The Use of Time-Frequency Moments as Inputs of LSTM Network for ECG Signal Classification
Abstract
:1. Introduction
1.1. Related Work in Machine Learning
1.2. Related Work in Reference to LSTM
1.3. Objective and Contribution
2. Materials and Methods
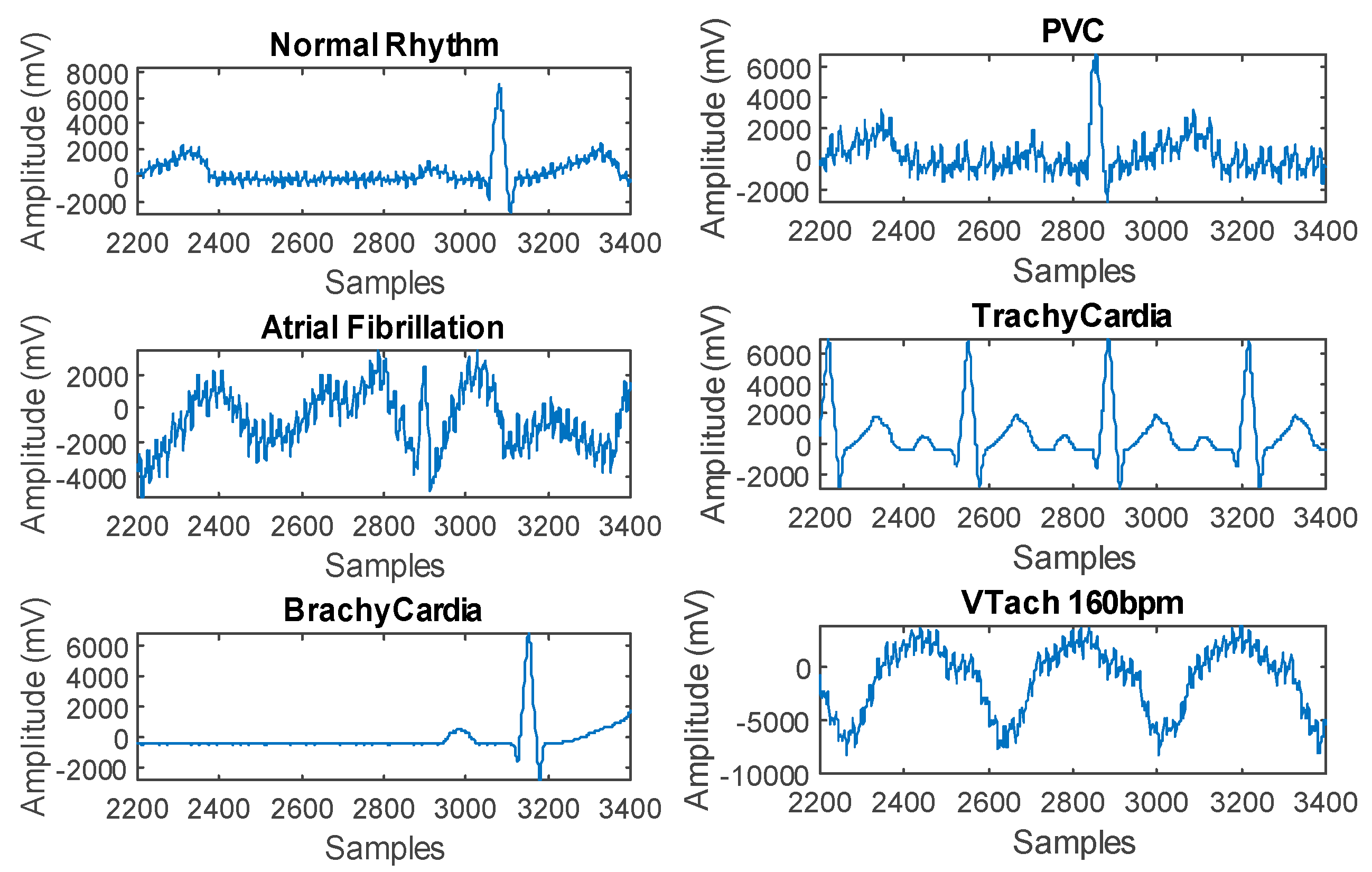
2.1. Problem Description
2.2. Dataset Acquisition
2.3. LSTM Architecture
2.4. Classification of Raw ECG Signal
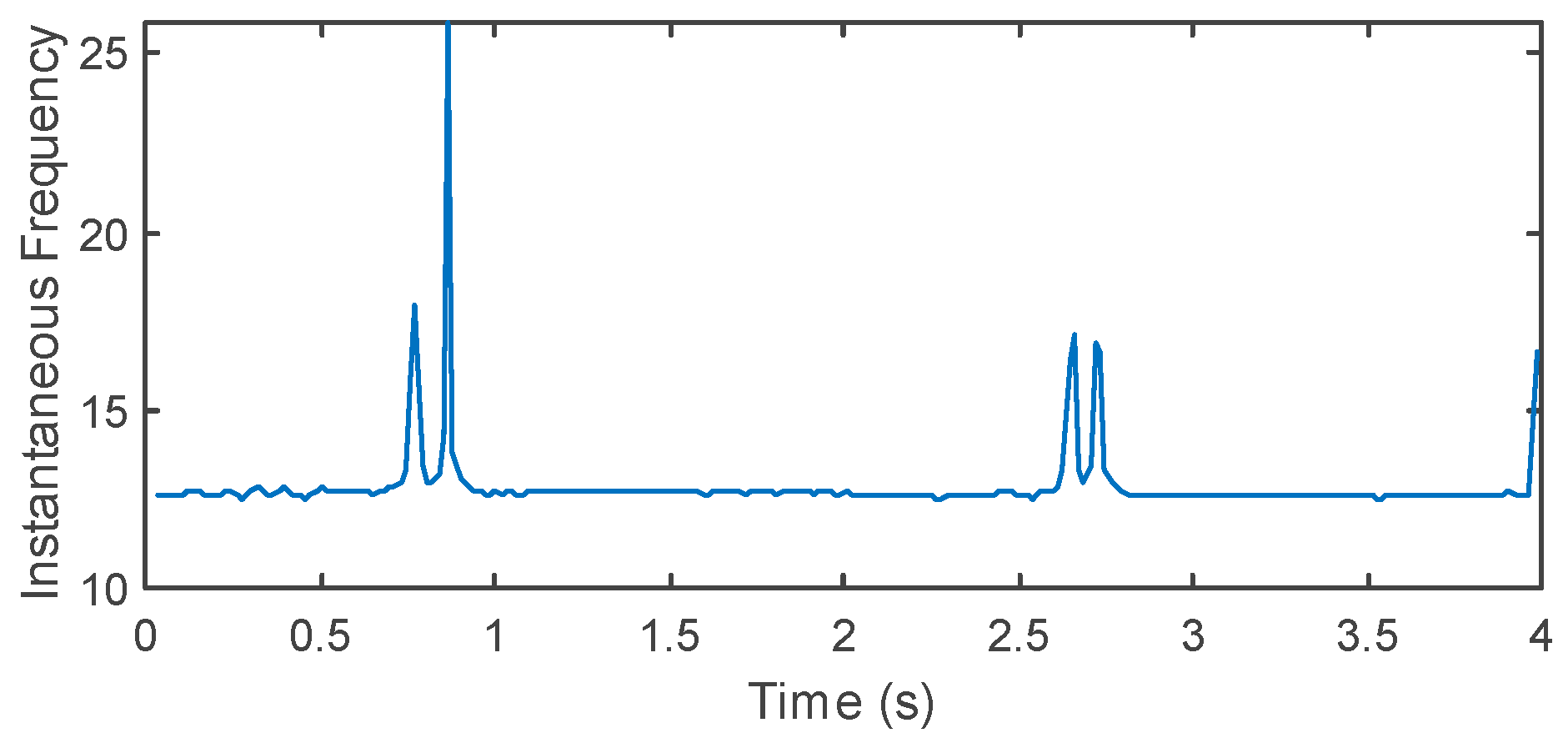
2.5. Spectral Feature Extraction
3. Results
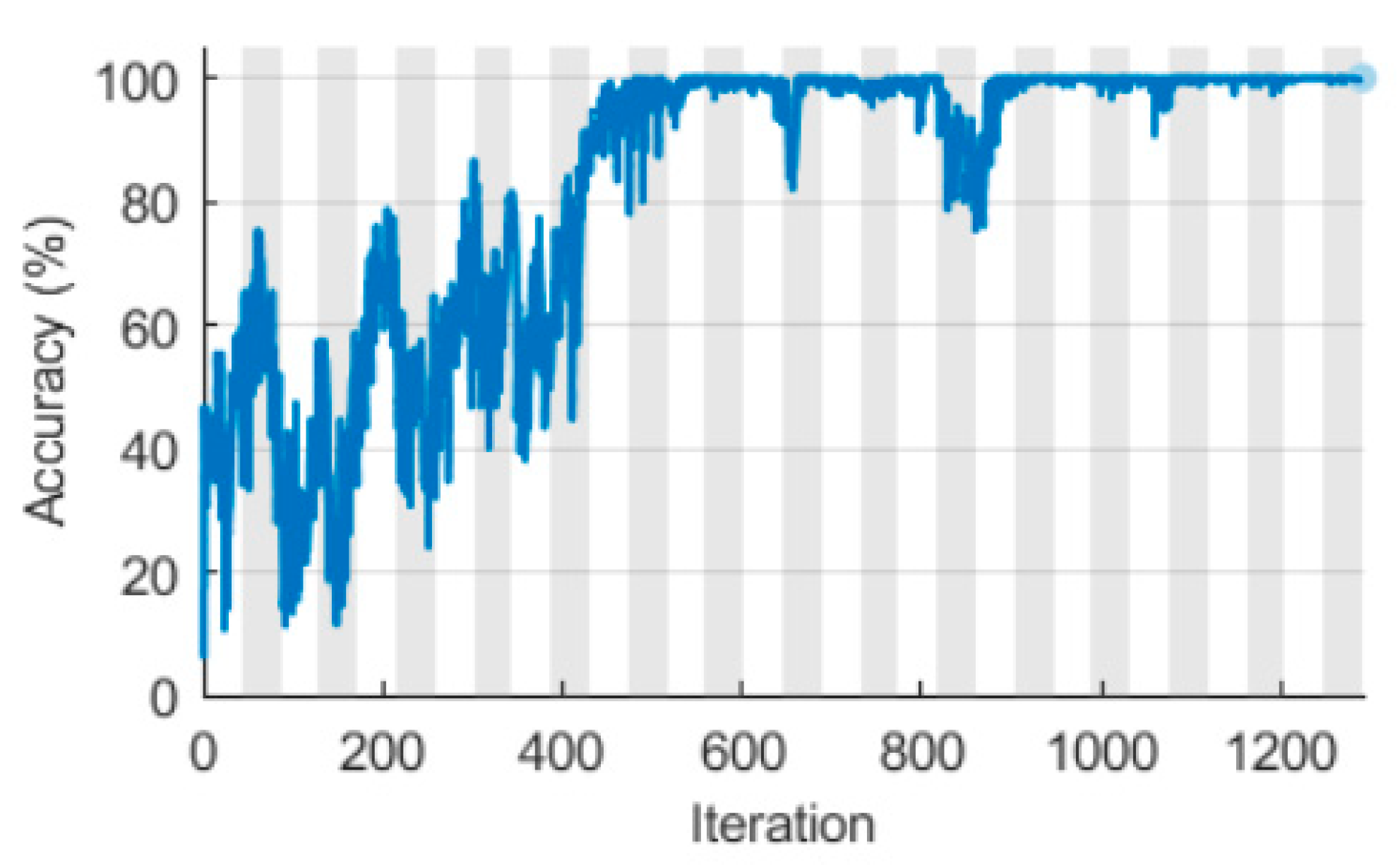
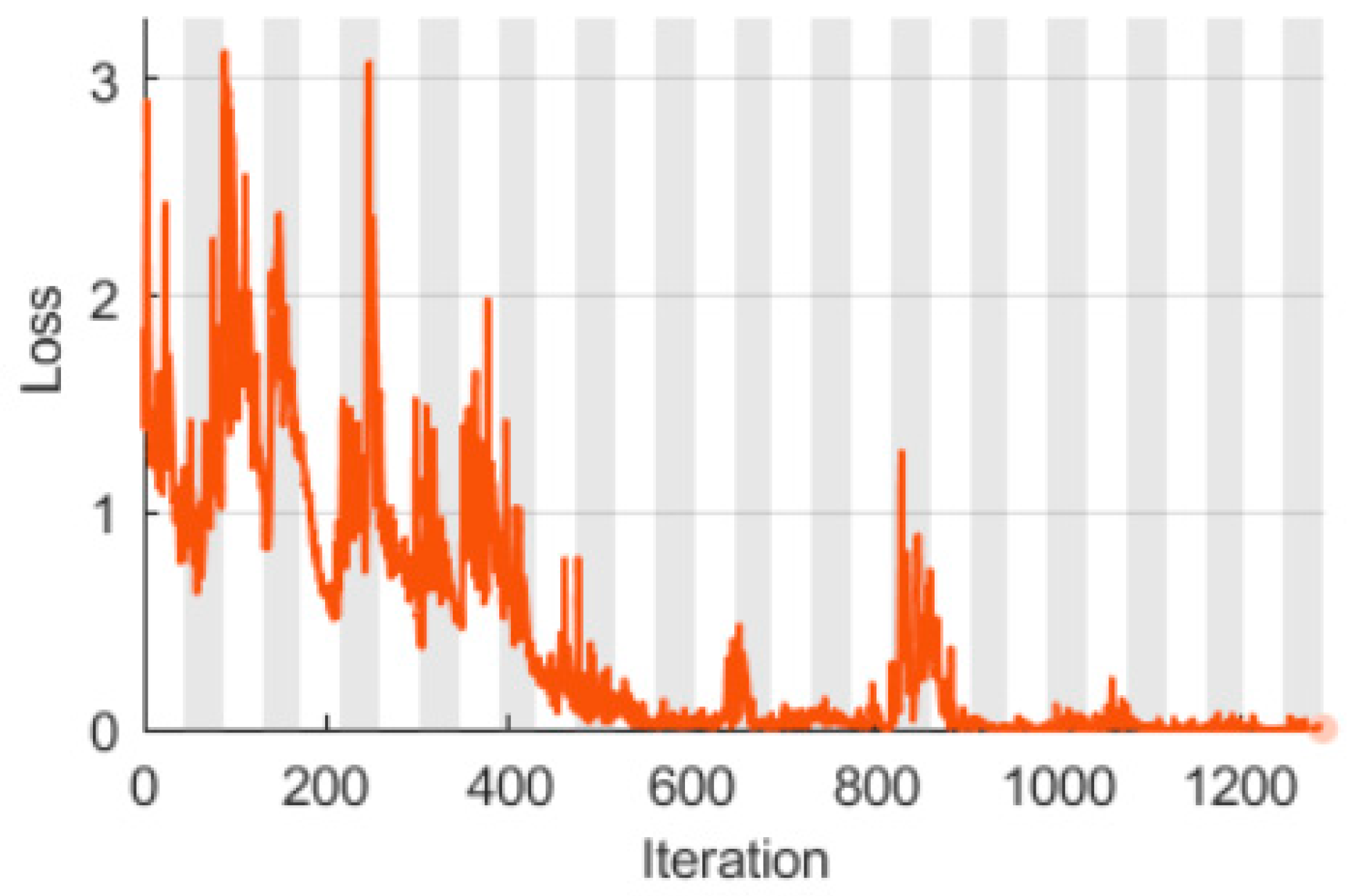
3.1. LSTM with a Singular Raw ECG Signal Input
3.2. LSTM with Double Spectral Input Features
3.3. Model Validation in Real Conditions
4. Discussion
5. Conclusions
- Very high effectiveness—100% accuracy was obtained on the testing set,
- It is possible to classify up to six categories of signals (five diseases and one for a healthy heart),
- Combination of the advantages of convolution neural networks for image classification and recursive networks having implemented memory mechanisms,
- Relatively low level of complexity—a single layer of BiLSTM with 200 hidden units was enough,
- Increase in the number of input signals from one raw ECG to two spectral inputs (TF moments)—instantaneous frequency (IF) and spectral entropy (SE),
- Converting raw ECG to two TF moments has reduced the amount of data needed to train the neural network. Thanks to this, the network learns not only more effectively but several times faster.
Author Contributions
Funding
Conflicts of Interest
References
- Heron, M. Deaths: Leading Causes for 2017. Natl. Vital Stat. Rep. 2019, 68, 1–77. [Google Scholar]
- Nicols, H.; Tavella, V.J. What Are the Leading Causes of Death in the US. Available online: www.medicalnewstoday.com/articles/282929 (accessed on 31 August 2020).
- Qiu, H.; Qiu, M.; Lu, Z. Selective encryption on ECG data in body sensor network based on supervised machine learning. Inf. Fus. 2020, 55, 59–67. [Google Scholar] [CrossRef]
- Kosinski, T.; Obaid, M.; Wozniak, P.W.; Fjeld, M.; Kucharski, J. A fuzzy data-based model for Human-Robot Proxemics. In Proceedings of the 25th IEEE International Symposium on Robot and Human Interactive Communication, RO-MAN 2016, New York, NY, USA, 26–31 August 2016; pp. 335–340. [Google Scholar]
- Rymarczyk, T.; Niderla, K.; Kozłowski, E.; Kłosowski, G.; Tchórzewski, P. The concept of the technological process control using a distributed industrial tomography system. Prz. Elektrotechniczny 2018, 94, 166–169. [Google Scholar] [CrossRef] [Green Version]
- Romanowski, A. Big Data-Driven Contextual Processing Methods for Electrical Capacitance Tomography. IEEE Trans. Ind. Inform. 2019, 15, 1609–1618. [Google Scholar] [CrossRef]
- Romanowski, A.; Łuczak, P.; Grudzień, K. X-ray Imaging Analysis of Silo Flow Parameters Based on Trace Particles Using Targeted Crowdsourcing. Sensors 2019, 19, 3317. [Google Scholar] [CrossRef] [Green Version]
- Fraczyk, A.; Kucharski, J. Surface temperature control of a rotating cylinder heated by moving inductors. Appl. Therm. Eng. 2017, 125, 767–779. [Google Scholar] [CrossRef]
- Kłosowski, G.; Rymarczyk, T.; Gola, A. Increasing the Reliability of Flood Embankments with Neural Imaging Method. Appl. Sci. 2018, 8, 1457. [Google Scholar] [CrossRef] [Green Version]
- Attia, Z.I.; Noseworthy, P.A.; Lopez-Jimenez, F.; Asirvatham, S.J.; Deshmukh, A.J.; Gersh, B.J.; Carter, R.E.; Yao, X.; Rabinstein, A.A.; Erickson, B.J.; et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: A retrospective analysis of outcome prediction. Lancet 2019, 394, 861–867. [Google Scholar] [CrossRef]
- Hasan, N.I.; Bhattacharjee, A. Deep Learning Approach to Cardiovascular Disease Classification Employing Modified ECG Signal from Empirical Mode Decomposition. Biomed. Signal Process. Control 2019, 52, 128–140. [Google Scholar] [CrossRef]
- Rahman, T.M.; Siddiqua, S.; Rabby, S.E.; Hasan, N.; Imam, M.H. Early detection of kidney disease using ECG signals through machine learning based modelling. In Proceedings of the 1st International Conference on Robotics, Electrical and Signal Processing Techniques (ICREST 2019), Dhaka, Bangladesh, 10–12 January 2019; pp. 319–323. [Google Scholar]
- Kim, M.G.; Pan, S.B. Deep Learning Based on 1-D Ensemble Networks Using ECG for Real-Time User Recognition. IEEE Trans. Ind. Inform. 2019, 15, 5656–5663. [Google Scholar] [CrossRef]
- Zhao, P.; Quan, D.; Yu, W.; Yang, X.; Fu, X. Towards deep learning-based detection scheme with raw ECG signal for wearable telehealth systems. In Proceedings of the Proceedings—International Conference on Computer Communications and Networks (ICCCN), Valencia, Spain, 29 July–1 August 2019. [Google Scholar]
- Zhao, W.; Hu, J.; Jia, D.; Wang, H.; Li, Z.; Yan, C.; You, T. Deep Learning Based Patient-Specific Classification of Arrhythmia on ECG signal. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBS), Berlin, Germany, 23–27 July 2019; pp. 1500–1503. [Google Scholar]
- Isasi, I.; Irusta, U.; Elola, A.; Aramendi, E.; Eftestol, T.; Kramer-Johansen, J.; Wik, L. A Robust Machine Learning Architecture for a Reliable ECG Rhythm Analysis during CPR. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBS), Berlin, Germany, 23–27 July 2019; pp. 1903–1907. [Google Scholar]
- Janghel, R.R.; Pandey, S. kumar Classification and Detection of Arrhythmia in ECG Signal Using Machine Learning Techniques. In Proceedings of the 2019 16th International Conference on Electrical Engineering/Electronics, Computer, Telecommunications and Information Technology (ECTI-CON), Pattaya, Thailand, 10–13 July 2019; pp. 101–104. [Google Scholar]
- Abdeldayem, S.S.; Bourlai, T. A Novel Approach for ECG-based Human Identification using Spectral Correlation and Deep Learning. IEEE Trans. Biom. Behav. Identity Sci. 2019, 2, 1–14. [Google Scholar] [CrossRef]
- Yıldırım, Ö.; Pławiak, P.; Tan, R.S.; Acharya, U.R. Arrhythmia detection using deep convolutional neural network with long duration ECG signals. Comput. Biol. Med. 2018, 102, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Izci, E.; Ozdemir, M.A.; Degirmenci, M.; Akan, A. Cardiac arrhythmia detection from 2d ecg images by using deep learning technique. In Proceedings of the TIPTEKNO 2019—Tip Teknolojileri Kongresi, Izmir, Turkey, 3–5 October 2019. [Google Scholar]
- Hermawan, I.; Anwar Ma’sum, M.A.; Riskyana Dewi Intan, P.; Jatmiko, W.; Wiweko, B.; Boediman, A.; Pradekso, B.K. Temporal feature and heuristics-based Noise Detection over Classical Machine Learning for ECG Signal Quality Assessment. In Proceedings of the 2019 International Workshop on Big Data and Information Security (IWBIS 2019), Nusa Dua, Indonesia, 11 October 2019; pp. 1–8. [Google Scholar]
- Porumb, M.; Stranges, S.; Pescapè, A.; Pecchia, L. Precision Medicine and Artificial Intelligence: A Pilot Study on Deep Learning for Hypoglycemic Events Detection based on ECG. Sci. Rep. 2020, 10, 170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheen, Y.T. 3D spectral analysis for vibration signals by wavelet-based demodulation. Mech. Syst. Signal Process. 2006, 20, 843–853. [Google Scholar] [CrossRef]
- Diker, A.; Avci, D.; Avci, E.; Gedikpinar, M. A new technique for ECG signal classification genetic algorithm Wavelet Kernel extreme learning machine. Optik 2019, 180, 46–55. [Google Scholar] [CrossRef]
- Lynn, H.M.; Pan, S.B.; Kim, P. A Deep Bidirectional GRU Network Model for Biometric Electrocardiogram Classification Based on Recurrent Neural Networks. IEEE Access 2019, 7, 145395–145405. [Google Scholar] [CrossRef]
- Salem, M.; Taheri, S.; Yuan, J.S. ECG Arrhythmia Classification Using Transfer Learning from 2- Dimensional Deep CNN Features. In Proceedings of the 2018 IEEE Biomedical Circuits and Systems Conference, BioCAS 2018—Proceedings, Cleveland, OH, USA, 17–19 October 2018. [Google Scholar]
- Zhu, H.; Cheng, C.; Yin, H.; Li, X.; Zuo, P.; Ding, J.; Lin, F.; Wang, J.; Zhou, B.; Li, Y.; et al. Automatic multilabel electrocardiogram diagnosis of heart rhythm or conduction abnormalities with deep learning: A cohort study. Lancet Digital Health 2020, 2, e348–e357. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Werth, J.; Radha, M.; Andriessen, P.; Aarts, R.M.; Long, X. Deep learning approach for ECG-based automatic sleep state classification in preterm infants. Biomed. Signal Process. Control 2020, 56, 101663. [Google Scholar] [CrossRef]
- Yildirim, Ö. A novel wavelet sequences based on deep bidirectional LSTM network model for ECG signal classification. Comput. Biol. Med. 2018, 96, 189–202. [Google Scholar] [CrossRef]
- Chen, Y.J.; Liu, C.L.; Tseng, V.S.; Hu, Y.F.; Chen, S.A. Large-scale classification of 12-lead ECG with deep learning. In Proceedings of the 2019 IEEE EMBS International Conference on Biomedical and Health Informatics, BHI 2019—Proceedings, Chicago, IL, USA, 19–22 May 2019. [Google Scholar]
- Zihlmann, M.; Perekrestenko, D.; Tschannen, M. Convolutional recurrent neural networks for electrocardiogram classification. In Proceedings of the Computing in Cardiology, Rennes, France, 24–27 September 2017; Volume 44, pp. 1–4. [Google Scholar]
- Attia, Z.I.; Kapa, S.; Lopez-Jimenez, F.; McKie, P.M.; Ladewig, D.J.; Satam, G.; Pellikka, P.A.; Enriquez-Sarano, M.; Noseworthy, P.A.; Munger, T.M.; et al. Screening for cardiac contractile dysfunction using an artificial intelligence–enabled electrocardiogram. Nat. Med. 2019, 25, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Berger, R.D.; Kasper, E.K.; Baughman, K.L.; Marban, E.; Calkins, H.; Tomaselli, G.F. Beat-to-beat QT interval variability: Novel evidence for repolarization lability in ischemic and nonischemic dilated cardiomyopathy. Circulation 1997, 96, 1557–1565. [Google Scholar] [CrossRef]
- Moskalenko, V.; Zolotykh, N.; Osipov, G. Deep learning for ECG segmentation. In Proceedings of the Studies in Computational Intelligence; Springer: Berlin/Heidelberg, Germany, 2020; Volume 856, pp. 246–254. [Google Scholar]
- Jeong, Y.-S.; Kang, A.R.; Jung, W.; Lee, S.J.; Lee, S.; Lee, M.; Chung, Y.H.; Koo, B.S.; Kim, S.H. Prediction of Blood Pressure after Induction of Anesthesia Using Deep Learning: A Feasibility Study. Appl. Sci. 2019, 9, 5135. [Google Scholar] [CrossRef] [Green Version]
- Arsene, C.T.C.; Hankins, R.; Yin, H. Deep learning models for denoising ECG signals. In Proceedings of the European Signal Processing Conference; European Signal Processing Conference, EUSIPCO, Coruña, Spain, 2–6 September 2019; Volume 2019. [Google Scholar]
- Ramya, E.; Prabha, R.; Jayageetha, J.; Keerthana, M.; Swetha, S.; Lakshmi, N. Envisaging Ventricular Arrhythmia from an ECG by Using Machine learning algorithm. In Proceedings of the 2019 5th International Conference on Advanced Computing and Communication Systems (ICACCS 2019), Tamil Nadu, India, 15–16 December 2019; pp. 991–994. [Google Scholar]
- Mostayed, A.; Luo, J.; Shu, X.; Wee, W. Classification of 12-Lead ECG Signals with Bi-Directional LSTM Network. Available online: http://arxiv.org/abs/1811.02090 (accessed on 2 February 2020).
- Saadatnejad, S.; Oveisi, M.; Hashemi, M. LSTM-Based ECG Classification for Continuous Monitoring on Personal Wearable Devices. IEEE J. Biomed. Heal. Inform. 2019, 24, 515–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Zhang, H.; Lu, P.; Wang, Z. An Effective LSTM Recurrent Network to Detect Arrhythmia on Imbalanced ECG Dataset. J. Healthc. Eng. 2019, 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, D.; Agarwal, S. Cardiac Arrhythmia Detection from Single-lead ECG using CNN and LSTM assisted by Oversampling. In Proceedings of the 2018 International Conference on Advances in Computing, Communications and Informatics (ICACCI 2018), Bangalore, India, 19–22 September 2018; pp. 14–17. [Google Scholar]
- Chang, Y.C.; Wu, S.H.; Tseng, L.M.; Chao, H.L.; Ko, C.H. AF Detection by Exploiting the Spectral and Temporal Characteristics of ECG Signals with the LSTM Model. In Proceedings of the Computing in Cardiology, Maastricht, Netherlands, 23–26 September 2018. [Google Scholar]
- Yuen, B.; Dong, X.; Lu, T. Inter-Patient CNN-LSTM for QRS Complex Detection in Noisy ECG Signals. IEEE Access 2019, 7, 169359–169370. [Google Scholar] [CrossRef]
- Beale, M.H.; Hagan, M.T.; Demuth, H.B. Deep Learning Toolbox User’s Guide; The Mathworks Inc.: Herborn, MA, USA, 2018. [Google Scholar]
- Glorot, X.; Yoshua, B. Understanding the difficulty of training deep feedfor-ward neural networks. In Proceedings of the Thirteenth International Conference on artificial intelligence and statistics, Sardinia, Italy, 13 May 2010; pp. 249–256. [Google Scholar]
- Kingma, D.P.; Ba, J.L. Adam: A method for stochastic optimization. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings. International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Mathworks. Signal Processing Toolbox User’s Guide; Mathworks Inc.: Natick, MA, USA, 2019. [Google Scholar]
- Sharma, V.; Parey, A. A Review of Gear Fault Diagnosis Using Various Condition Indicators. Procedia Engineering; Elsevier Ltd.: Amsterdam, The Netherlands, 2016; Volume 144, pp. 253–263. [Google Scholar]
- Kłosowski, G.; Rymarczyk, T.; Kania, K.; Świć, A.; Cieplak, T. Maintenance of industrial reactors supported by deep learning driven ultrasound tomography. Eksploat. i Niezawodn. Maint. Reliab. 2020, 22, 138–147. [Google Scholar] [CrossRef]
- Korzeniewska, E.; Krawczyk, A.; Mróz, J.; Wyszyńska, E.; Zawiślak, R. Applications of smart textiles in post-stroke rehabilitation. Sensors 2020, 20, 2370. [Google Scholar] [CrossRef] [Green Version]
- Korzeniewska, E.; Krawczyk, A. Applications of smart textiles in electromedicine. In Proceedings of the 2019 19th International Symposium on Electromagnetic Fields in Mechatronics, Electrical and Electronic Engineering (ISEF 2019), Nancy, France, 29–31 August 2019; Institute of Electrical and Electronics Engineers Inc.: Nancy, France, 2019; pp. 1–2. [Google Scholar]
- Rymarczyk, T.; Kłosowski, G. Innovative methods of neural reconstruction for tomographic images in maintenance of tank industrial reactors. Eksploat. i Niezawodn. Maint. Reliab. 2019, 21, 261–267. [Google Scholar] [CrossRef]
- Romanowski, A. Contextual Processing of Electrical Capacitance Tomography Measurement Data for Temporal Modeling of Pneumatic Conveying Process. In Proceedings of the 2018 Federated Conference on Computer Science and Information Systems (FedCSIS), Poznań, Poland, 9–12 September 2018; pp. 283–286. [Google Scholar]
- Wajman, R.; Fiderek, P.; Fidos, H.; Jaworski, T.; Nowakowski, J.; Sankowski, D.; Banasiak, R. Metrological evaluation of a 3D electrical capacitance tomography measurement system for two-phase flow fraction determination. Meas. Sci. Technol. 2013, 24, 065302. [Google Scholar] [CrossRef]
- Kozłowski, E.; Mazurkiewicz, D.; Kowalska, B.; Kowalski, D. Binary Linear Programming as a Decision-Making Aid for Water Intake Operators; Springer: Cham, Switzerland, 2018; pp. 199–208. [Google Scholar]
- Vališ, D.; Mazurkiewicz, D. Application of selected Levy processes for degradation modelling of long range mine belt using real-time data. Arch. Civ. Mech. Eng. 2018, 18, 1430–1440. [Google Scholar] [CrossRef]















| Authors | Feature Extraction Methods | Classification Models | Maximum Accuracy Obtained (%) |
|---|---|---|---|
| Chen et al., 2019 [31] | No feature extraction - raw data | Convolutional Neural Network + Long Short-Term Memory (LSTM) | 81.0 |
| Zihlmann, Perekrestenko, and Tschannen, 2017 [32] | Logarithmic Transform | Long Short Term Memory | 82.3 |
| Attia et al., 2019 [33] | No feature extraction - raw data | Convolutional Neural Network | 83.3 |
| P. Zhao et al., 2019 [14] | Wavelet transforms using a db3 wavelet filter | Convolutional Neural Network | 87.8 |
| Diker et al., 2019 [24] | Morphological and statistical features | Extreme Learning Machine | 95 |
| Qiu, Qiu and Lu, 2020 [3] | Auto-Regressive model coefficients, Shannon Entropy values, and Multi- Fractal Wavelet Leader Estimation | Support Vector Machine | 96.3 |
| Isasi et al., 2019 [16] | Stationary Wavelet Transform | Artificial Neural Network, Support Vector Machine, Kernel Logistic Regression, Boosting of Decision Trees | 96.7 |
| Salem, Taheri, and Yuan, 2018 [26] | Fourier Transform Spectrograms | Long Short Term Memory | 97.2 |
| Rahman et al., 2019 [12] | QT interval and the RR interval extracted using Berger’s algorithm [34] | Support Vector Machine | 97.6 |
| W. Zhao et al., 2019 [15] | Wavelet Transform and Independent Component Analysis | Convolutional Neural Network | 98.6 |
| Yildirim Özal, 2018 [30] | Daubechies dB6 wavelet member of the wavelet family | Long Short Term Memory | 99.4 |
| Hasan and Bhattacharjee, 2019 [11] | Empirical Mode Decomposition and higher order Intrinsic Mode Functions | Convolutional Neural Network | 99.7 |
| Moskalenko, Zolotykh, and Osipov, 2020 [35] | Cubic Spline | UNet-like full-convolutional neural network | 99.9 |
| Kim and Pan, 2019 [13] | Multi-Layer Perceptron (MLP) layer | Convolutional Neural Network, 1-D Ensemble Network | 100 |
| Abdeldayem and Bourlai, 2019 [18] | Cyclic Autocorrelation, Spectral Correlation | Convolutional Neural Network | 100 |
| Proposed Method | Instantaneous Frequency and Spectral Entropy | Long Short Term Memory | 100 |
| Cardiac Dysfunctions | The Original Number of Signals | The Number of Signals After Leveling | |
|---|---|---|---|
| Training | Testing | ||
| AFib | 600 | 1080 | 60 |
| Brachycardia | 60 | 1080 | 60 |
| Normal ECG | 1140 | 1080 | 60 |
| Premature ventricular contraction (PVC) | 600 | 1080 | 60 |
| Trachycardia | 121 | 1080 | 60 |
| VTach 160 bpm | 600 | 1080 | 60 |
| # | Layer Description | Activations | Learnable Parameters (Weights and Biases) |
|---|---|---|---|
| 1 | Sequence input with 1 dimension | 1 | - |
| 2 | BiLSTM with 200 hidden units | 400 | Input weights: 1600 × 1; Recurrent Weights: 1600 × 200; Bias: 1600 × 1 |
| 3 | Fully connected layer | 6 | Weights: 6 × 400; Bias: 6 × 1 |
| 4 | Softmax | 6 | - |
| 5 | Classification output (cross entropy) | - | - |
| # | Layer Description | Activations | Learnable Parameters (Weights and Biases) |
|---|---|---|---|
| 1 | Sequence input with two dimensions | 2 | - |
| 2 | BiLSTM with 200 hidden units | 400 | Input weights: 1600 × 2, Recurrent Weights: 1600 × 200, Bias: 1600 × 1 |
| 3 | Fully connected layer | 6 | Weights: 6 × 400, Bias: 6 × 1 |
| 4 | Softmax | 6 | - |
| 5 | Classification output (crossentropy) | - | - |
| ECG Signal Categories | Probability (%) | |
|---|---|---|
| Volunteer No. 1 | Volunteer No. 2 | |
| AFib | ||
| Brachycardia | ||
| Normal ECG | ||
| PVC | ||
| Trachycardia | ||
| VTach 160 bpm | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kłosowski, G.; Rymarczyk, T.; Wójcik, D.; Skowron, S.; Cieplak, T.; Adamkiewicz, P. The Use of Time-Frequency Moments as Inputs of LSTM Network for ECG Signal Classification. Electronics 2020, 9, 1452. https://doi.org/10.3390/electronics9091452
Kłosowski G, Rymarczyk T, Wójcik D, Skowron S, Cieplak T, Adamkiewicz P. The Use of Time-Frequency Moments as Inputs of LSTM Network for ECG Signal Classification. Electronics. 2020; 9(9):1452. https://doi.org/10.3390/electronics9091452
Chicago/Turabian StyleKłosowski, Grzegorz, Tomasz Rymarczyk, Dariusz Wójcik, Stanisław Skowron, Tomasz Cieplak, and Przemysław Adamkiewicz. 2020. "The Use of Time-Frequency Moments as Inputs of LSTM Network for ECG Signal Classification" Electronics 9, no. 9: 1452. https://doi.org/10.3390/electronics9091452
APA StyleKłosowski, G., Rymarczyk, T., Wójcik, D., Skowron, S., Cieplak, T., & Adamkiewicz, P. (2020). The Use of Time-Frequency Moments as Inputs of LSTM Network for ECG Signal Classification. Electronics, 9(9), 1452. https://doi.org/10.3390/electronics9091452






