strategies. Front. Microbiol. 2017, 8, 1566. [Google Scholar] [CrossRef]
- McLellan, L.K.; Hunstad, D.A. Urinary Tract Infection: Pathogenesis and Outlook. Trends Mol. Med. 2016, 22, 946–957. [Google Scholar] [CrossRef] [PubMed]
- Kodner, C.; Gupton, E.K.T. Recurrent Urinary Tract Infections in Women: Diagnosis and Management. Am. Fami. Physic. 2010, 82, 638–643. [Google Scholar]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef]
- Yari, A.R.; Mohammadi, M.J.; Geravandi, S.; Doosti, Z.; Matboo, S.A.; Jang, S.A.; Nazari, S. Assessment of microbial quality of household water output from desalination systems by the heterotrophic plate count method. J. Water Health. 2018, 16, 930–937. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Answer, R.; Sehrawat, A.; Sehrawat, N.; Yadav, M.; Sharma, A.K. Isolation and characterization of pathogenic bacteria from drinking water in North India. Int. J. Environ. Sci. Technol. 2021. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; Clinical and Laboratory Standards Institute: Wayne, IL, USA, 2021. [Google Scholar]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Rodríguez-Sánchez, B.; Cercenado, E.; Coste, A.T.; Greub, G. Review of the impact of MALDI-TOF MS in public health and hospital hygiene, 2018. Euro Surveill. 2019, 24, 1800193. [Google Scholar] [CrossRef]
- Zhou, C.; Hu, B.; Zhang, X.; Huang, S.; Shan, Y.; Ye, X. The value of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in identifying clinically relevant bacteria: A comparison with automated microbiology system. J. Thorac. Dis. 2014, 6, 545–552. [Google Scholar] [PubMed]
- Zboromyrska, Y.; Rubio, E.; Alejo, I.; Vergara, A.; Mons, A.; Campo, I.; Bosch, J.; Marco, F.; Vila, J. Development of a new protocol for rapid bacterial identification and susceptibility testing directly from urine samples. Clin. Microbiol. Infect. 2016, 22, 561.e1–561.e6. [Google Scholar] [CrossRef] [PubMed]
- Florio, W.; Baldeschi, L.; Rizzato, C.; Tavanti, A.; Ghelardi, E.; Lupetti, A. Detection of Antibiotic-Resistance by MALDI-TOF Mass Spectrometry: An Expanding Area. Front. Cell. Infect. Microbiol. 2020, 10, 572909. [Google Scholar] [CrossRef] [PubMed]
- Burckhardt, I.; Zimmermann, S. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J. Clin. Microbiol. 2011, 49, 3321–3324. [Google Scholar] [CrossRef]
- Johansson, A.; Nagy, E.; Soki, J.; ESGAI (ESCMID Study Group on Anaerobic Infections). Detection of carbapenemase activities of Bacteroides fragilis strains with matrix-assisted laser desorption ionization– time of flight mass spectrometry (MALDI-TOF MS). Anaerobe 2014, 26, 49–52. [Google Scholar] [CrossRef]
- Chang, K.C.; Chung, C.Y.; Yeh, C.H.; Hsu, K.H.; Chin, Y.C.; Huan, S.S. Direct detection of carbapenemase-associated proteins of Acinetobacter baumannii using nanodiamonds coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Microbiol. Methods. 2018, 147, 36–42. [Google Scholar] [CrossRef]
- Alqasim, A.; Jaffal, A.A.; Alyousef, A.A. Prevalence and molecular characterization of sequence type 131 clone among clinical uropathogenic Escherichia coli isolates in Riyadh, Saudi Arabia. Saudi J. Biol. Sci. 2020, 27, 296–302. [Google Scholar] [CrossRef]
- Yasir, M.; Ajlan, A.M.; Shakil, S.; Jiman-Fatani, A.A.; Almasaudi, S.B.; Farman, M.; Baazeem, Z.M.; Baabdullah, R.; Alawi, M.; Al-Abdullah, N.; et al. Molecular characterization, antimicrobial resistance and clinic-bioinformatics approaches to address the problem of extended-spectrum β-lactamase-producing Escherichia coli in western Saudi Arabia. Sci. Rep. 2018, 8, 14847. [Google Scholar] [CrossRef]
| S. No. | Isolate ID | MALDI Results | Source | MALDI Score | Nitrofurantoine | Cotrimoxazole | Tetracycline | Erythromycin | Amikacin | Gentamicin | Tobramycin |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | B-1 | E. coli | Urine | 2.26 | S (26) | S (31) | S (24) | R | S (24) | S (27) | S (32) |
| 2 | B-15 | E. coli | Urine | 1.9 | S (21) | R | R | R | S (21) | I (14) | S (15) |
| 3 | B-16 | E. coli | Urine | 1.56 | S (23) | S (30) | R | R | S (25) | S (26) | S (19) |
| 4 | B-3 | E. coli | Urine | 2 | S (31) | R | R | R | S (23) | S (25) | S (25) |
| 5 | B-5 | E. coli | Urine | 2.1 | S (31) | R | R | R | S (24) | I (14) | S (17) |
| 6 | H-1 | E. coli | Urine | 1.89 | R | R | S (19) | R | S (21) | S (26) | S (27) |
| 7 | H-10 | Enterococcus faecalis | Urine | 1.9 | S (31) | S (31) | R | S (30) | R | S (26) | S (24) |
| 8 | H-4 | E. coli | Urine | 1.87 | S (26) | R | R | R | S (24) | S (25) | S (25) |
| 9 | H-5 | Klebsiella pneumoniae | Urine | 1.77 | S (26) | S (34) | S (25) | R | S (31) | S (29) | R |
| 10 | H-6 | E. coli | Urine | 1.41 | S (21) | R | R | R | S (21) | S (21) | S (21) |
| 11 | H-7 | Klebsiella pneumoniae | Urine | 1.5 | S (24) | S (31) | S (17) | R | S (27) | S (27) | S (27) |
| 12 | H-8 | E. coli | Urine | 1.4 | S (27) | S (35) | R | R | S (29) | S (27) | S (27) |
| 13 | H-9 | E. coli | Urine | 2 | S (27) | R | R | R | S (26) | S (26) | S (26) |
| 14 | R-5 | Aeromonas caviae | Urine | 2.15 | S (25) | S (25) | S (17) | R | S (26) | S (27) | S (26) |
| 15 | R-8 | E. coli | Urine | 1.89 | S (25) | S (27) | S (17) | R | S (24) | S (25) | S (26) |
| 16 | R-10 | E. coli | Urine | 1.63 | S (27) | R | S (20) | R | S (25) | S (25) | S (26) |
| 17 | S-3 | Klebsiella pneumoniae | Urine | 1.4 | S (18) | S (28) | S (20) | R | S (24) | S (26) | S (26) |
| 18 | S-4 | E. coli | Urine | 1.69 | S (24) | S (28) | S (17) | R | S (25) | S (26) | S (25) |
| S. No. | E. coli ID | MLST Allele Profiles of Each Gene | ST | ||||||
|---|---|---|---|---|---|---|---|---|---|
| adk | fumC | gyrB | icd | mdh | purA | recA | |||
| 1 | B1 | 6 | 6 | 15 | 10 | 9 | 7 | 14 | 2033 |
| 2 | B15 | 6 | 6 | 15 | 10 | 9 | 7 | 14 | 2033 |
| 3 | B16 | 6 | 4 | 4 | 16 | 24 | 5 | 14 | 1126 |
| 4 | B3 | 6 | 4 | 14 | 16 | 24 | 13 | 14 | 1015 |
| 5 | B5 | 6 | 4 | 14 | 16 | 24 | 13 | 14 | 1015 |
| 6 | H1 | 6 | 4 | 5 | 18 | 11 | 8 | 14 | 533 |
| 7 | H4 | 6 | 23 | 3 | 26 | 9 | 7 | 7 | 129 |
| 8 | H6 | 6 | 23 | 3 | 26 | 9 | 7 | 7 | 129 |
| 9 | H8 | 6 | 23 | 3 | 26 | 9 | 7 | 7 | 129 |
| 10 | H9 | 6 | 65 | 4 | 18 | 24 | 8 | 14 | 1432 |
| 11 | R8 | 6 | 4 | 4 | 18 | 176 | 8 | 14 | 1493 |
| 12 | R10 | 6 | 6 | 15 | 56 | 8 | 26 | 6 | 1987 |
| 13 | S4 | 6 | 11 | 4 | 16 | 11 | 7 | 2 | 1773 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anwer, R.; Darami, H.; Almarri, F.K.; Albogami, M.A.; Alahaydib, F. MALDI-TOF MS for Rapid Analysis of Bacterial Pathogens Causing Urinary Tract Infections in the Riyadh Region. Diseases 2022, 10, 78. https://doi.org/10.3390/diseases10040078
Anwer R, Darami H, Almarri FK, Albogami MA, Alahaydib F. MALDI-TOF MS for Rapid Analysis of Bacterial Pathogens Causing Urinary Tract Infections in the Riyadh Region. Diseases. 2022; 10(4):78. https://doi.org/10.3390/diseases10040078
Chicago/Turabian StyleAnwer, Razique, Hassan Darami, Firas K. Almarri, Mazen A. Albogami, and Faisal Alahaydib. 2022. "MALDI-TOF MS for Rapid Analysis of Bacterial Pathogens Causing Urinary Tract Infections in the Riyadh Region" Diseases 10, no. 4: 78. https://doi.org/10.3390/diseases10040078
APA StyleAnwer, R., Darami, H., Almarri, F. K., Albogami, M. A., & Alahaydib, F. (2022). MALDI-TOF MS for Rapid Analysis of Bacterial Pathogens Causing Urinary Tract Infections in the Riyadh Region. Diseases, 10(4), 78. https://doi.org/10.3390/diseases10040078








