Hyperspectral Image Denoising and Classification Using Multi-Scale Weighted EMAPs and Extreme Learning Machine
Abstract
1. Introduction
2. Introductions of Related Works
2.1. Normalization
2.2. Attribute Profiles (APs)
2.3. Extreme Learning Machine (ELM)
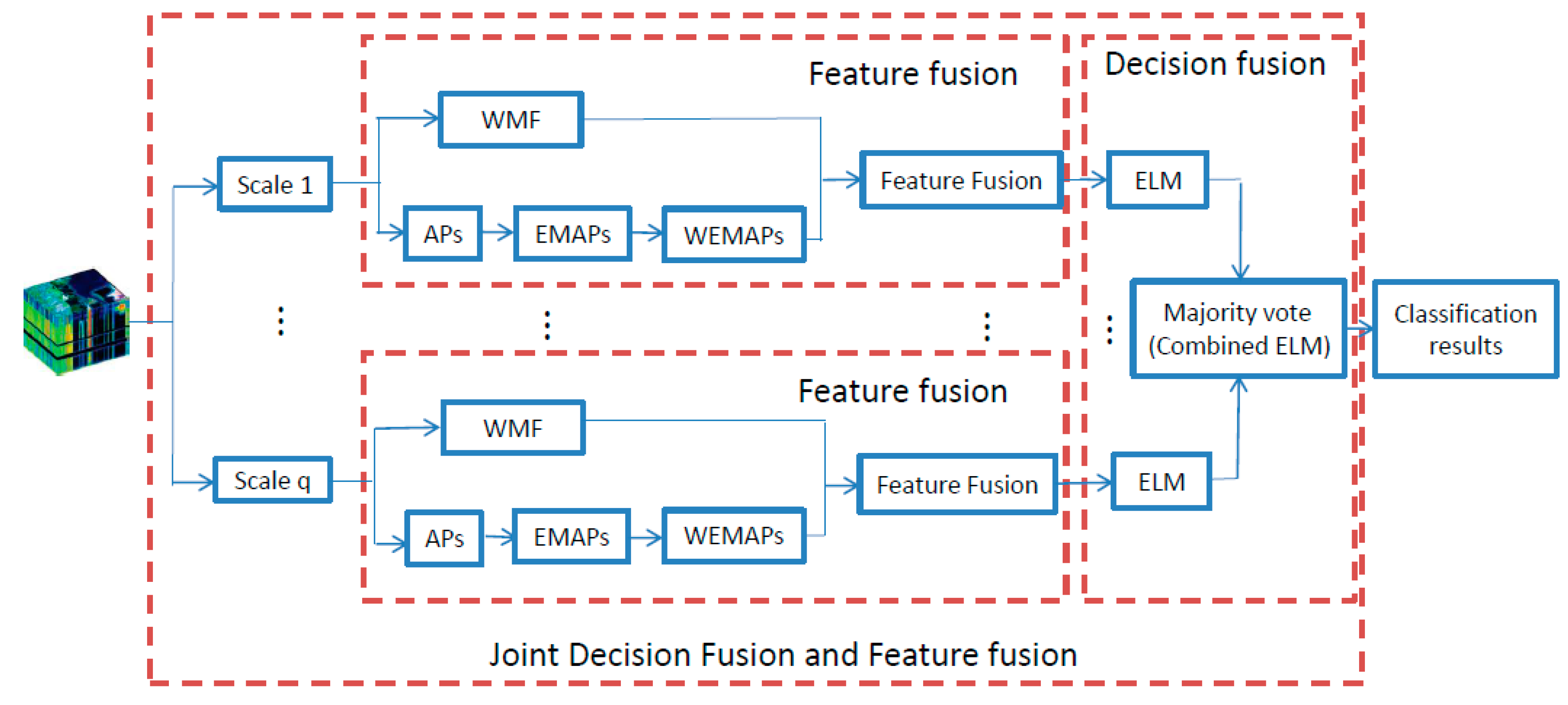
3. The Proposed Joint Decision Fusion and Feature Fusion (JDFFF) Framework
3.1. Weighted Mean Filters (WMFs)
3.2. The Proposed Weighted Extended Multi-Attribute Profiles (WEMAPs)
3.3. Feature Fusion (FF)
3.4. Joint Decision and Feature Fusion (JDFFF)
4. Experimental Results and Discussion
4.1. Evaluation Criteria and Parameter Settings
4.2. Investigation on the Effect of Different Strategies
4.3. Investigation on the Suitability of Different Datasets
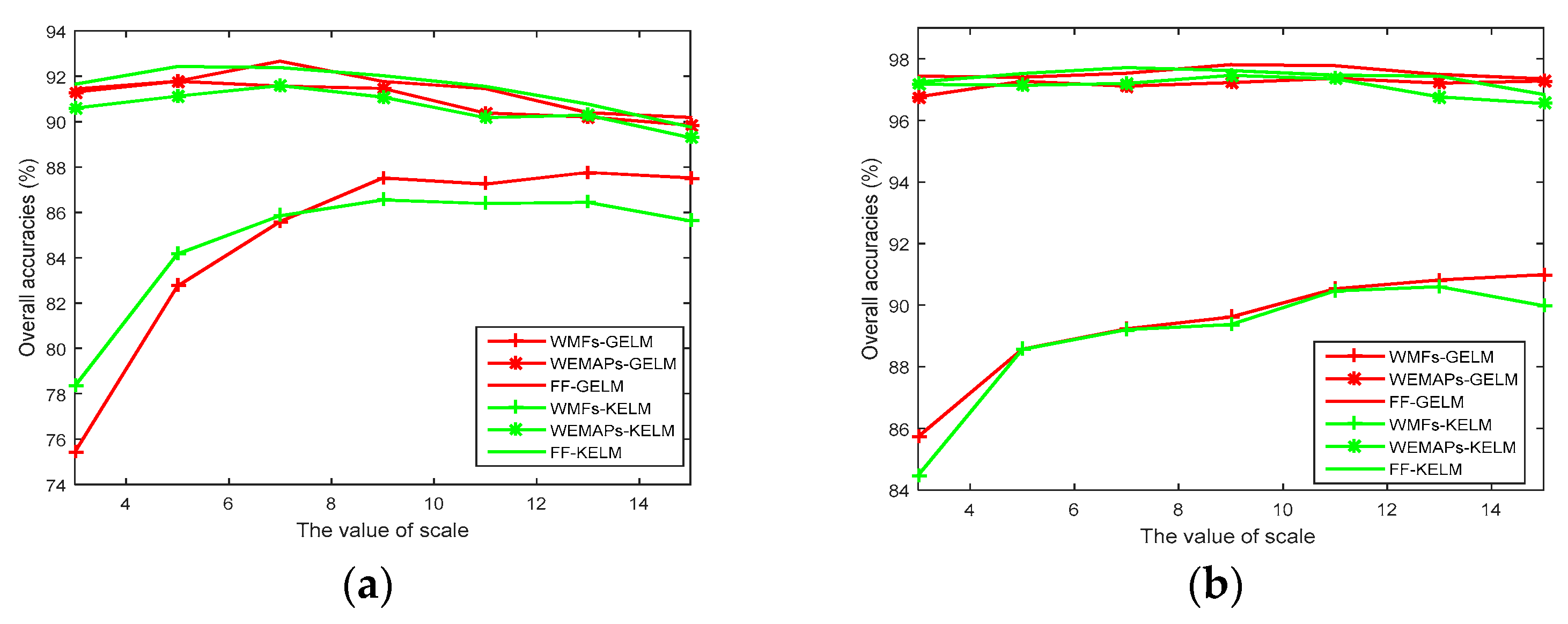
4.4. Investigation on the Effect of Scales
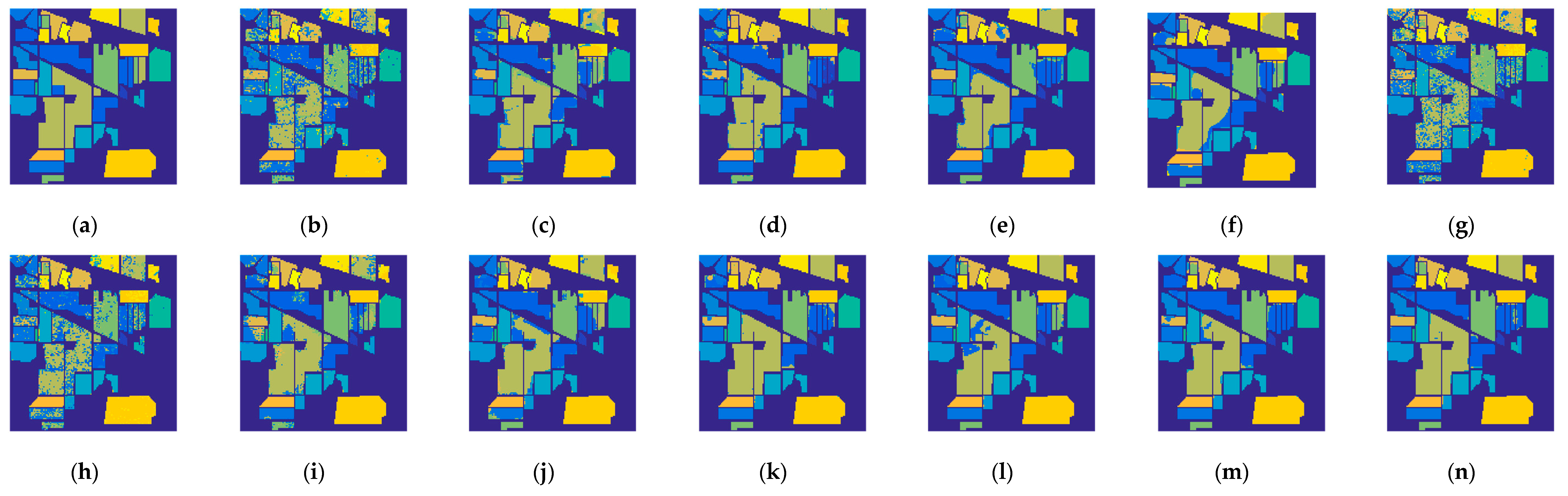
4.5. Classification Results and Comparisons on the Two Datasets
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Deng, C.; Liu, X. Active multi-kernel domain adaptation for hyperspectral image classification. Pattern Recognit. 2018, 77, 306–315. [Google Scholar] [CrossRef]
- Torti, E.; Leon, R.; La, M. Parallel classification pipelines for skin cancer detection exploiting hyperspectral imaging on hybrid systems. Electronics 2020, 9, 1053. [Google Scholar] [CrossRef]
- Zhou, Y.; Peng, J.; Chen, C.L.P. Dimension reduction using spatial and spectral regularized local discriminant embedding for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 1082–1095. [Google Scholar] [CrossRef]
- Plaza, A.; Benediktsson, J.A.; Boardman, J.W. Recent advances in techniques for hyperspectral image processing. Remote Sens. Environ. 2019, 113, S110–S122. [Google Scholar] [CrossRef]
- Hughes, G.F. On the mean accuracy of statistical pattern recognizers. IEEE Trans. Inform. Theory. 1968, 14, 55–63. [Google Scholar] [CrossRef]
- Song, X.; Wu, L.; Hao, H. Hyperspectral image denoising based on spectral dictionary learning and sparse coding. Electronics 2019, 8, 86. [Google Scholar] [CrossRef]
- Lin, L.; Chen, C.; Yang, J. Deep transfer HSI classification method based on information measure and optimal neighborhood noise reduction. Electronics 2019, 8, 1112. [Google Scholar] [CrossRef]
- Kang, X.; Xiang, X.; Li, S.; Benediktsson, J.A. PCA-based edge-preserving features for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 7140–7151. [Google Scholar] [CrossRef]
- Zabalza, J.; Ren, J.; Yang, M. Novel folded-PCA for improved feature extraction and data reduction with hyperspectral imaging and SAR in remote sensing. ISPRS J. Photogramm. Remote Sens. 2014, 93, 112–122. [Google Scholar] [CrossRef]
- Mura, M.D.; Benediktsson, J.A.; Waske, B. Morphological attribute profiles for the analysis of very high resolution images. IEEE Trans. Geosci. Remote Sens. 2010, 48, 3747–3762. [Google Scholar] [CrossRef]
- Mura, M.D.; Benediktsson, J.A.; Waske, B. Extended profiles with morphological attribute filters for the analysis of hyperspectral data. Int. J. Remote Sens. 2010, 31, 5975–5991. [Google Scholar] [CrossRef]
- Jerome, H.F. Regularized discriminant analysis. J. Am. Stat. Assoc. 1989, 84, 165–175. [Google Scholar]
- Kuo, B.C.; Landgrebe, D.A. Nonparametric weighted feature extraction for classification. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1096–1105. [Google Scholar]
- Zabalza, J.; Ren, J.; Zheng, J.; Zhao, H. Novel segmented stacked autoencoder for effective dimensionality reduction and feature extraction in hyperspectral imaging. Neurocomputing 2016, 185, 1–10. [Google Scholar] [CrossRef]
- Yu, H.; Gao, L.; Li, J. Spectral-spatial hyperspectral image classification using subspace based support vector machines and adaptive markov random fields. Remote Sens. 2016, 8, 355. [Google Scholar] [CrossRef]
- Samat, A.; Du, P.; Liu, S. Ensemble extreme learning machines for hyperspectral image classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 1060–1069. [Google Scholar] [CrossRef]
- Li, J.; Huang, X.; Gamba, P. Multiple feature learning for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 1592–1606. [Google Scholar] [CrossRef]
- Cao, F.; Yang, Z.; Ren, J.; Ling, W.K. Extreme Sparse Multinomial Logistic Regression: A Fast and Robust Framework for Hyperspectral Image Classification. Remote Sens. 2017, 9, 1255. [Google Scholar] [CrossRef]
- Torti, E.; Fontanella, A.; Plaza, A. Hyperspectral Image Classification Using Parallel Autoencoding Diabolo Networks on Multi-Core and Many-Core Architectures. Electronics 2018, 7, 411. [Google Scholar] [CrossRef]
- Pesaresi, M.; Benediktsson, J.A. A new approach for the morphological segmentation of high-resolution satellite imagery. IEEE Trans. Geosci. Remote Sens. 2001, 39, 309–320. [Google Scholar] [CrossRef]
- Benediktsson, J.A.; Palmason, J.A.; Sveinsson, J.R. Classification of hyperspectral data from urban areas based on extended morphological profiles. IEEE Trans. Geosci. Remote Sens. 2005, 43, 480–491. [Google Scholar] [CrossRef]
- Xia, J.; Mura, M.D.; Chanussot, J. Random subspace ensembles for hyperspectral image classification with extended morphological attribute profiles. IEEE Trans. Geosci. Remote Sens. 2015, 53, 4768–4786. [Google Scholar] [CrossRef]
- Mura, M.D.; Villa, A.; Benediktsson, J.A. Classification of hyperspectral images by using extended morphological attribute profiles and independent component analysis. IEEE Geosci. Remote Sens. Lett. 2011, 8, 542–546. [Google Scholar] [CrossRef]
- Li, J.; Marpu, P.R.; Plaza, A. Generalized composite kernel framework for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2013, 51, 4816–4829. [Google Scholar] [CrossRef]
- Li, W.; Prasad, S.; Fowler, J.E. Decision fusion in kernel-induced spaces for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2014, 52, 3399–3411. [Google Scholar] [CrossRef]
- Li, W.; Chen, C.; Su, H. Local binary patterns and extreme learning machine for hyperspectral imagery classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 3681–3693. [Google Scholar] [CrossRef]
- Prasad, S.; Bruce, L.M. Decision fusion with confidence-based weight assignment for hyperspectral target recognition. IEEE Trans. Geosci. Remote Sens. 2008, 46, 1448–1456. [Google Scholar] [CrossRef]
- Lee, K.H. Combining Feature Fusion and Decision Fusion in Multimodal Biometric Authentication. J. Korea Inst. Inf Secur. Cryptol. 2010, 20, 133–138. [Google Scholar]
- Sun, B.; Li, L.; Wu, X. Combining feature-level and decision-level fusion in a hierarchical classifier for emotion recognition in the wild. J. Multimodal User In. 2015, 10, 125–137. [Google Scholar] [CrossRef]
- Liao, W.; Bellens, R.; Pizurica, A. Combining feature fusion and decision fusion for classification of hyperspectral and LiDAR data. In Proceedings of the 2014 IEEE Geoscience and Remote Sensing Symposium, Quebec City, QC, Canada, 13–18 July 2014; pp. 1241–1244. [Google Scholar]
- Zhou, Y.; Peng, J.; Chen, C.L.P. Region-kernel-based support vector machines for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 4810–4824. [Google Scholar]
- Soille, P. Morphological image analysis: Principles and applications. Sensor Rev. 1999, 28, 800–801. [Google Scholar]
- Marpu, P.R.; Pedergnana, M.; Mura, M.D. Automatic generation of standard deviation attribute profiles for spectral–spatial classification of remote sensing data. IEEE Geosci. Remote Sens. Lett. 2013, 10, 293–297. [Google Scholar] [CrossRef]
- Huang, G.B.; Zhu, Q.Y.; Siew, C.K. Extreme learning machine: Theory and applications. Neurocomputing 2006, 70, 489–501. [Google Scholar] [CrossRef]
- Cao, F.; Yang, Z.; Ren, J.; Ling, W.K. Sparse Representation-Based Augmented Multinomial Logistic Extreme Learning Machine with Weighted Composite Features for Spectral-Spatial Classification of Hyperspectral Images. IEEE Trans. Geosci. Remote Sens. 2018, 56, 6263–6279. [Google Scholar] [CrossRef]
- Huang, G.B.; Zhou, H.; Ding, X. Extreme learning machine for regression and multiclass classification. IEEE Trans. Syst. Man Cybern. B Cybern. 2012, 42, 513–529. [Google Scholar] [CrossRef]
- Banerjee, K.S. Generalized Inverse of Matrices and its Applications; Wiley: Hoboken, HJ, USA, 1971. [Google Scholar]
- Camps-Valls, G.; Gomez-Chova, L.; Muñoz-Marí, J. Composite kernels for hyperspectral image classification. IEEE Geosci. Remote Sens. Lett. 2006, 3, 93–97. [Google Scholar] [CrossRef]
- Fletcher, R. Practical Methods of Optimization; Wiley: Hoboken, HJ, USA, 1980. [Google Scholar]
- Huang, X.; Zhang, L. An SVM ensemble approach combining spectral, structural, and semantic features for the classification of high-resolution remotely sensed imagery. IEEE Trans. Geosci. Remote Sens. 2013, 51, 257–272. [Google Scholar] [CrossRef]
- Ham, J.; Chen, Y.C.; Ghosh, J. Investigation of the random forest framework for classification of hyperspectral data. IEEE Geosci. Remote Sens. 2015, 43, 492–501. [Google Scholar] [CrossRef]
- Bao, R.; Xia, J.; Du, P. Combining morphological attribute profiles via an ensemble method for hyperspectral image classification. IEEE Geosci. Remote Sens. Lett. 2016, 13, 359–363. [Google Scholar] [CrossRef]
- Zhou, Y.; Peng, J.; Chen, C.L.P. Extreme learning machine with composite kernels for hyperspectral image classification. IEEE J. Sel. Topics Appl. Earth Observ. Remote Sens. 2015, 8, 2351–2360. [Google Scholar] [CrossRef]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM 2011, 2, 1–39. [Google Scholar] [CrossRef]
- Sun, L.; Wu, Z.; Liu, J. Supervised spectral–spatial hyperspectral image classification with weighted Markov random fields. IEEE Trans. Geosci. Remote Sens. 2015, 53, 1490–1503. [Google Scholar] [CrossRef]
- Chen, W.; Yang, Z. Dimensionality Reduction Based on Determinantal Point Process and Singular Spectrum Analysis for Hyperspectral Images. IET Image Process. 2018, 13, 299–306. [Google Scholar] [CrossRef]
- Li, J.; Bioucas-Dias, J.M.; Plaza, A. Robust collaborative nonnegative matrix factorization for hyperspectral unmixing. IEEE Trans. Geosci. Remote Sens. 2016, 54, 6076–6090. [Google Scholar] [CrossRef]

| Abbreviations List | |||
|---|---|---|---|
| AA | average accuracies | K | kappa coefficient |
| APs | attribute profiles | KELM | kernel extreme learning machine |
| CKs | composite kernels | KELM-CKs | kernel extreme learning machine-composite kernels |
| DF | decision fusion | KSVM | kernel support vector machine |
| ELM | extreme learning machine | LDA | the linear discriminate analysis |
| EMAPs | extended multi-attribute profiles | MWEMAPs | multiscale weighted extended multi-attribute profiles |
| FF | feature-level fusion | MV | majority voting |
| FF-KSVM | feature-level fusion-kernel support vector machine | MPs | morphological profiles |
| FF-GELM | feature-level fusion-generalized extreme learning machine | OA | overall accuracies |
| FF-KELM | feature-level fusion-kernel extreme learning machine | PCA | principal component analysis |
| GELM | generalized extreme learning machine | SVM | support vector machine |
| GELM-CKs | generalized extreme learning machine-composite kernels | SVM-CKs | support vector machine-composite kernels |
| HSI | hyperspectral image | SMLR | sparse multinomial logistic regression |
| JDFFF | joint decision fusion and feature fusion | SMLR-SPATV | sparse multinomial logistic regression-weighed Markov random fields |
| JDFFF-KSVM | joint decision fusion and feature fusion-kernel support vector machine | SSA | singular spectrum analysis |
| JDFFF-GELM | joint decision fusion and feature fusion-generalized extreme learning machine | WEMAPs | weighted extended multi-attribute profiles |
| JDFFF-KELM | joint decision fusion and feature fusion-kernel extreme learning machine | WMFs | weighted mean filters |
| Datasets | Strategies | GELM | GELM (Noise 0.02) | GELM (Noise 0.04) | GELM (Noise 0.06) | KELM | KELM (Noise 0.02) | KELM (Noise 0.04) | KELM (Noise 0.06) |
|---|---|---|---|---|---|---|---|---|---|
| Indian Pines | raw data | 61.02 ± 1.52 | 55.45 ± 1.64 | 50.21 ± 1.36 | 44.85 ± 1.89 | 66.93 ± 2.45 | 60.07 ± 2.37 | 54.37 ± 2.11 | 47.98 ± 2.33 |
| WMFs | 75.35 ± 2.07 | 72.26 ± 2.78 | 69.42 ± 2.15 | 66.32 ± 2.45 | 78.35 ± 3.09 | 75.12 ± 1.61 | 72.31 ± 2.51 | 69.21 ± 2.63 | |
| EMAPs | 88.34 ± 2.02 | 88.25 ± 2.64 | 88.12 ± 2.51 | 88.09 ± 2.74 | 88.93 ± 1.73 | 88.54 ± 2.24 | 88.29 ± 2.66 | 88.17 ± 2.62 | |
| WEMAPs | 90.51 ± 1.77 | 90.42 ± 1.91 | 90.32 ± 2.12 | 90.31 ± 2.35 | 91.25 ± 1.95 | 91.14 ± 1.62 | 91.12 ± 2.24 | 91.15 ± 2.33 | |
| FF | 91.86 ± 2.02 | 91.77 ± 1.46 | 91.68 ± 1.72 | 91.71 ± 1.42 | 92.22 ± 1.37 | 92.18 ± 1.65 | 92.15 ± 2.14 | 92.08 ± 2.26 | |
| JDFFF | 92.74 ± 1.23 | 92.68 ± 1.61 | 92.57 ± 1.87 | 92.61 ± 1.54 | 93.09 ± 0.86 | 92.97 ± 1.18 | 92.92 ± 1.64 | 92.87 ± 1.67 | |
| Pavia University | raw data | 74.48 ± 3.67 | 63.58 ± 3.45 | 58.45 ± 3.12 | 54.26 ± 3.58 | 74.20 ± 5.12 | 63.72 ± 3.41 | 58.63 ± 3.09 | 54.62 ± 4.35 |
| WMFs | 85.07 ± 3.69 | 76.02 ± 3.56 | 68.91 ± 3.68 | 66.12 ± 3.46 | 84.21 ± 5.20 | 75.55 ± 5.51 | 68.31 ± 3.36 | 65.58 ± 2.04 | |
| EMAPs | 95.90 ± 1.85 | 95.85 ± 2.23 | 95.88 ± 2.31 | 95.78 ± 2.18 | 95.41 ± 3.21 | 95.34 ± 1.75 | 95.36 ± 1.28 | 95.31 ± 1.68 | |
| WEMAPs | 96.97 ± 1.15 | 96.88 ± 1.45 | 96.85 ± 1.96 | 96.79 ± 1.84 | 97.11 ± 1.06 | 97.05 ± 1.32 | 97.02 ± 2.62 | 96.95 ± 1.86 | |
| FF | 97.31 ± 1.15 | 97.26 ± 1.78 | 97.28 ± 1.79 | 97.18 ± 2.03 | 97.15 ± 0.74 | 97.11 ± 1.58 | 97.08 ± 1.05 | 97.04 ± 1.63 | |
| JDFFF | 97.98 ± 1.10 | 97.96 ± 1.21 | 97.93 ± 1.36 | 97.95 ± 1.58 | 98.16 ± 0.98 | 98.15 ± 0.85 | 98.08 ± 0.83 | 98.06 ± 1.15 |
| Datasets | Strategies | GELM | Time(s) | KELM | Time(s) |
|---|---|---|---|---|---|
| Indian Pines | raw data | 61.02 ± 1.52 | 0.43 | 66.93 ± 2.45 | 0.41 |
| WEMAPs | 90.51 ± 1.77 | 0.56 | 91.25 ± 1.95 | 0.52 | |
| FF | 91.86 ± 2.02 | 1.25 | 92.22 ± 1.37 | 2.08 | |
| JDFFF | 92.74 ± 1.23 | 17.15 | 93.09 ± 0.86 | 21.32 | |
| Pavia University | raw data | 74.48 ± 3.67 | 0.69 | 74.20 ± 5.12 | 0.57 |
| WEMAPs | 96.97 ± 1.15 | 0.76 | 97.11 ± 1.06 | 0.62 | |
| FF | 97.31 ± 1.15 | 5.35 | 97.15 ± 0.74 | 5.16 | |
| JDFFF | 97.98 ± 1.10 | 74.05 | 98.16 ± 0.98 | 72.67 | |
| Kennedy Space Center | raw data | 87.43 ± 1.23 | 0.85 | 87.76 ± 0.98 | 0.74 |
| WEMAPs | 88.32 ± 0.95 | 0.91 | 88.65 ± 0.75 | 0.83 | |
| FF | 93.75 ± 1.12 | 7.54 | 93.93 ± 1.06 | 7.02 | |
| JDFFF | 93.86 ± 1.09 | 87.52 | 93.94 ± 0.83 | 84.26 |
| Q | Index | KSVM | SVM-CKs | FF-KSVM | JDFFF-KSVM | SMLR-SPATV | GELM | KELM | GELM-CKs | KELM-CKs | FF-GELM | FF-KELM | JDFFF-GELM | JDFFF-KELM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | OA | 54.1 ± 4.1 | 59.18 ± 5.13 | 71.6 ± 2.9 | 74.5 ± 1.9 | 69.8 ± 5.7 | 50.8 ± 4.4 | 55.2 ± 3.8 | 62.3 ± 1.6 | 73.1 ± 2.8 | 80.3 ± 2.1 | 81.3 ± 2.1 | 82.3 ± 2.3 | 82.5 ± 2.7 |
| AA | 66.1 ± 2.2 | 71.1 ± 2.8 | 80.9 ± 3.1 | 83.1 ± 1.4 | 80.8 ± 0.6 | 64.8 ± 3.5 | 68.3 ± 1.5 | 75.9 ± 1.6 | 83.1 ± 2.2 | 86.6 ± 1.8 | 87.4 ± 1.4 | 88.2 ± 1.3 | 88.3 ± 1.8 | |
| k | 48.8 ± 4.4 | 54.4 ± 5.5 | 68.2 ± 3.2 | 71.4 ± 2.1 | 66.3 ± 6.1 | 45.7 ± 0.2 | 50.3 ± 3.8 | 58.1 ± 1.8 | 69.8 ± 3.1 | 77.8 ± 2.4 | 78.9 ± 2.3 | 80.1 ± 2.6 | 80.3 ± 3.1 | |
| 10 | OA | 64.3 ± 2.8 | 71.1 ± 4.1 | 79.3 ± 2.5 | 81.5 ± 2.8 | 77.8 ± 5.3 | 57.6 ± 2.9 | 63.3 ± 3.1 | 70.5 ± 1.7 | 80.9 ± 2.3 | 86.8 ± 1.9 | 88.2 ± 1.4 | 88.3 ± 2.1 | 88.8 ± 1.9 |
| AA | 74.8 ± 2.1 | 81.5 ± 2.6 | 87.5 ± 0.9 | 89.4 ± 1.2 | 88.1 ± 1.9 | 70.8 ± 2.2 | 74.5 ± 2.2 | 82.8 ± 1.1 | 89.2 ± 1.5 | 92.1 ± 1.2 | 92.8 ± 0.9 | 93.1 ± 1.3 | 93.3 ± 1.1 | |
| k | 59.9 ± 3.1 | 67.7 ± 4.6 | 76.6 ± 2.8 | 79.2 ± 3.1 | 75.1 ± 5.7 | 52.6 ± 3.1 | 58.8 ± 3.2 | 67.1 ± 1.9 | 78.5 ± 2.5 | 85.1 ± 2.2 | 86.6 ± 1.6 | 86.7 ± 2.2 | 87.3 ± 2.1 | |
| 15 | OA | 67.1 ± 2.4 | 78.7 ± 3.5 | 86.1 ± 1.9 | 86.9 ± 1.9 | 83.1 ± 2.4 | 61.1 ± 1.3 | 65.7 ± 3.3 | 77.7 ± 2.1 | 85.3 ± 2.1 | 91.8 ± 1.5 | 91.6 ± 2.2 | 92.5 ± 1.5 | 93.1 ± 1.1 |
| AA | 77.6 ± 1.8 | 86.7 ± 1.9 | 91.5 ± 1.3 | 92.2 ± 1.2 | 91.6 ± 0.7 | 75.1 ± 0.9 | 77.9 ± 2.5 | 87.4 ± 0.8 | 92.1 ± 1.6 | 95.2 ± 0.9 | 95.1 ± 1.4 | 95.6 ± 0.8 | 96.1 ± 0.7 | |
| k | 63.2 ± 2.6 | 76.1 ± 3.9 | 84.3 ± 2.1 | 85.1 ± 2.1 | 81.1 ± 2.6 | 56.6 ± 1.3 | 61.7 ± 3.6 | 75.1 ± 2.2 | 83.4 ± 2.4 | 90.7 ± 1.7 | 90.5 ± 2.5 | 91.4 ± 1.7 | 92.1 ± 1.1 | |
| 20 | OA | 71.1 ± 2.2 | 83.0 ± 2.4 | 88.1 ± 2.1 | 89.5 ± 1.2 | 85.9 ± 2.4 | 63.5 ± 1.4 | 70.8 ± 2.6 | 82.7 ± 1.7 | 89.6 ± 1.1 | 92.8 ± 1.2 | 93.5 ± 0.9 | 94.2 ± 0.9 | 94.1 ± 1.4 |
| AA | 80.9 ± 1.3 | 89.6 ± 1.2 | 92.8 ± 1.2 | 93.8 ± 0.7 | 92.5 ± 1.0 | 77.2 ± 0.9 | 81.9 ± 1.0 | 91.1 ± 1.1 | 94.6 ± 0.7 | 95.7 ± 0.9 | 96.2 ± 0.6 | 96.6 ± 0.6 | 96.6 ± 0.9 | |
| k | 67.4 ± 2.5 | 80.8 ± 2.6 | 86.5 ± 2.2 | 88.1 ± 1.4 | 84.1 ± 2.6 | 59.3 ± 1.4 | 67.2 ± 2.8 | 80.5 ± 1.8 | 88.2 ± 1.1 | 91.8 ± 1.4 | 92.6 ± 1.1 | 93.4 ± 1.1 | 93.3 ± 1.6 | |
| 25 | OA | 72.6 ± 1.8 | 85.5 ± 1.5 | 90.1 ± 1.1 | 90.5 ± 1.1 | 87.8 ± 2.1 | 66.5 ± 1.7 | 73.1 ± 2.3 | 85.9 ± 1.3 | 90.8 ± 1.1 | 93.1 ± 1.4 | 93.8 ± 1.1 | 93.8 ± 0.9 | 94.5 ± 1.1 |
| AA | 82.5 ± 1.6 | 91.8 ± 1.1 | 94.1 ± 0.7 | 94.6 ± 0.8 | 93.8 ± 0.9 | 79.5 ± 0.8 | 83.8 ± 0.9 | 92.6 ± 0.9 | 95.5 ± 0.5 | 96.1 ± 1.1 | 96.5 ± 0.6 | 96.8 ± 0.6 | 97.1 ± 0.6 | |
| k | 69.1 ± 1.9 | 83.6 ± 1.7 | 88.7 ± 1.1 | 89.2 ± 1.2 | 86.2 ± 2.2 | 62.5 ± 1.7 | 69.7 ± 2.5 | 84.1 ± 1.5 | 89.6 ± 1.2 | 92.1 ± 1.6 | 92.9 ± 1.3 | 92.9 ± 1.1 | 93.8 ± 1.3 | |
| 30 | OA | 74.4 ± 1.8 | 85.9 ± 1.8 | 90.6 ± 1.4 | 91.5 ± 1.1 | 89.1 ± 1.5 | 66.8 ± 0.6 | 74.3 ± 1.4 | 88.8 ± 1.6 | 91.9 ± 0.9 | 94.2 ± 1.1 | 94.9 ± 1.1 | 94.9 ± 1.1 | 95.1 ± 1.1 |
| AA | 83.8 ± 1.2 | 92.1 ± 0.9 | 94.5 ± 0.6 | 95.2 ± 0.8 | 94.9 ± 5.6 | 79.7 ± 0.9 | 85.3 ± 1.1 | 94.3 ± 1.1 | 96.1 ± 0.7 | 96.8 ± 0.6 | 97.2 ± 0.5 | 97.3 ± 0.6 | 97.4 ± 0.6 | |
| k | 71.1 ± 1.9 | 84.1 ± 2.1 | 89.3 ± 1.6 | 90.3 ± 1.3 | 87.6 ± 1.6 | 63.1 ± 0.6 | 71.2 ± 1.5 | 87.3 ± 1.8 | 90.8 ± 1.1 | 93.4 ± 1.3 | 94.2 ± 1.1 | 94.2 ± 1.2 | 94.4 ± 1.3 | |
| Time (s) | 23.98 | 41.80 | 37.51 | 152.47 | 30.3 | 0.45 | 1.05 | 1.86 | 9.59 | 1.83 | 2.69 | 19.58 | 23.77 |
| Q | Index | KSVM | SVM-CKs | FF-KSVM | JDFFF-KSVM | SMLR-SPATV | GELM | KELM | GELM-CKs | KELM-CKs | FF-GELM | FF-KELM | JDFFF-GELM | JDFFF-KELM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | OA | 61.7 ± 10.7 | 63.3 ± 4.5 | 81.2 ± 2.6 | 84.76 ± 3.87 | 67.46 ± 6.94 | 60.9 ± 8.1 | 59.9 ± 6.7 | 63.2 ± 3.7 | 63.7 ± 8.1 | 88.8 ± 4.5 | 89.3 ± 5.4 | 91.61 ± 4.03 | 90.1 ± 3.1 |
| AA | 73.1 ± 4.9 | 73.4 ± 3.3 | 86.4 ± 0.9 | 87.7 ± 2.1 | 76.1 ± 5.5 | 70.6 ± 4.6 | 71.9 ± 3.4 | 71.5 ± 2.2 | 72.5 ± 2.8 | 91.2 ± 2.2 | 91.5 ± 2.5 | 92.7 ± 1.8 | 91.9 ± 1.5 | |
| k | 53.1 ± 10.8 | 54.7 ± 4.7 | 76.1 ± 2.9 | 80.3 ± 4.6 | 59.3 ± 7.2 | 52.1 ± 8.3 | 51.1 ± 6.7 | 54.4 ± 3.5 | 55.2 ± 8.7 | 85.6 ± 5.5 | 86.27 ± 6.7 | 89.1 ± 5.1 | 87.2 ± 3.7 | |
| 10 | OA | 71.2 ± 4.4 | 74.2 ± 6.7 | 87.7 ± 4.8 | 89.8 ± 4.2 | 77.4 ± 4.5 | 69.8 ± 4.3 | 67.6 ± 3.3 | 71.6 ± 3.5 | 75.7 ± 6.5 | 92.1 ± 3.3 | 92.00 ± 3.1 | 93.3 ± 3.7 | 93.7 ± 3.1 |
| AA | 79.1 ± 1.9 | 79.9 ± 2.8 | 92.3 ± 2.4 | 93.5 ± 1.9 | 85.1 ± 2.4 | 77.9 ± 1.4 | 78.3 ± 1.9 | 77.2 ± 1.6 | 80.9 ± 3.2 | 95.4 ± 1.6 | 95.48 ± 1.9 | 95.9 ± 3.1 | 95.9 ± 3.1 | |
| k | 63.8 ± 4.8 | 67.3 ± 7.8 | 84.3 ± 5.9 | 86.9 ± 5.2 | 71.5 ± 4.8 | 62.2 ± 4.5 | 59.8 ± 3.5 | 63.9 ± 4.2 | 69.2 ± 7.9 | 89.7 ± 4.1 | 89.6 ± 3.8 | 91.3 ± 4.6 | 91.8 ± 3.7 | |
| 15 | OA | 74.1 ± 6.4 | 85.7 ± 2.6 | 96.2 ± 1.1 | 97.1 ± 0.8 | 79.3 ± 3.1 | 74.7 ± 3.8 | 72.8 ± 4.3 | 76.1 ± 3.2 | 86.2 ± 3.4 | 97.4 ± 1.1 | 97.4 ± 0.8 | 97.9 ± 1.1 | 98.3 ± 0.9 |
| AA | 81.1 ± 3.1 | 87.3 ± 1.3 | 96.7 ± 0.7 | 97.2 ± 0.6 | 86.1 ± 1.6 | 79.7 ± 1.2 | 81.2 ± 1.8 | 79.8 ± 1.8 | 87.2 ± 0.9 | 98.1 ± 0.8 | 97.9 ± 0.6 | 98.4 ± 0.5 | 98.5 ± 0.7 | |
| k | 67.2 ± 7.4 | 81.4 ± 3.3 | 95.1 ± 1.4 | 96.2 ± 1.2 | 73.8 ± 3.6 | 67.6 ± 4.2 | 65.7 ± 4.9 | 69.2 ± 3.7 | 82.1 ± 4.1 | 96.60 ± 1.5 | 96.6 ± 1.1 | 97.3 ± 1.4 | 97.7 ± 1.2 | |
| 20 | OA | 75.9 ± 3.6 | 86.1 ± 2.9 | 96.1 ± 1.7 | 97.1 ± 2.1 | 85.3 ± 2.9 | 74.4 ± 3.1 | 75.3 ± 3.8 | 78.3 ± 1.1 | 86.1 ± 2.9 | 97.5 ± 1.6 | 97.5 ± 1.5 | 98.4 ± 1.5 | 98.3 ± 1.8 |
| AA | 82.6 ± 1.1 | 88.2 ± 1.5 | 97.5 ± 0.5 | 97.9 ± 0.7 | 87.9 ± 2.6 | 80.4 ± 1.6 | 82.6 ± 1.6 | 82.4 ± 1.1 | 87.5 ± 2.1 | 98.3 ± 0.8 | 98.4 ± 0.5 | 98.9 ± 0.6 | 98.9 ± 0.6 | |
| k | 69.5 ± 4.1 | 81.9 ± 3.5 | 94.8 ± 2.2 | 96.2 ± 2.6 | 81.1 ± 3.6 | 67.6 ± 3.6 | 68.8 ± 4.3 | 72.1 ± 1.3 | 81.8 ± 3.7 | 96.7 ± 2.1 | 96.7 ± 2.1 | 97.9 ± 1.9 | 97.7 ± 2.3 | |
| 25 | OA | 80.6 ± 2.3 | 87.2 ± 2.1 | 97.1 ± 1.0 | 98.1 ± 1.1 | 86.3 ± 5.3 | 77.3 ± 2.5 | 78.4 ± 3.2 | 79.5 ± 4.9 | 88.8 ± 1.8 | 98.6 ± 0.6 | 98.6 ± 0.7 | 99.2 ± 0.2 | 99.1 ± 0.4 |
| AA | 85.5 ± 1.2 | 89.1 ± 1.3 | 97.9 ± 0.5 | 98.2 ± 0.8 | 90.2 ± 2.2 | 82.7 ± 1.4 | 84.9 ± 1.1 | 83.4 ± 1.6 | 89.9 ± 1.3 | 98.9 ± 0.4 | 99.1 ± 0.3 | 99.3 ± 0.2 | 99.3 ± 0.2 | |
| k | 75.1 ± 2.7 | 83.3 ± 2.6 | 96.2 ± 1.3 | 97.4 ± 1.4 | 82.5 ± 6.4 | 71.1 ± 2.8 | 72.6 ± 3.7 | 73.8 ± 5.7 | 85.3 ± 2.3 | 98.2 ± 0.8 | 98.1 ± 0.9 | 99.1 ± 0.3 | 98.9 ± 0.5 | |
| 30 | OA | 81.8 ± 1.3 | 89.4 ± 1.8 | 97.7 ± 1.1 | 98.5 ± 0.6 | 90.1 ± 2.5 | 79.1 ± 1.7 | 80.9 ± 2.4 | 83.9 ± 1.9 | 90.9 ± 1.8 | 99.1 ± 0.5 | 98.7 ± 1.7 | 99.4 ± 0.3 | 99.3 ± 0.4 |
| AA | 86.1 ± 0.8 | 90.6 ± 0.8 | 98.3 ± 0.6 | 98.6 ± 0.3 | 92.1 ± 1.9 | 83.6 ± 0.5 | 85.8 ± 0.8 | 85.6 ± 0.6 | 91.4 ± 1.2 | 99.3 ± 0.2 | 99.1 ± 0.8 | 99.4 ± 0.3 | 99.4 ± 0.1 | |
| k | 76.6 ± 1.5 | 86.1 ± 2.3 | 97.1 ± 1.3 | 98.1 ± 0.8 | 87.1 ± 3.1 | 73.1 ± 2.1 | 75.5 ± 2.9 | 79.1 ± 2.3 | 88.1 ± 2.4 | 98.8 ± 0.6 | 98.3 ± 2.2 | 99.2 ± 0.4 | 99.1 ± 0.5 | |
| Time (s) | 4.98 | 16.48 | 14.62 | 109.95 | 106.6 | 0.84 | 0.62 | 6.15 | 7.86 | 6.41 | 6.25 | 78.63 | 76.31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.; Cao, F.; Yang, Z.; Hong, X.; Huang, Y. Hyperspectral Image Denoising and Classification Using Multi-Scale Weighted EMAPs and Extreme Learning Machine. Electronics 2020, 9, 2137. https://doi.org/10.3390/electronics9122137
Liu M, Cao F, Yang Z, Hong X, Huang Y. Hyperspectral Image Denoising and Classification Using Multi-Scale Weighted EMAPs and Extreme Learning Machine. Electronics. 2020; 9(12):2137. https://doi.org/10.3390/electronics9122137
Chicago/Turabian StyleLiu, Meizhuang, Faxian Cao, Zhijing Yang, Xiaobin Hong, and Yuezhen Huang. 2020. "Hyperspectral Image Denoising and Classification Using Multi-Scale Weighted EMAPs and Extreme Learning Machine" Electronics 9, no. 12: 2137. https://doi.org/10.3390/electronics9122137
APA StyleLiu, M., Cao, F., Yang, Z., Hong, X., & Huang, Y. (2020). Hyperspectral Image Denoising and Classification Using Multi-Scale Weighted EMAPs and Extreme Learning Machine. Electronics, 9(12), 2137. https://doi.org/10.3390/electronics9122137





