Extended Segmented Beat Modulation Method for Cardiac Beat Classification and Electrocardiogram Denoising
Abstract
1. Introduction
2. Methodology
2.1. Preprocessing
2.2. Review of Segmented Beat Modulation Method
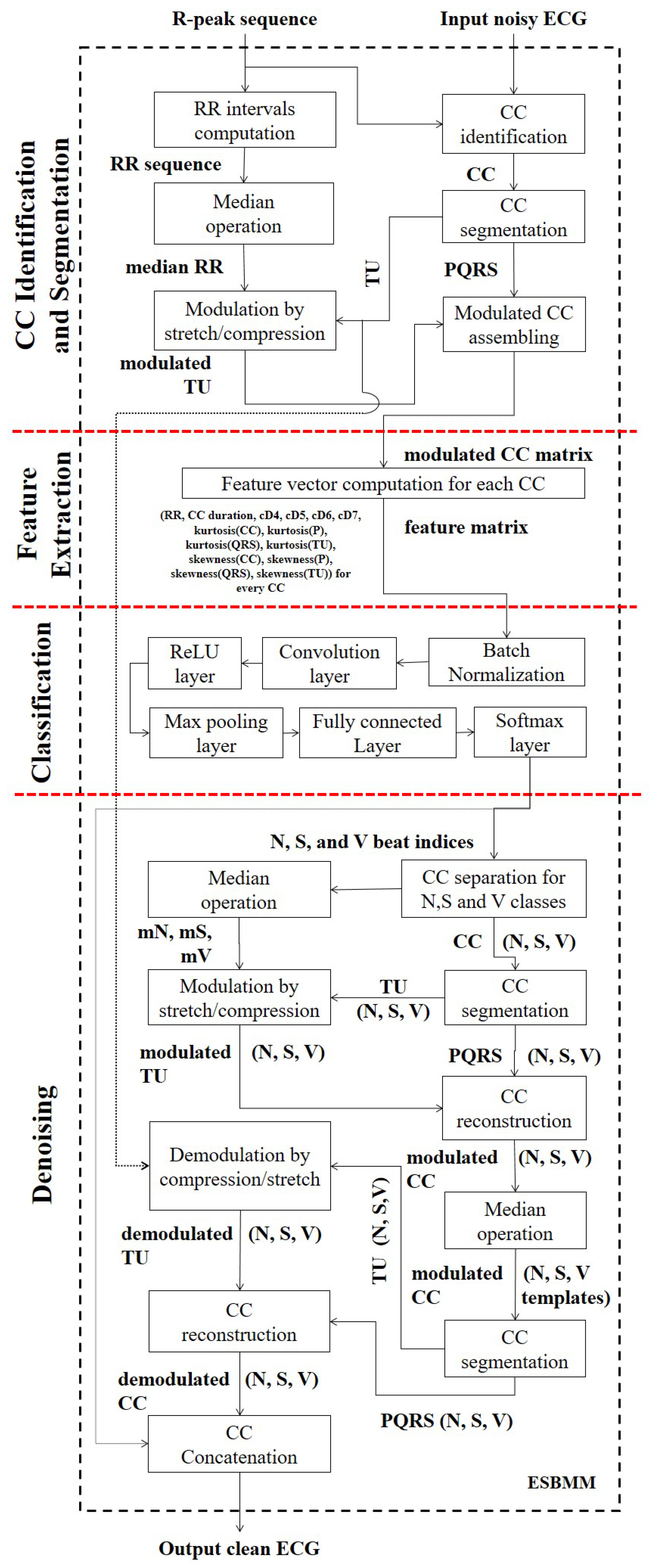
2.3. Extended Segmented Beat Modulation Method (ESBMM)
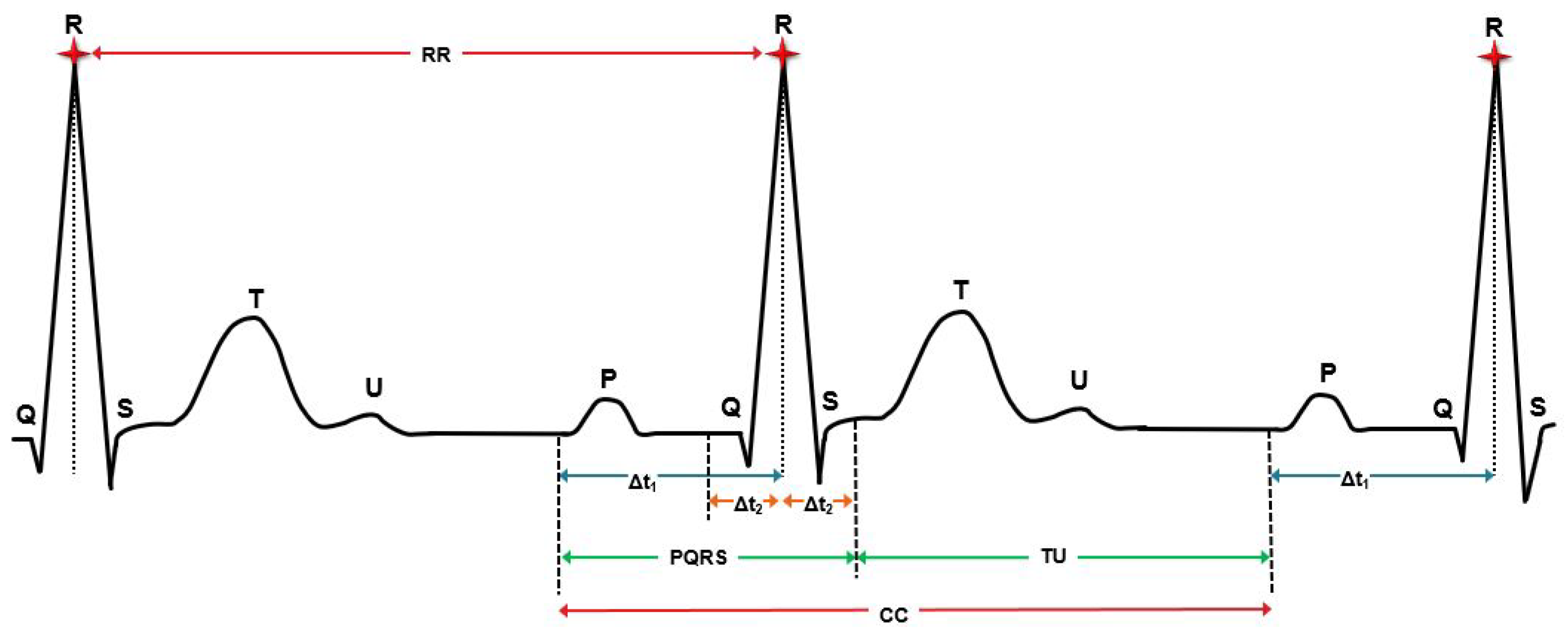
2.3.1. Cardiac Cycle Identification and Segmentation
2.3.2. Feature Extraction
2.3.3. Convolutional Neural Network Classification
2.3.4. Denoising
2.4. Data
2.5. Validation Study
2.5.1. Classification
2.5.2. Denoising
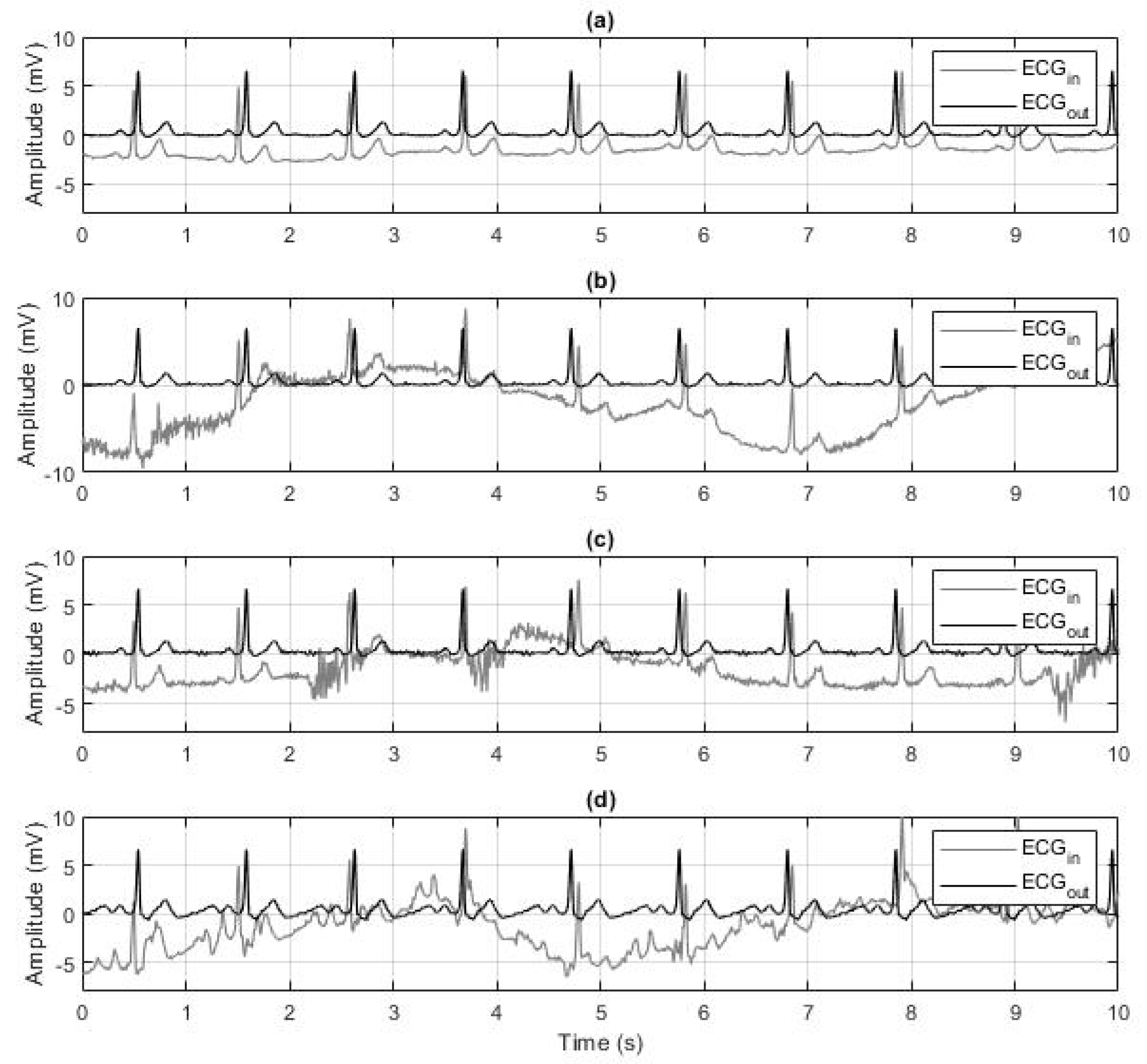
2.6. Robustness to Noise Evaluation
3. Results
3.1. Validation Study
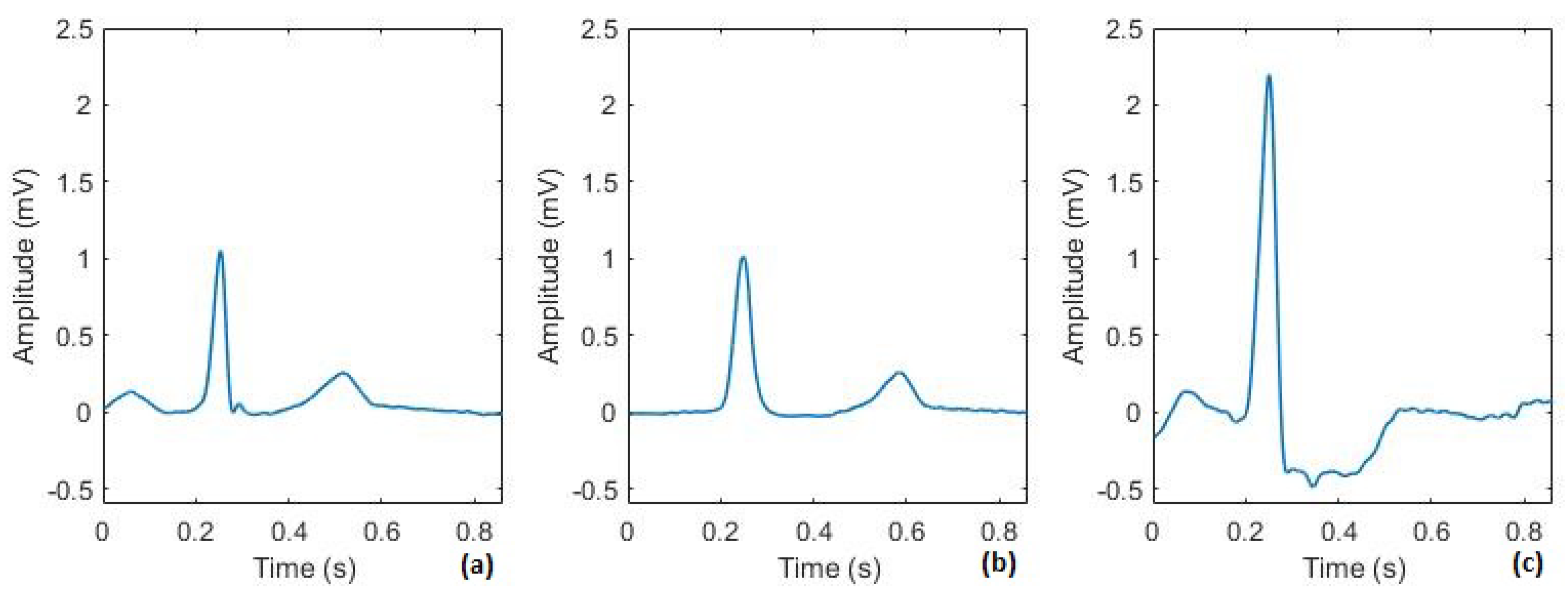
3.1.1. Classification
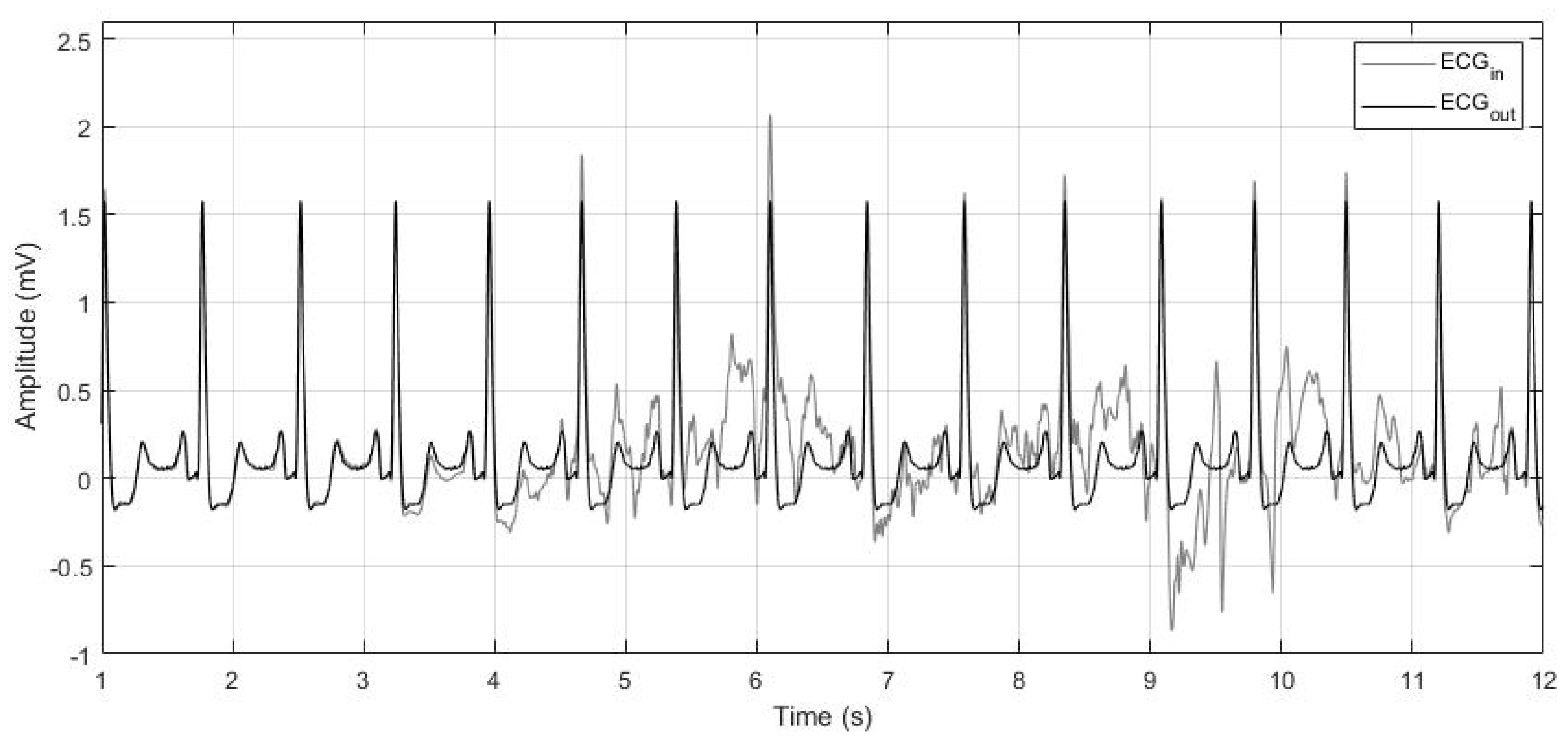
3.1.2. Denoising
3.2. Robustness to Noise Evaluation
4. Discussion
- A new ECG segmentation procedure that separates repolarization waveforms from depolarization waveforms;
- A proposed feature vector composed of spectral, RR interval, and higher-order statistical features;
- A convolutional neural network to classify cardiac beats into N, S, and V classes;
- A denoising algorithm designed to separately construct median templates for N, S, and V beats and reconstruct the original ECG recording including arrhythmic beats to match the original beat duration and morphology.
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AAMI | Association for the Advancement of Medical Instrumentation |
| ANSI | American National Standards Institute |
| CC | Cardiac Cycle |
| CNN | Convolutional Neural Network |
| ECG | Electrocardiogram |
| ESBMM | Extended Segmented Beat Modulation Method |
| N | Normal sinus beat |
| S | Supraventricular beat |
| SBMM | Segmented Beat Modulation Method |
| V | Ventricular beat |
References
- Meier, T.; Gräfe, K.; Senn, F.; Sur, P.; Stangl, G.I.; Dawczynski, C.; März, W.; Kleber, M.E.; Lorkowski, S. Cardiovascular mortality attributable to dietary risk factors in 51 countries in the WHO European Region from 1990 to 2016: A systematic analysis of the Global Burden of Disease Study. Eur. J. Epidemiol. 2019, 34, 37–55. [Google Scholar] [CrossRef] [PubMed]
- Naghavi, M.; Abajobir, A.A.; Abbafati, C.; Abbas, K.M.; Abd-Allah, F.; Abera, S.F.; Aboyans, V.; Adetokunboh, O.; Afshin, A.; Agrawal, A.; et al. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1151–1210. [Google Scholar] [CrossRef]
- Arrhythmia. Available online: https://www.nhlbi.nih.gov/health-topics/arrhythmia (accessed on 22 November 2019).
- Luz, E.J.D.S.; Schwartz, W.R.; Cámara-Chávez, G.; Menotti, D. ECG-based heartbeat classification for arrhythmia detection: A survey. Comput. Methods Programs Biomed. 2016, 127, 144–164. [Google Scholar] [CrossRef] [PubMed]
- Hoefman, E.; Bindels, P.J.; van Weert, H.C. Efficacy of diagnostic tools for detecting cardiac arrhythmias: Systematic literature search. Neth. Heart J. 2010, 18, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Kutlu, Y.; Kuntalp, D. A multi-stage automatic arrhythmia recognition and classification system. Comput. Biol. Med. 2011, 41, 37–45. [Google Scholar] [CrossRef]
- Sahab, A.; Gilmalek, Y.M. An automatic diagnostic machine for ECG arrhythmias classification based on wavelet transformation and neural networks. Int. J. Circuits Syst. Signal Process. 2011, 5, 255–262. [Google Scholar]
- Vijayavanan, M.; Rathikarani, V.; Dhanalakshmi, P. Automatic classification of ECG signal for heart disease diagnosis using morphological features. Int. J. Comput. Sci. Eng. Technol. 2014, 5, 449–455. [Google Scholar]
- Alickovic, E.; Subasi, A. Medical decision support system for diagnosis of heart arrhythmia using DWT and random forests classifier. J. Med. Syst. 2016, 40, 108. [Google Scholar] [CrossRef]
- Yıldırım, Ö.; Pławiak, P.; Tan, R.S.; Acharya, U.R. Arrhythmia detection using deep convolutional neural network with long duration ECG signals. Comput. Biol. Med. 2018, 102, 411–420. [Google Scholar] [CrossRef]
- Shang, Z.; Zhao, Y.; Lian, Y. Removal of Baseline Wander Noise in ECG Signal Using Asymmetrical Frequency-Response Masking Bandpass Filters. In Proceedings of the 40th Annual International Conference of Engineering in Medicine and Biology Society (EMBC 2018), Honolulu, HI, USA, 18–21 July 2018; pp. 6002–6005. [Google Scholar]
- Da Poian, G.; Rozell, C.J.; Bernardini, R.; Rinaldo, R.; Clifford, G.D. Matched filtering for heart rate estimation on compressive sensing ECG measurements. IEEE Trans. Biomed. Eng. 2017, 65, 1349–1358. [Google Scholar] [CrossRef]
- Goel, S.; Tomar, P.; Kaur, G. An optimal wavelet approach for ECG noise cancellation. Int. J. Bio-Sci. Bio-Technol. 2016, 8, 39–52. [Google Scholar] [CrossRef]
- Jenkal, W.; Latif, R.; Toumanari, A.; Dliou, A.; El B’charri, O.; Maoulainine, F.M. An efficient algorithm of ECG signal denoising using the adaptive dual threshold filter and the discrete wavelet transform. Biocybern. Biomed. Eng. 2016, 36, 499–508. [Google Scholar] [CrossRef]
- Duhan, M. ECG De-noising Techniques and Optimal Feature Selection Using Principle Component Analysis. J. Telecommun. Switch. Syst. Netw. 2018, 5, 14–20. [Google Scholar]
- Artoni, F.; Delorme, A.; Makeig, S. Applying dimension reduction to EEG data by Principal Component Analysis reduces the quality of its subsequent Independent Component decomposition. NeuroImage 2018, 175, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Xiong, P.; Wang, H.; Liu, M.; Liu, X. Denoising autoencoder for eletrocardiogram signal enhancement. J. Med. Imaging Health Inform. 2015, 5, 1804–1810. [Google Scholar] [CrossRef]
- Agostinelli, A.; Giuliani, C.; Burattini, L. Extracting a clean ECG from a noisy recording: A new method based on segmented-beat modulation. In Proceedings of the Computing in Cardiology Conference (CinC 2014), Cambridge, MA, USA, 7–10 September 2014; pp. 49–52. [Google Scholar]
- Agostinelli, A.; Sbrollini, A.; Giuliani, C.; Fioretti, S.; Di Nardo, F.; Burattini, L. Segmented beat modulation method for electrocardiogram estimation from noisy recordings. Med. Eng. Phys. 2016, 38, 560–568. [Google Scholar] [CrossRef] [PubMed]
- Agostinelli, A.; Sbrollini, A.; Burattini, L.; Fioretti, S.; Di Nardo, F.; Burattini, L. Noninvasive fetal electrocardiography part II: Segmented-Beat Modulation Method for signal denoising. Open Biomed. Eng. J. 2017, 11, 25–35. [Google Scholar] [CrossRef]
- Sbrollini, A.; Strazza, A.; Candelaresi, S.; Marcantoni, I.; Morettini, M.; Fioretti, S.; Di Nardo, F.; Burattini, L. Surface electromyography low-frequency content: Assessment in isometric conditions after electrocardiogram cancellation by the Segmented-Beat Modulation Method. Inform. Med. Unlocked 2018, 13, 71–80. [Google Scholar] [CrossRef]
- Agostinelli, A.; Giuliani, C.; Fioretti, S.; Di Nardo, F.; Burattini, L. Robustness of the segmented-beat modulation method to noise. In Proceedings of the 2015 Computing in Cardiology Conference (CinC), Nice, France, 6–9 September 2015; pp. 205–208. [Google Scholar]
- Nasim, A.; Della Santa, E.; Tanchi, D.; Sbrollini, A.; Marcantoni, I.; Morettini, M.; Burattini, L. GPU-Based Segmented-Beat Modulation Method for Denoising Athlete Electrocardiograms During Training. In Proceedings of the Computing in Cardiology Conference (CinC 2018), Maastricht, The Netherlands, 23–26 September 2018; Volume 45, pp. 1–4. [Google Scholar]
- Nasim, A.; Sbrollini, A.; Marcantoni, I.; Morettini, M.; Burattini, L. Compressed Segmented Beat Modulation Method using Discrete Cosine Transform. In Proceedings of the 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 2273–2276. [Google Scholar]
- Nasim, A.; Pinti, F.; Gentili, A.; Belli, A.; Palma, L.; Pierleoni, P. Dynamic Segmented Beat Modulation Method for Denoising ECG Data from Wearable Sensors. In Proceedings of the International Conference on Sensing and Instrumentation in IoT Era (ISSI 2019), Lisbon, Portugal, 29–30 August 2019; pp. 1–4. [Google Scholar]
- EC57, A.A. Testing and Reporting Performance Results of Cardiac Rhythm and ST Segment Measurement Algorithms; Association for the Advancement of Medical Instrumentation: Arlington, VA, USA, 1998. [Google Scholar]
- Malik, M.; Hnatkova, K.; Sisakova, M.; Schmidt, G. Subject-specific heart rate dependency of electrocardiographic QT, PQ, and QRS intervals. J. Electrocardiol. 2008, 41, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Moody, G.B.; Mark, R.G. The impact of the MIT–BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef]
- Asl, B.M.; Setarehdan, S.K.; Mohebbi, M. Support vector machine-based arrhythmia classification using reduced features of heart rate variability signal. Artif. Intell. Med. 2008, 44, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef]
- Chen, S.; Hua, W.; Li, Z.; Li, J.; Gao, X. Heartbeat classification using projected and dynamic features of ECG signal. Biomed. Signal Process. Control 2017, 31, 165–173. [Google Scholar] [CrossRef]
- Moody, G.B.; Muldrow, W.; Mark, R.G. A noise stress test for arrhythmia detectors. Comput. Cardiol. 1984, 11, 381–384. [Google Scholar]
- Zhang, Z.; Dong, J.; Luo, X.; Choi, K.S.; Wu, X. Heartbeat classification using disease-specific feature selection. Comput. Biol. Med. 2014, 46, 79–89. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Sun, L.; Rong, J.; Wang, H.; Zhang, Y. A pyramid-like model for heartbeat classification from ECG recordings. PLoS ONE 2018, 13, e0206593. [Google Scholar] [CrossRef] [PubMed]
- Martis, R.J.; Acharya, U.R.; Min, L.C. ECG beat classification using PCA, LDA, ICA and discrete wavelet transform. Biomed. Signal Process. Control 2013, 8, 437–448. [Google Scholar] [CrossRef]
- Ye, C.; Kumar, B.V.; Coimbra, M.T. Heartbeat classification using morphological and dynamic features of ECG signals. IEEE Trans. Biomed. Eng. 2012, 59, 2930–2941. [Google Scholar] [PubMed]
- Korürek, M.; Doğan, B. ECG beat classification using particle swarm optimization and radial basis function neural network. Expert Syst. Appl. 2010, 37, 7563–7569. [Google Scholar] [CrossRef]
- Yu, S.N.; Chou, K.T. Integration of independent component analysis and neural networks for ECG beat classification. Expert Syst. Appl. 2008, 34, 2841–2846. [Google Scholar] [CrossRef]
- De Chazal, P.; Reilly, R.B. A patient-adapting heartbeat classifier using ECG morphology and heartbeat interval features. IEEE Trans. Biomed. Eng. 2006, 53, 2535–2543. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, S.; Kanungo, B.; Behera, S.; Sabut, S. Multiresolution wavelet transform based feature extraction and ECG classification to detect cardiac abnormalities. Measurement 2017, 108, 55–66. [Google Scholar] [CrossRef]
- De Chazal, P.; O’Dwyer, M.; Reilly, R.B. Automatic classification of heartbeats using ECG morphology and heartbeat interval features. IEEE Trans. Biomed. Eng. 2004, 51, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
| AAMI | MIT–BIH | Beat Description |
|---|---|---|
| N | N or . | Normal beat |
| L | Left bundle branch block | |
| R | Right bundle branch block | |
| e | Atrial escape beat | |
| j | Nodal (junctional) escape beat | |
| S | S | Supraventricular premature beat |
| A | Atrial premature beat | |
| a | Aberrated atrial premature beat | |
| J | Nodal (junctional) premature beat | |
| V | V | Premature ventricular contraction |
| E | Ventricular escape beat | |
| ! | Ventricular flutter wave |
| Dataset/Class | N | S | V | Total |
|---|---|---|---|---|
| Training | 41,020 | 1646 | 4057 | 46,723 |
| Testing | 27,346 | 1097 | 2704 | 31,147 |
| Total | 68,366 | 2743 | 6761 | 77,870 |
| True/Predicted | N | S | V | Total | |
|---|---|---|---|---|---|
| Training | N | 39,205 | 765 | 1050 | 41,020 |
| S | 15 | 1604 | 27 | 1646 | |
| V | 202 | 342 | 3513 | 4057 | |
| Total | 39,422 | 2711 | 4590 | 46,723 | |
| Testing | N | 25,388 | 722 | 1236 | 27,346 |
| S | 51 | 1019 | 27 | 1097 | |
| V | 391 | 198 | 2115 | 2704 | |
| Total | 25,830 | 1939 | 3378 | 31,147 | |
| Total | N | 64,603 | 1487 | 2276 | 68,366 |
| S | 66 | 2623 | 54 | 2743 | |
| V | 593 | 523 | 5645 | 6761 | |
| Total | 65,262 | 4633 | 7975 | 77,870 |
| Dataset | Acc | PP(N) | PP(S) | PP(V) |
|---|---|---|---|---|
| (%) | (%) | (%) | (%) | |
| Training | 94.86 | 85.57 | 97.44 | 86.59 |
| Testing | 91.57 | 92.84 | 92.89 | 78.21 |
| Overall | 93.58 | 94.49 | 95.62 | 83.49 |
| Noise Type | Acc | PP(N) | PP(S) | PP(V) | |
|---|---|---|---|---|---|
| (%) | (%) | (%) | (%) | (dB) | |
| No additional noise | 93.38 | 96.48 | 76.33 | 78.60 | 1.71 [0.46; 2.97] |
| Baseline wander | 91.36 | 95.88 | 75.44 | 62.33 | 2.67 [1.76; 3.57] |
| Muscle activity | 91.52 | 96.45 | 73.11 | 60.00 | 3.86 [3.01; 4.68] |
| Electrode motion artifacts | 91.11 | 95.85 | 52.88 | 65.43 | 5.02 [3.30; 6.71] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasim, A.; Sbrollini, A.; Morettini, M.; Burattini, L. Extended Segmented Beat Modulation Method for Cardiac Beat Classification and Electrocardiogram Denoising. Electronics 2020, 9, 1178. https://doi.org/10.3390/electronics9071178
Nasim A, Sbrollini A, Morettini M, Burattini L. Extended Segmented Beat Modulation Method for Cardiac Beat Classification and Electrocardiogram Denoising. Electronics. 2020; 9(7):1178. https://doi.org/10.3390/electronics9071178
Chicago/Turabian StyleNasim, Amnah, Agnese Sbrollini, Micaela Morettini, and Laura Burattini. 2020. "Extended Segmented Beat Modulation Method for Cardiac Beat Classification and Electrocardiogram Denoising" Electronics 9, no. 7: 1178. https://doi.org/10.3390/electronics9071178
APA StyleNasim, A., Sbrollini, A., Morettini, M., & Burattini, L. (2020). Extended Segmented Beat Modulation Method for Cardiac Beat Classification and Electrocardiogram Denoising. Electronics, 9(7), 1178. https://doi.org/10.3390/electronics9071178








