GMDNet: Grouped Encoder-Mixer-Decoder Architecture Based on the Role of Modalities for Brain Tumor MRI Image Segmentation
Abstract
1. Introduction
- (1)
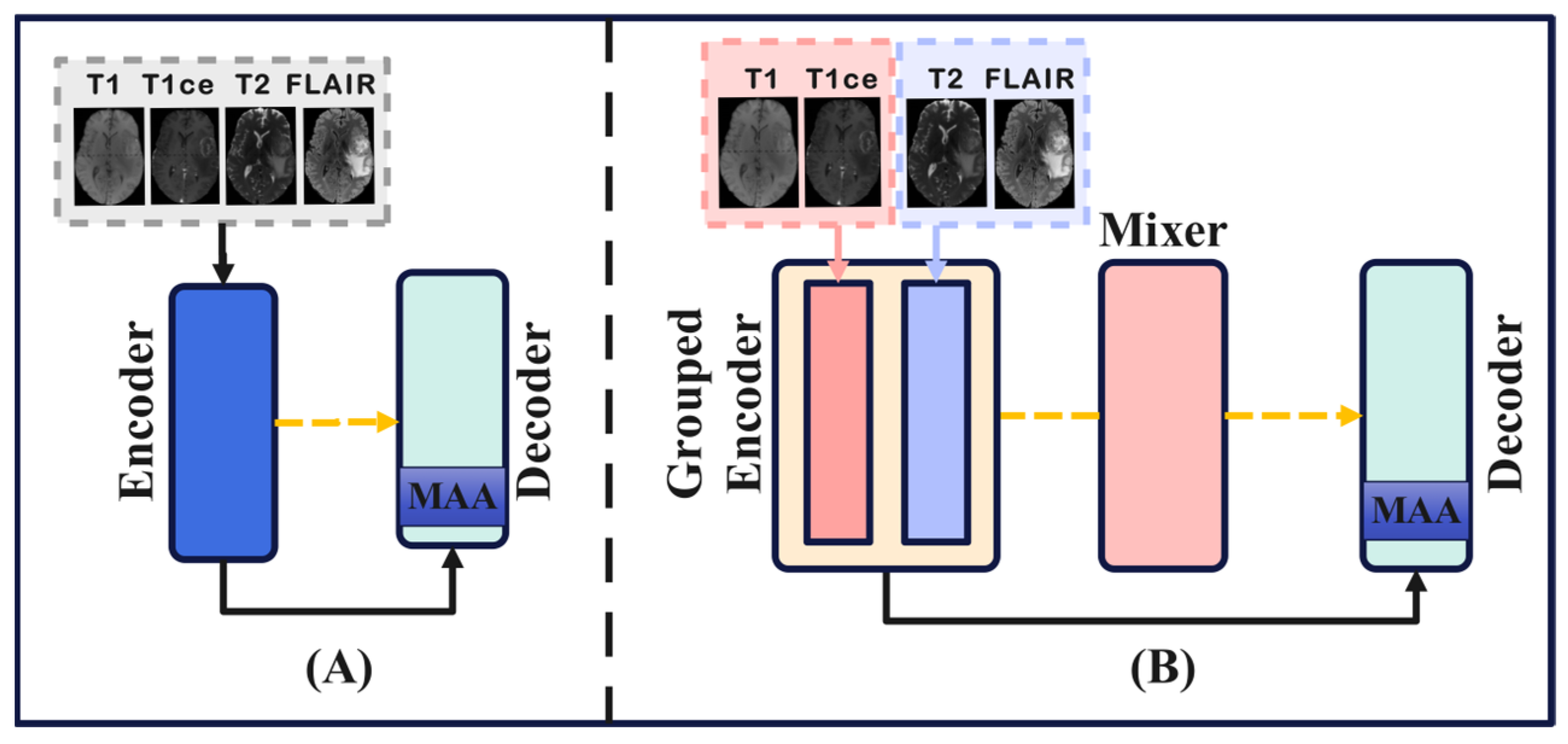
- This paper, for the first time, proposes a novel network architecture for brain tumor segmentation, consisting of Grouped Encoder, Mixer, and Decoder. This architecture effectively leverages the characteristics of the four MRI modalities in brain tumor imaging, thereby enhancing the segmentation performance.
- (2)
- Building on the architecture of GMD, we introduce a new brain tumor segmentation network called GMDNet (Grouped Encoder-Mixer-Decoder Network). On the BraTS 2018 dataset, the Dice scores for WT, TC, and ET were 91.21%, 87.11%, and 80.97%, respectively. On the BraTS 2021 dataset, the segmentation results for the ET, TC, and WT regions achieved Dice scores of 83.16%, 87.25%, and 91.87%, respectively.
- (3)
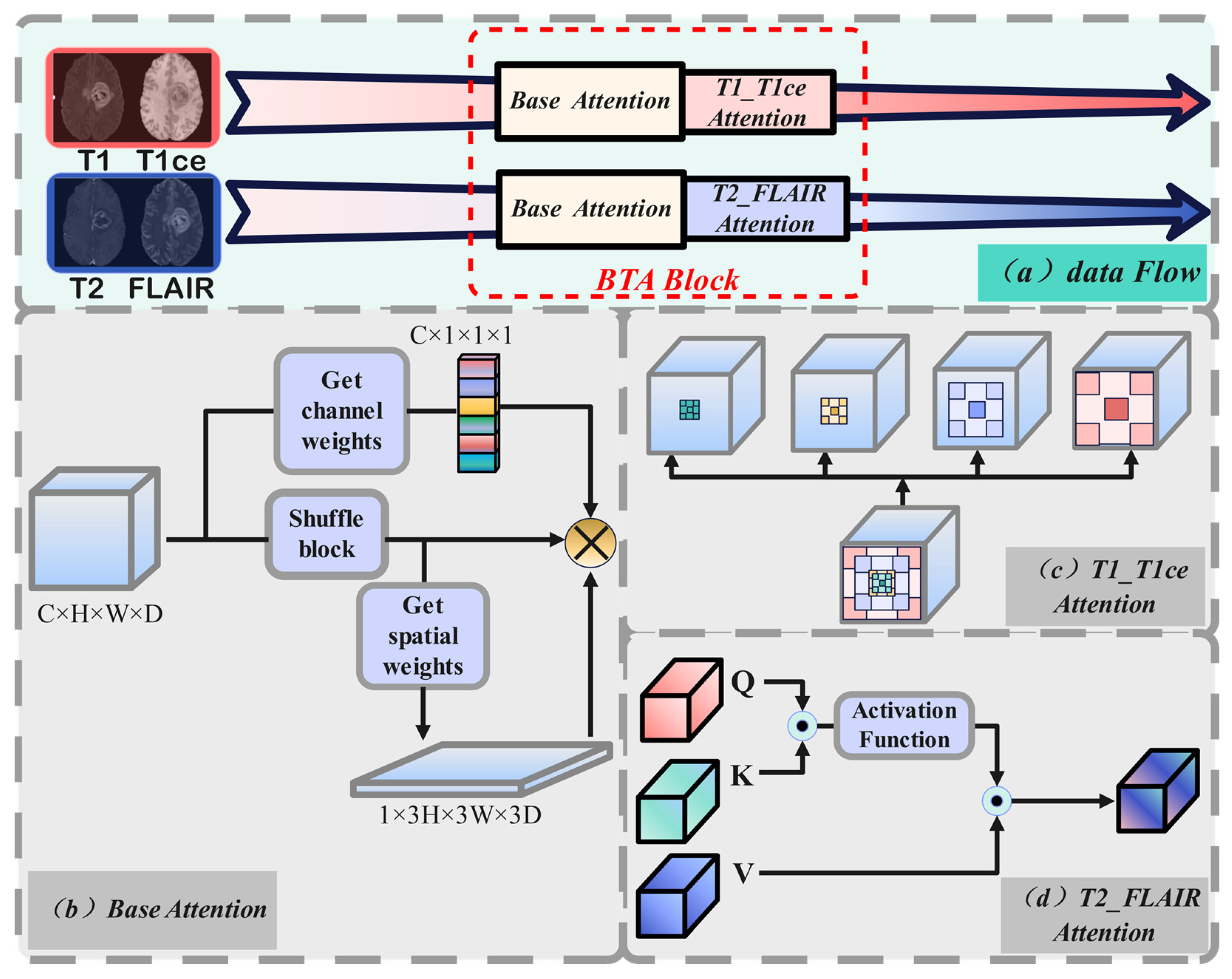
- The GMDNet architecture includes BTA (Base Attention and T1_T1ce and T2_FLAlR Modality Group Attention), MAA (Multi-Scale Axial Attention), and FMA (Feature Mixer Attention). Base attention and modality group attention employ different methods tailored to the characteristics of each modality. MAA extracts detailed information from the images at multiple scales and dimensions. FMA processes the mixed modality data by extracting features and then passes them to the decoder to aid in image reconstruction.
- (4)
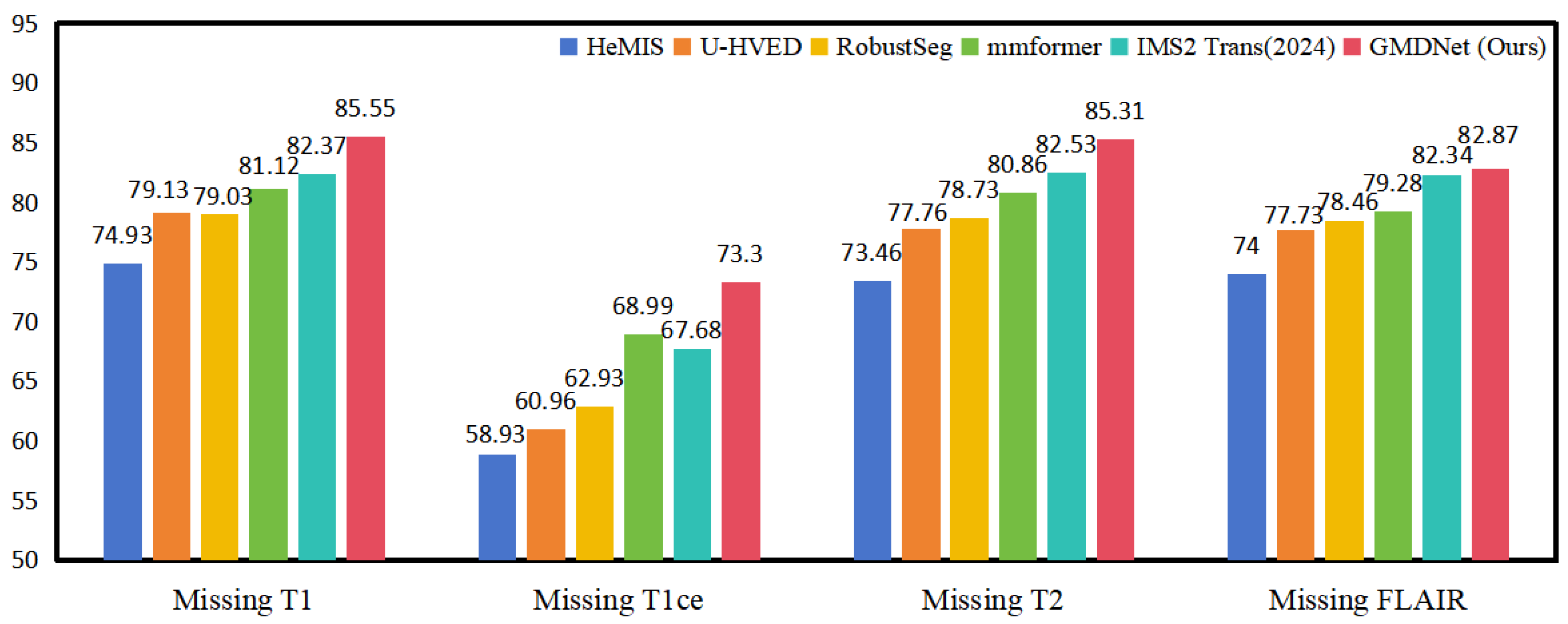
- Experiments conducted on brain tumor MRI images with incomplete modality demonstrate that GMDNet outperforms the compared networks in segmentation accuracy.
- (5)
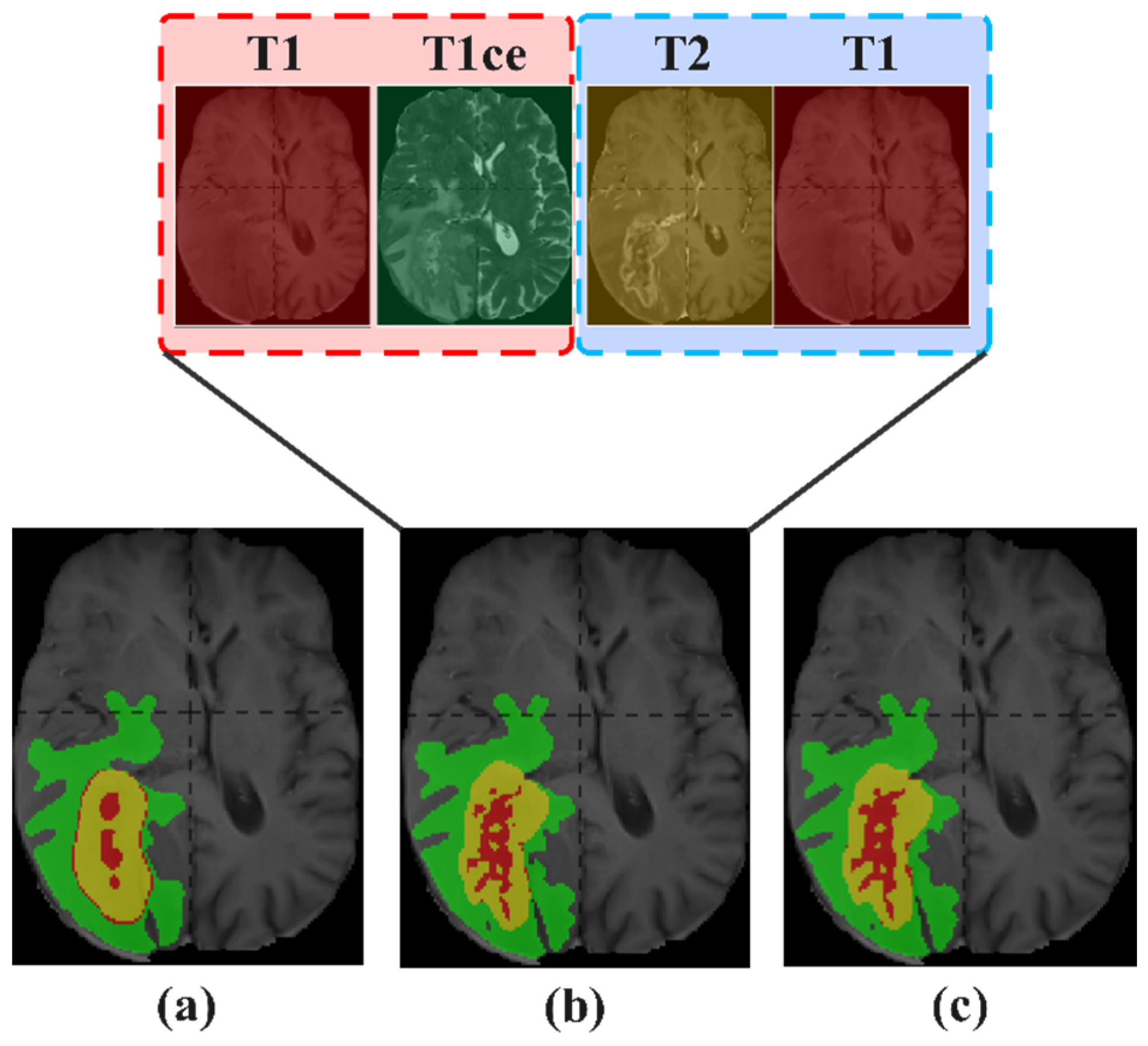
- To further improve the segmentation performance of brain tumor MRI images with incomplete modality, we propose for the first time a reuse modality strategy to enhance the overall segmentation precision of the model, laying the foundation for future research in this field.
- (6)
- Extensive experiments on the BraTS 2018 and BraTS 2021 datasets show that GMDNet achieves SOTA (State-of-The-Art) performance in brain tumor segmentation in both complete and incomplete modality, compared to the other networks evaluated in this study.
2. Related Research
2.1. Medical Image Segmentation Methods Based on Traditional Deep Learning
2.2. Medical Image Segmentation Methods Based on Modality Fusion
2.3. Medical Segmentation Methods Based on Incomplete Modality
3. Methodology
3.1. GMDNet Network Architecture
3.2. Grouped Encoder
3.3. Mixer
3.4. Decoder
4. Experiments
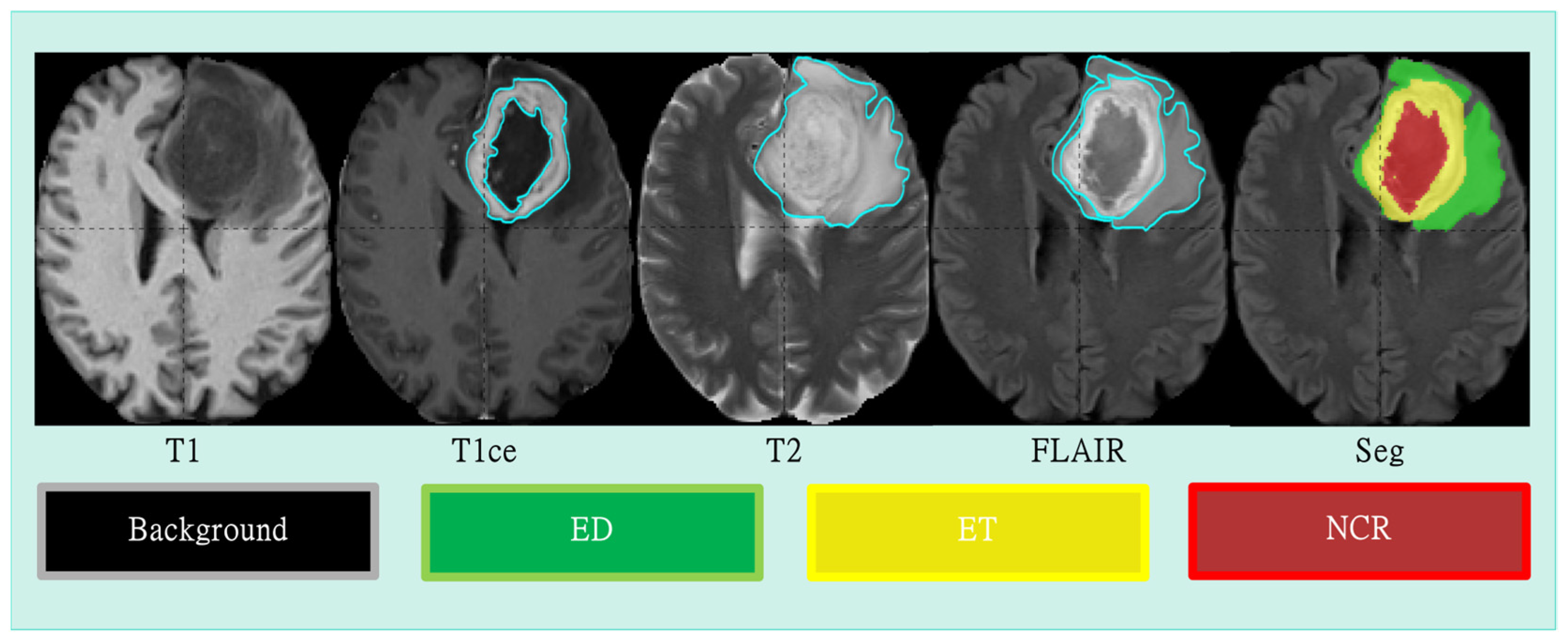
4.1. Datasets and Preprocessing
4.2. Implementation Details and Loss Function
4.3. Evaluation Metrics
5. Results and Analysis
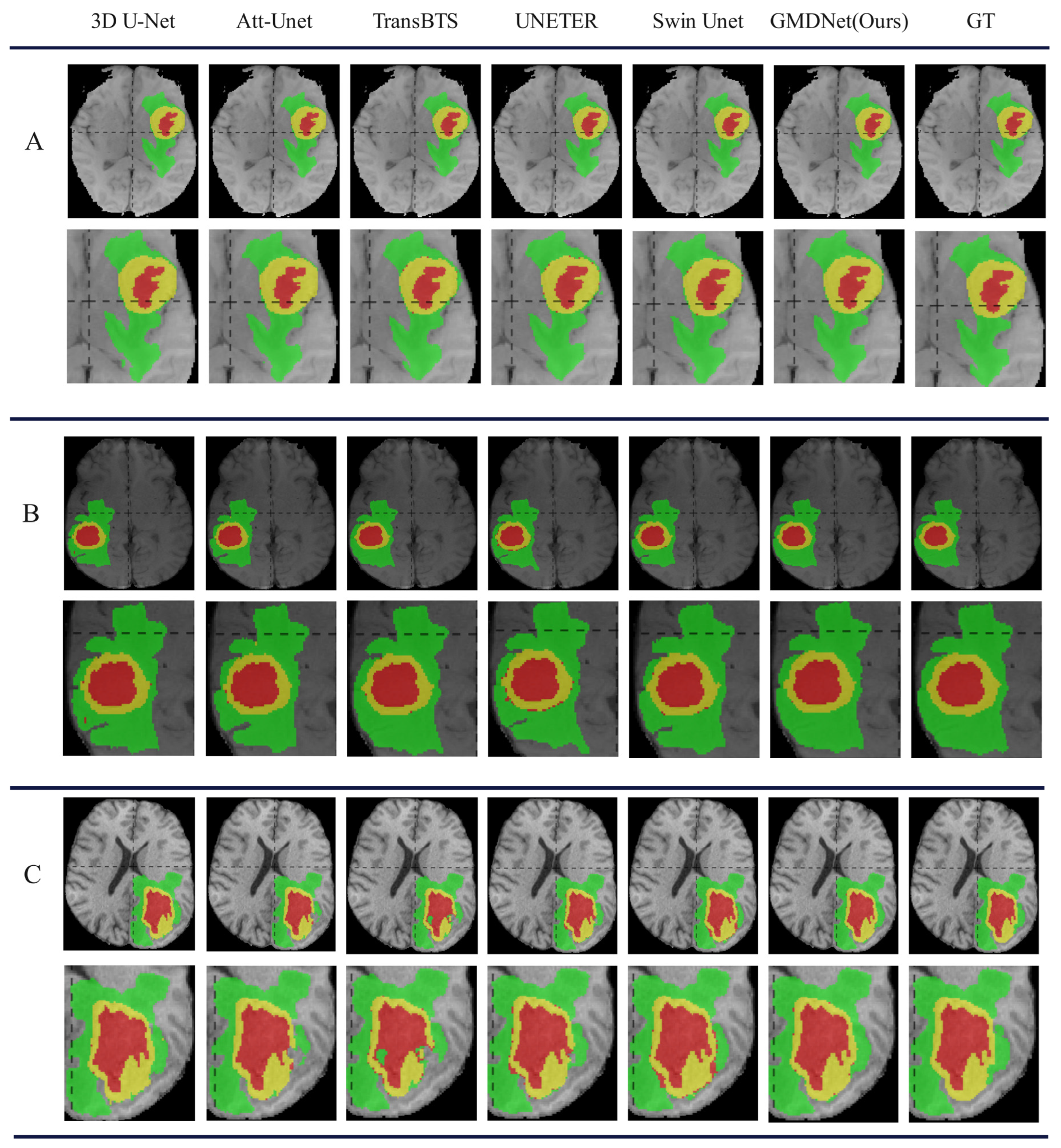
5.1. Complete Modality
5.1.1. Comparison with Methods in Complete Modality
5.1.2. Ablation Study of Each Component in GMDNet
5.1.3. Research on GMD Architecture
5.2. Incomplete Modality
5.2.1. Comparison with Methods in Incomplete Modality
5.2.2. Study on Single Modality
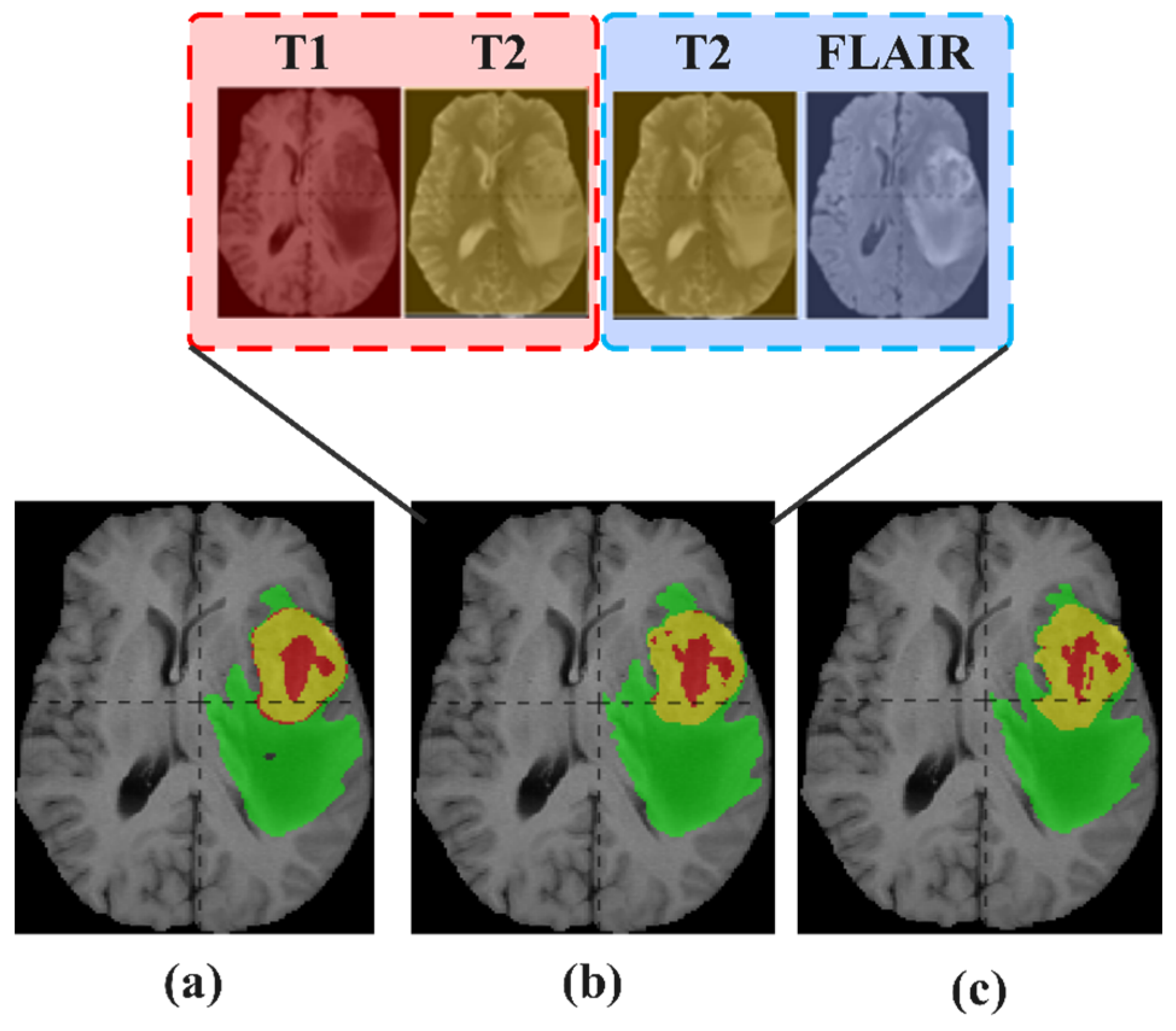
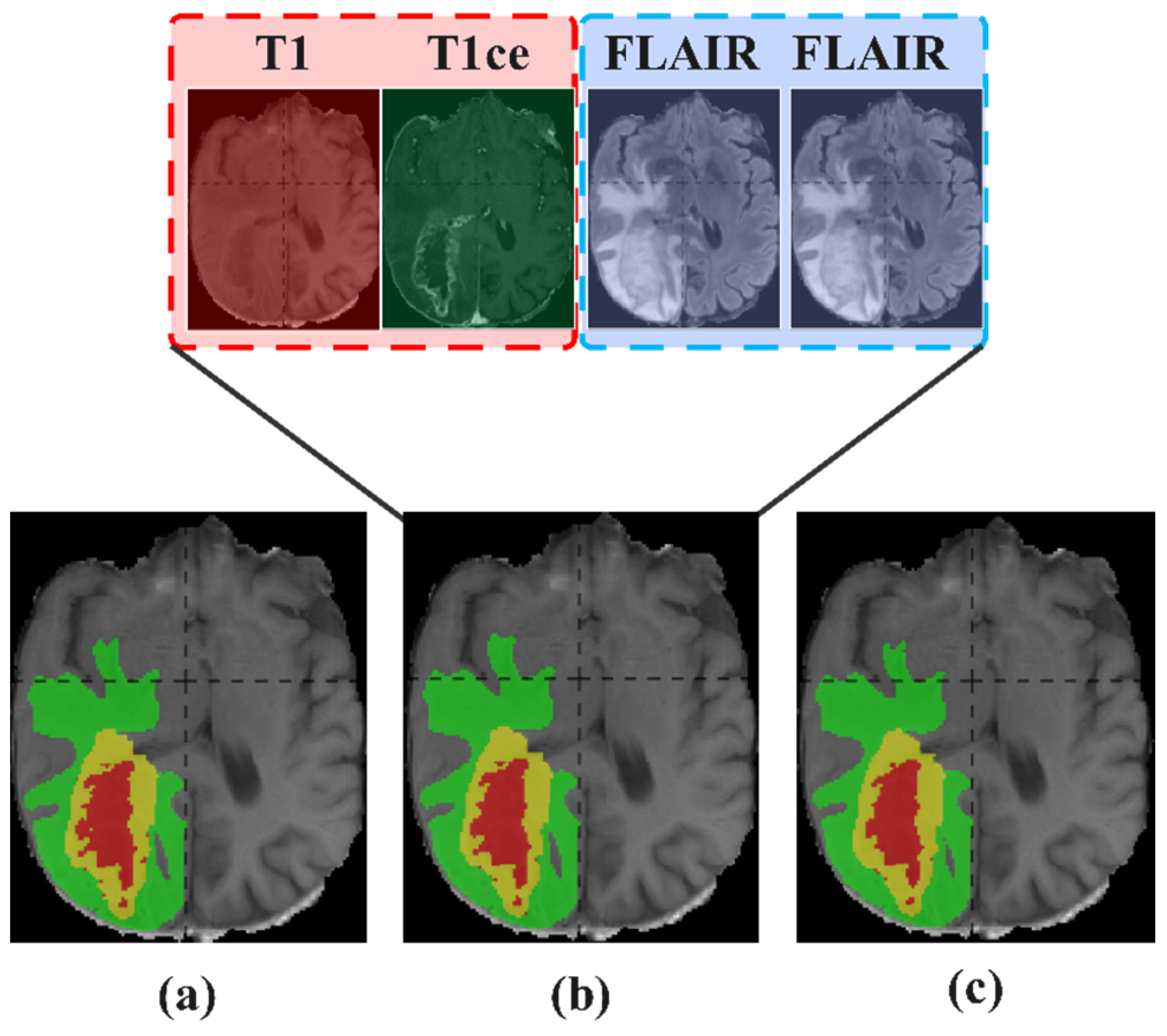
5.3. Reuse Modality Strategy
5.3.1. Study on Reuse Modality Performance of Missing T1
5.3.2. Study on Reuse Modality Performance of Missing T1ce
5.3.3. Study on Reuse Modality Performance of Missing T2
5.3.4. Study on Reuse Modality Performance of Missing FLAIR
5.3.5. Summary of Reuse Modality Strategy
6. Discussion and Conclusions
7. Limitations and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kleihues, P.; Burger, P.C.; Scheithauer, B.W. The new WHO classification of brain tumours. Brain Pathol. 1993, 3, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Rice, J.M. Inducible and transmissible genetic events and pediatric tumors of the nervous system. J. Radiat. Res. 2006, 47, B1–B11. [Google Scholar] [CrossRef] [PubMed]
- Persaud-Sharma, D.; Burns, J.; Trangle, J.; Sabyasachi, M. Disparities in brain cancer in the United States: A literature review of gliomas. Med. Sci. 2017, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Giese, A.; Westphal, M. Glioma invasion in the central nervous syste. Neurosurgery 1996, 39, 235–252. [Google Scholar] [CrossRef]
- Schneider, T.; Mawrin, C.; Scherlach, C.; Skalej, M.; Firsching, R. Gliomas in adult. Dtsch. Ärzteblatt Int. 2010, 107, 799–808. [Google Scholar] [CrossRef]
- Khalighi, S.; Reddy, K.; Midya, A.; Pandav, K.B.; Madabhushi, A.; Abedalthagafi, A. Artificial intelligence in neuro-oncology: Advances and challenges in brain tumor diagnosis, prognosis, and precision treatmen. NPJ Precis. Oncol. 2024, 8, 80. [Google Scholar] [CrossRef]
- Iqbal, S.; Khan, M.U.G.; Saba, T.; Rehman, A. Computer-assisted brain tumor type discrimination using magnetic resonance imaging feature. Biomed. Eng. Lett. 2018, 8, 5–28. [Google Scholar] [CrossRef]
- Najjar, R. Clinical applications, safety profiles, and future developments of contrast agents in modern radiology: A comprehensive review. iRADIOLOGY 2024, 2, 430–468. [Google Scholar] [CrossRef]
- Farhan, A.S.; Khalid, M.; Manzoor, U. XAI-MRI: An ensemble dual-modality approach for 3D brain tumor segmentation using magnetic resonance imagin. Front. Artif. Intell. 2025, 8, 1525240. [Google Scholar] [CrossRef]
- Howarth, C.; Hutton, C.; Deichmann, R. Improvement of the image quality of T1-weighted anatomical brain scans. Neuroimage 2006, 29, 930–937. [Google Scholar] [CrossRef]
- Lei, Y.; Xu, L.; Wang, X.; Zheng, B. IFGAN: Pre-to Post-Contrast Medical Image Synthesis Based on Interactive Frequency GA. Electronics 2024, 13, 4351. [Google Scholar] [CrossRef]
- Liu, Z.; Wei, J.; Li, R.; Zhou, J. Learning multi-modal brain tumor segmentation from privileged semi-paired MRI images with curriculum disentanglement learnin. Comput. Biol. Med. 2023, 159, 106927. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, V.M.; Piechnik, S.K.; Dall’Armellina, E.; Karamitsos, T.D.; Francis, J.M.; Choudhury, R.P.; Friedrich, M.J.; Robson, M.D.; Neubauer, S. Non-contrast T1-map** detects acute myocardial edema with high diagnostic accuracy: A comparison to T2-weighted cardiovascular magnetic resonanc. J. Cardiovasc. Magn. Reson. 2012, 14, 53. [Google Scholar] [CrossRef] [PubMed]
- Sati, P.; George, I.C.; Shea, C.D.; Gaitán, M.I.; Reich, D.S. FLAIR*: A combined MR contrast technique for visualizing white matter lesions and parenchymal veins. Radiology 2012, 265, 926–932. [Google Scholar] [CrossRef]
- Usuzaki, T.; Takahashi, K.; Inamori, R.; Morishita1, Y.; Shizukuishi1, T.; Takagi1, H.; Ishikur, M.; Obar, T.; Takase, K. Identifying key factors for predicting O6-Methylguanine-DNA methyltransferase status in adult patients with diffuse glioma: A multimodal analysis of demographics, radiomics, and MRI by variable Vision Transformer. Neuroradiology 2024, 66, 761–773. [Google Scholar] [CrossRef]
- Jiang, Z.; Capellán-Martín, D.; Parida, A.; Liu, X.; Ledesma-Carbayo, M.J.; Anwar, S.M.; Linguraru, M.G. Enhancing generalizability in brain tumor segmentation: Model ensemble with adaptive post-processing. In Proceedings of the 2024 IEEE International Symposium on Biomedical Imaging, Athens, Greece, 27–30 May 2024; pp. 1–4. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, C.; She, W. A multi-plane 2D medical image segmentation method combined with transformers. In Proceedings of the International Conference on Remote Sensing, Mapping, and Image Processing (RSMIP 2024), Xiamen, China, 21 June 2024; pp. 721–727. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016: 19th International Conference, Athens, Greece, 17–21 October 2016; pp. 424–432. [Google Scholar]
- Bukhari, S.T.; Mohy-ud-Din, H. E1D3 U-Net for brain tumor segmentation: Submission to the RSNA-ASNR-MICCAI BraTS 2021 challenge. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Proceedings of the International MICCAI Brainlesion Workshop, Virtual Event, 27 September 2021; Springer: Cham, Switzerland, 2021; pp. 276–288. [Google Scholar] [CrossRef]
- Jiao, C.; Yang, T.; Yan, Y.; Yang, A. Rftnet: Region–attention fusion network combined with dual-branch vision transformer for multimodal brain tumor image segmentatio. Electronics 2023, 13, 77. [Google Scholar] [CrossRef]
- Liu, X.; Song, L.; Liu, S.; Zhang, Y. A review of deep-learning-based medical image segmentation method. Sustainability 2021, 13, 1224. [Google Scholar] [CrossRef]
- Cai, L.; Gao, J.; Zhao, D. A review of the application of deep learning in medical image classification and segmentatio. Ann. Transl. Med. 2020, 8, 713. [Google Scholar] [CrossRef]
- Thakur, G.K.; Thakur, A.; Kulkarni, S.; Khan, N.; Khan, S. Deep learning approaches for medical image analysis and diagnosi. Cureus 2024, 16, e59507. [Google Scholar] [CrossRef]
- Rasool, N.; Bhat, J.I. Unveiling the complexity of medical imaging through deep learning approache. Chaos Theory Appl. 2023, 5, 267–280. [Google Scholar] [CrossRef]
- Mistry, J. Automated Knowledge Transfer for Medical Image Segmentation Using Deep Learnin. J. Xidian Univ. 2024, 18, 601–610. [Google Scholar]
- Sahiner, B.; Chan, H.P.; Petrick, N.; Wei, D.; Helvie, M.A.; Adler, D.D.; Goodsitt, M.M. Classification of mass and normal breast tissue: A convolution neural network classifier with spatial domain and texture image. IEEE Trans. Med. Imaging 1996, 15, 598–610. [Google Scholar] [CrossRef]
- Rawat, W.; Wang, Z. Deep convolutional neural networks for image classification: A comprehensive revie. Neural Comput. 2017, 29, 2352–2449. [Google Scholar] [CrossRef]
- Chen, L.; Li, S.; Bai, Q.; Yang, J.; Jiang, S.; Miao, Y. Review of image classification algorithms based on convolutional neural networks. Remote Sens. 2021, 13, 4712. [Google Scholar] [CrossRef]
- Weng, Y.; Zhou, T.; Li, Y.; Qiu, X. Nas-unet: Neural architecture search for medical image segmentatio. IEEE Access 2019, 7, 44247–44257. [Google Scholar] [CrossRef]
- Yasrab, R.; Gu, N.; Zhang, X. An encoder-decoder based convolution neural network (CNN) for future advanced driver assistance system (ADAS). Appl. Sci. 2017, 7, 312. [Google Scholar] [CrossRef]
- Zhang, J.; Luan, Z.; Qi, L.; Gong, X. MSDANet: A multi-scale dilation attention network for medical image segmentatio. Biomed. Signal Process. Control 2024, 90, 105889. [Google Scholar] [CrossRef]
- Nie, D.; Lu, J.; Zhang, H.; Adeli, E.; Wang, J.; Yu, Z.; Liu, L.Y.; Wang, Q.; Shen, D. Multi-channel 3D deep feature learning for survival time prediction of brain tumor patients using multi-modal neuroimages. Sci. Rep. 2019, 9, 1103. [Google Scholar] [CrossRef]
- Zucchet, N.; Orvieto, A. Recurrent neural networks: Vanishing and exploding gradients are not the end of the stor. Adv. Neural Inf. Process. Syst. 2024, 37, 139402–139443. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support Proceedings of the 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Granada, Spain, 20 September 2018; Held in Conjunction with MICCAI 2018; Springer: Cham, Switzerland, 2018; pp. 3–11. [Google Scholar] [CrossRef]
- Huang, X.; Deng, Z.; Li, D.; Yuan, X. Missformer: An effective medical image segmentation transformer. arXiv 2021, arXiv:2109.07162. [Google Scholar] [CrossRef]
- Jia, Q.; Shu, H. Bitr-unet: A cnn-transformer combined network for mri brain tumor segmentation. In Proceedings of the International MICCAI Brainlesion Workshop, Virtual Event, 27 September 2021; pp. 3–14. [Google Scholar] [CrossRef]
- Yu, M.; Han, M.; Li, X.; Wei, X.; Jiang, H.; Chen, H.; Yu, R. Adaptive soft erasure with edge self-attention for weakly supervised semantic segmentation: Thyroid ultrasound image case study. Comput. Biol. Med. 2022, 144, 105347. [Google Scholar] [CrossRef]
- Wang, W.; Chen, C.; Ding, M.; Yu, H.; Zha, S.; Li, J. TransBTS: Multimodal brain tumor segmentation using transformer. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Virtual Event, 27 September–1 October 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 109–119. [Google Scholar]
- Cai, Y.; Long, Y.; Han, Z.; Liu, M.; Zheng, Y.; Yang, W.; Chen, L. Swin Unet3D: A three-dimensionsal medical image segmentation network combining vision transformer and convolution. BMC Med. Inform. Decis. Mak. 2023, 23, 33. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, X.; Zhou, C.; Peng, H.; Zheng, Z.; Chen, J.; Ding, W. A review of cancer data fusion methods based on deep learning. Inf. Fusion 2024, 108, 102361. [Google Scholar] [CrossRef]
- Wang, J.; Li, X.; Ma, Z. Multi-Scale Three-Path Network (MSTP-Net): A new architecture for retinal vessel segmentation. Measurement 2025, 250, 117100. [Google Scholar] [CrossRef]
- Lin, J.; Lin, J.; Lu, C.; Chen, H.; Lin, H.; Zhao, B.; Shi, Z.; Qiu, B.; Pan, X.; Xu, Z. CKD-TransBTS: Clinical knowledge-driven hybrid transformer with modality-correlated cross-attention for brain tumor segmentation. IEEE Trans. Med. Imaging 2023, 42, 2451–2461. [Google Scholar] [CrossRef]
- Zhuang, Y.; Liu, H.; Song, E.; Hung, C.C. A 3D cross-modality feature interaction network with volumetric feature alignment for brain tumor and tissue segmentation. IEEE J. Biomed. Health Inf. 2022, 27, 75–86. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Hou, F.; Liu, Y.; Tian, J.; Zhong, C.; Zhang, Y.; He, Z. Modality-pairing learning for brain tumor segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Proceedings of the 6th International Workshop, BrainLes 2020, Lima, Peru, 4 October 2020; Held in Conjunction with MICCAI 2020, Revised Selected Papers, Part I 6; Springer: Cham, Switzerland, 2021; pp. 230–240. [Google Scholar] [CrossRef]
- Guo, B.; Cao, N.; Yang, P.; Zhang, R. SSGNet: Selective Multi-Scale Receptive Field and Kernel Self-Attention Based on Group-Wise Modality for Brain Tumor Segmentation. Electronics 2024, 13, 1915. [Google Scholar] [CrossRef]
- Zhou, T.; Canu, S.; Vera, P.; Ruan, S. Latent correlation representation learning for brain tumor segmentation with missing MRI modalitie. IEEE Trans. Image Process. 2021, 30, 4263–4274. [Google Scholar] [CrossRef]
- Yang, Q.; Guo, X.; Chen, Z.; Woo, P.Y.; Yuan, Y. D 2-Net: Dual disentanglement network for brain tumor segmentation with missing modalities. IEEE Trans. Med. Imaging 2022, 41, 2953–2964. [Google Scholar] [CrossRef]
- Zhou, T. Feature fusion and latent feature learning guided brain tumor segmentation and missing modality recovery networ. Pattern Recognit. 2023, 141, 109665. [Google Scholar] [CrossRef]
- Havaei, M.; Guizard, N.; Chapados, N.; Bengio, Y. Hemis: Hetero-modal image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016, Proceedings of the 19th International Conference, Athens, Greece, 17–21 October 2016; Proceedings, Part II 19; Springer: Cham, Switzerland, 2016; pp. 469–477. [Google Scholar] [CrossRef]
- Dorent, R.; Joutard, S.; Modat, M.; Ourselin, S.; Vercauteren, T. Hetero-modal variational encoder-decoder for joint modality completion and segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019, Proceedings of the 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part II 22; Springer: Cham, Switzerland, 2019; pp. 74–82. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, C.; Chen, T.; Chen, W.; Shen, Y. Scalable Swin Transformer network for brain tumor segmentation from incomplete MRI modalities. Artif. Intell. Med. 2024, 149, 102788. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Y.; Li, H.; Chai, Y.; Yang, Y. Deformation-aware and reconstruction-driven multimodal representation learning for brain tumor segmentation with missing modalities. Biomed. Signal Process. Control 2024, 91, 106012. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, G.; Zhang, Y.; Yue, H.; Liu, A.; Ou, Y.; Gong, J.; Sun, X. Tmformer: Token merging transformer for brain tumor segmentation with missing modalities. Proc. AAAI Conf. Artif. Intell. 2024, 38, 7414–7422. [Google Scholar] [CrossRef]
- Myronenko, A. 3D MRI brain tumor segmentation using autoencoder regularization. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Proceedings of the 4th International Workshop, BrainLes 2018, Granada, Spain, 16 September 2018; Held in Conjunction with MICCAI 2018, Revised Selected Papers, Part II 4; Springer: Cham, Switzerland, 2019; pp. 311–320. [Google Scholar] [CrossRef]
- Baumgartner, C.F.; Tezcan, K.C.; Chaitanya, K.; Hötker, A.M.; Muehlematter, U.J.; Schawkat, K.; Becker, A.S.; Donati, O.; Konukoglu, E. Phiseg: Capturing uncertainty in medical image segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019, Proceedings of the 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part II 22; Springer: Cham, Switzerland, 2019; pp. 119–127. [Google Scholar] [CrossRef]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar]
- Baid, U.; Ghodasara, S.; Mohan, S.; Bilello, M.; Calabrese, E.; Colak, E.; Farahani, K.; Kalpathy-Cramer, J.; Kitamura, F.C.; Pati, S. The rsna-asnr-miccai brats 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv 2021, arXiv:2107.02314. [Google Scholar] [CrossRef]
- Liu, L.; Jiang, H.; He, P.; Chen, W.; Liu, X.; Gao, J.; Han, J. On the variance of the adaptive learning rate and beyond. arXiv 2019, arXiv:1908.03265. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Kim, I.-S.; McLean, W. Computing the Hausdorff distance between two sets of parametric curves. Commun. Korean Math. Soc. 2013, 28, 833–850. [Google Scholar] [CrossRef]
- Aydin, O.U.; Taha, A.A.; Hilbert, A.; Khalil, A.A.; Galinovic, I.; Fiebach, J.B.; Frey, D.; Madai, V.I. On the usage of average Hausdorff distance for segmentation performance assessment: Hidden error when used for ranking. Eur. Radiol. Exp. 2021, 5, 4. [Google Scholar] [CrossRef]
- Chen, C.; Liu, X.; Ding, M.; Zheng, J.; Li, J. 3D dilated multi-fiber network for real-time brain tumor segmentation in MRI. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019, Proceedings of the 22nd International Conference, Shenzhen, China, 13–17 October 2019; Springer: Cham, Switzerland, 2019; pp. 184–192. [Google Scholar]
- Luo, Z.; Jia, Z.; Yuan, Z.; Peng, J. HDC-Net: Hierarchical decoupled convolution network for brain tumor segmentation. IEEE J. Biomed. Health Inf. 2020, 25, 737–745. [Google Scholar] [CrossRef]
- Zhang, Y.; He, N.; Yang, J.; Li, Y.; Wei, D.; Huang, Y.; Zhang, Y.; He, Z.; Zheng, Y. mmformer: Multimodal medical transformer for incomplete multimodal learning of brain tumor segmentation. In Medical Image Computing and Computer-Assisted Intervention, Proceedings of the 25th International Conference, Singapore, 18–22 September 2022; Springer Nature: Cham, Switzerland, 2022; pp. 107–117. [Google Scholar]
- Guan, X.; Zhao, Y.; Nyatega, C.O.; Li, Q. Brain tumor segmentation network with multi-view ensemble discrimination and kernel-sharing dilated convolution. Brain Sci. 2023, 13, 650. [Google Scholar] [CrossRef]
- Li, X.; Jiang, Y.; Li, M.; Zhang, J.; Yin, S.; Luo, H. MSFR-Net: Multi-modality and single-modality feature recalibration network for brain tumor segmentation. Med. Phys. 2023, 50, 2249–2262. [Google Scholar] [CrossRef]
- Liu, H.; Huang, J.; Li, Q.; Guan, X.; Tseng, M. A deep convolutional neural network for the automatic segmentation of glioblastoma brain tumor: Joint spatial pyramid module and attention mechanism network. Artif. Intell. Med. 2024, 148, 102776. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar] [CrossRef]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. Unetr: Transformers for 3d medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 574–584. [Google Scholar]
- Peiris, H.; Hayat, M.; Chen, Z.; Egan, G.; Harandi, M. A robust volumetric transformer for accurate 3D tumor segmentation. In Medical Image Computing and Computer-Assisted Intervention, Proceedings of the 25th International Conference, Singapore, 18–22 September 2022; Springer: Cham, Switzerland, 2022; pp. 162–172. [Google Scholar] [CrossRef]
- Tian, W.; Li, D.; Lv, M.; Huang, P. Axial attention convolutional neural network for brain tumor segmentation with multi-modality MRI scans. Brain Sci. 2022, 13, 12. [Google Scholar] [CrossRef]
- Wu, Q.; Pei, Y.; Cheng, Z.; Hu, X.; Wang, C. SDS-Net: A lightweight 3D convolutional neural network with multi-branch attention for multimodal brain tumor accurate segmentation. Math. Biosci. Eng. 2023, 20, 17384–17406. [Google Scholar] [CrossRef]
- Håversen, A.H.; Bavirisetti, D.P.; Kiss, G.H.; Lindseth, F. QT-UNet: A self-supervised self-querying all-Transformer U-Net for 3D segmentation. IEEE Access 2024, 12, 62664–62676. [Google Scholar] [CrossRef]
- Akbar, A.S.; Fatichah, C.; Suciati, N.; Za’in, C. Yaru3DFPN: A lightweight modified 3D UNet with feature pyramid network and combine thresholding for brain tumor segmentation. Neural Comput. Appl. 2024, 36, 7529–7544. [Google Scholar] [CrossRef]
- Chen, C.; Dou, Q.; Jin, Y.; Chen, H.; Qin, J.; Heng, P.A. Robust multimodal brain tumorsegmentation via feature disentanglement and gated fusion. In Medical Image Computing and Computer Assisted Intervention-MICCAI 2019, Proceedings of the 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part II 22; Springer: Cham, Switzerland, 2019; pp. 447–456. [Google Scholar] [CrossRef]









| Basic Configuration | Value |
|---|---|
| PyTorch Version | 1.10.0 |
| Python | 3.8.10 |
| GPU | NVIDIA GeForce RTX 4090 GPU (24 G) |
| Cuda | 11.3 |
| Learning Rate | 1 × 10−4 |
| Optimizer | Ranger |
| Batch Size | 1 |
| Methods | Dice (%) | HD 95 (mm) | ||||||
|---|---|---|---|---|---|---|---|---|
| WT | TC | ET | AVG | WT | TC | ET | AVG | |
| 3D U-Net [18] | 88.53 | 71.77 | 75.96 | 78.75 | 17.10 | 11.62 | 6.04 | 11.59 |
| V-Net [61] | 89.60 | 81.00 | 76.60 | 82.40 | 6.54 | 7.82 | 7.21 | 7.19 |
| DMFNet [65] | 89.90 | 83.50 | 78.10 | 83.83 | 4.86 | 7.74 | 3.38 | 5.33 |
| HDCNet [66] | 88.50 | 84.80 | 76.60 | 83.30 | 7.89 | 7.09 | 7.21 | 7.40 |
| TransUNet (2022) [38] | 89.95 | 82.04 | 78.38 | 83.46 | 7.11 | 7.67 | 4.28 | 6.35 |
| mmformer (2022) [67] | 89.64 | 85.78 | 77.61 | 84.34 | 4.43 | 8.04 | 3.27 | 5.25 |
| MVKS-Net (2023) [68] | 90.00 | 83.39 | 79.88 | 84.42 | 3.95 | 7.63 | 2.31 | 4.63 |
| MSFR-Net (2023) [69] | 90.90 | 85.80 | 80.70 | 85.80 | 4.24 | 6.72 | 2.73 | 4.82 |
| RFTNet (2024) [20] | 90.30 | 82.15 | 80.24 | 84.23 | 5.97 | 6.41 | 3.16 | 5.18 |
| SPA-Net (2024) [70] | 89.63 | 85.89 | 79.90 | 85.14 | 4.79 | 5.40 | 2.77 | 4.32 |
| GMDNet (Ours) | 91.21 | 87.11 | 80.97 | 86.43 | 4.43 | 5.57 | 2.63 | 4.21 |
| Methods | Dice (%) | HD 95 (mm) | ||||||
|---|---|---|---|---|---|---|---|---|
| WT | TC | ET | AVG | WT | TC | ET | AVG | |
| 3D U-Net [18] | 88.02 | 76.17 | 76.20 | 80.13 | 9.97 | 21.57 | 25.48 | 19.01 |
| Att-Unet [71] | 89.74 | 81.59 | 79.60 | 83.64 | 8.09 | 14.68 | 19.37 | 14.05 |
| UNETR [72] | 90.89 | 83.73 | 80.93 | 85.18 | 4.71 | 13.38 | 21.39 | 13.16 |
| TransBTS [38] | 90.45 | 83.49 | 81.17 | 85.03 | 6.77 | 10.14 | 18.94 | 11.95 |
| VT-UNet [73] | 91.66 | 84.41 | 80.75 | 85.61 | 4.11 | 13.20 | 15.08 | 10.80 |
| AABTS-Net (2022) [74] | 92.20 | 86.10 | 83.00 | 87.10 | 4.00 | 11.18 | 17.73 | 10.97 |
| E1D3 UNet (2022) [19] | 92.40 | 86.50 | 82.20 | 87.03 | 4.23 | 9.61 | 19.73 | 11.25 |
| Swin Unet3D (2023) [39] | 90.50 | 86.60 | 83.40 | 86.83 | - | - | - | - |
| SDS-Net (2023) [75] | 91.80 | 86.80 | 82.50 | 87.03 | 21.07 | 11.99 | 13.13 | 15.40 |
| QT-UNet-B (2024) [76] | 91.24 | 83.20 | 79.99 | 84.81 | 4.44 | 12.95 | 17.19 | 11.53 |
| Yaru3DFPN (2024) [77] | 92.02 | 86.27 | 80.90 | 86.40 | 4.09 | 8.43 | 21.91 | 11.48 |
| GMDNet (Ours) | 91.87 | 87.25 | 83.16 | 87.42 | 5.16 | 8.22 | 18.27 | 10.55 |
| Methods | WT | TC | ET | |||
|---|---|---|---|---|---|---|
| %Subjects | p | %Subjects | p | %Subjects | p | |
| GMDNet vs. 3D U-Net | 79.28 | 1.20 × 10−8 | 84.06 | 1.35 × 10−7 | 80.08 | 0.00129 |
| GMDNet vs. Att-Unet | 78.49 | 3.04 × 10−6 | 83.27 | 1.75 × 10−6 | 80.08 | 0.00419 |
| GMDNet vs. TransBTS | 84.06 | 1.75 × 10−6 | 85.26 | 2.60 × 10−7 | 83.27 | 1.83 × 10−6 |
| GMDNet vs. UNETR | 78.09 | 1.47 × 10−5 | 85.66 | 1.51 × 10−9 | 80.08 | 0.00187 |
| GMDNet vs. SwinUnet3D | 71.31 | 0.00724 | 81.27 | 0.00029 | 78.49 | 0.00296 |
| Experiment. | BTA | FMA | MAA | DP | Dice (%) | |||
|---|---|---|---|---|---|---|---|---|
| WT | TC | ET | Avg | |||||
| A (w/o Group) | 89.55 | 81.66 | 76.22 | 82.47 | ||||
| B | 91.65 | 84.70 | 76.14 | 84.16 | ||||
| C | √ | 91.80 | 86.17 | 82.66 | 86.87 | |||
| D | √ | 91.24 | 85.18 | 82.91 | 86.44 | |||
| E | √ | 89.91 | 85.08 | 81.36 | 85.45 | |||
| F | √ | √ | √ | 91.95 | 86.31 | 82.90 | 87.05 | |
| G (GMDNet) | √ | √ | √ | √ | 91.87 | 87.25 | 83.16 | 87.42 |
| Experiment | FLOPs | Parameter | Dice (%) | |||
|---|---|---|---|---|---|---|
| WT | TC | ET | Avg | |||
| A | 746.371 G | 48.128 M | 89.91 | 85.92 | 82.41 | 86.08 |
| B (GMDNet) | 989.921 G | 35.416 M | 91.87 | 87.25 | 83.16 | 87.42 |
| T1 | T1ce | T2 | FLAIR | Methods | Dice (%) | |||
|---|---|---|---|---|---|---|---|---|
| WT | TC | ET | Avg | |||||
| O | ● | ● | ● | HeMIS | 85.7 | 72.9 | 66.2 | 74.93 |
| U-HVED | 88.2 | 77.5 | 71.7 | 79.13 | ||||
| RobustSeg | 88.2 | 80.3 | 68.6 | 79.03 | ||||
| mmformer | 88.14 | 79.55 | 75.67 | 81.12 | ||||
| IMS2 Trans(2024) | 89.47 | 81.47 | 76.19 | 82.37 | ||||
| GMDNet (Ours) | 90.76 | 86.12 | 79.77 | 85.55 | ||||
| ● | O | ● | ● | HeMIS | 85.9 | 58.0 | 32.9 | 58.93 |
| U-HVED | 87.4 | 62.1 | 33.4 | 60.96 | ||||
| RobustSeg | 87.6 | 65.6 | 35.6 | 62.93 | ||||
| mmformer | 87.75 | 71.52 | 47.70 | 68.99 | ||||
| IMS2 Trans(2024) | 88.77 | 71.70 | 42.59 | 67.68 | ||||
| GMDNet (Ours) | 90.67 | 75.16 | 54.07 | 73.3 | ||||
| ● | ● | O | ● | HeMIS | 83.0 | 71.1 | 66.3 | 73.46 |
| U-HVED | 86.3 | 77.1 | 69.9 | 77.76 | ||||
| RobustSeg | 87.7 | 77.9 | 70.6 | 78.73 | ||||
| mmformer | 87.33 | 79.80 | 75.47 | 80.86 | ||||
| IMS2 Trans(2024) | 89.02 | 82.55 | 76.03 | 82.53 | ||||
| GMDNet (Ours) | 89.61 | 86.20 | 80.14 | 85.31 | ||||
| ● | ● | ● | O | HeMIS | 81.2 | 72.6 | 68.2 | 74 |
| U-HVED | 82.9 | 77.8 | 72.5 | 77.73 | ||||
| RobustSeg | 85.9 | 80.1 | 69.4 | 78.46 | ||||
| mmformer | 82.71 | 80.39 | 74.75 | 79.28 | ||||
| IMS2 Trans(2024) | 88.44 | 82.42 | 76.16 | 82.34 | ||||
| GMDNet (Ours) | 86.46 | 81.95 | 80.22 | 82.87 | ||||
| Modality | Dice (%) | |||
|---|---|---|---|---|
| WT | TC | ET | Avg | |
| T1 | 80.65 | 67.41 | 47.89 | 65.31 |
| T1ce | 80.19 | 78.53 | 79.47 | 79.39 |
| T2 | 87.93 | 72.07 | 54.03 | 71.34 |
| FLAIR | 91.04 | 70.70 | 48.93 | 70.22 |
| Modality | WT | TC | ET |
|---|---|---|---|
| T1 | Low | Low | Low |
| T1ce | Low | High | High |
| T2 | High | Medium | Low |
| FLAIR | High | Low | Low |
| Reuse Modality→Missing Modality | Dice (%) | |||
|---|---|---|---|---|
| WT | TC | ET | Avg | |
| Missing T1 | 91.83 | 86.28 | 81.3 | 86.47 |
| T1ce→T1 | 91.92 | 86.61 | 80.7 | 86.41 |
| T2→T1 | 91.88 | 85.28 | 80.71 | 85.95 |
| FLAIR→T1 | 92.01 | 86.31 | 83.2 | 87.17 |
| Reuse Modality→Missing Modality | Dice (%) | |||
|---|---|---|---|---|
| WT | TC | ET | Avg | |
| Missing T1ce | 90.46 | 75.2 | 54.23 | 73.29 |
| T1→T1ce | 91.51 | 75.67 | 56.86 | 74.68 |
| T2→T1ce | 91.48 | 76.32 | 57.42 | 75.07 |
| FLAIR→T1ce | 91.25 | 74.18 | 55.57 | 73.66 |
| Reuse Modality→Missing Modality | Dice (%) | |||
|---|---|---|---|---|
| WT | TC | ET | Avg | |
| Missing T2 | 91.55 | 85.83 | 82.00 | 86.46 |
| T1→T2 | 91.66 | 85.64 | 83.11 | 86.80 |
| T1ce→T2 | 91.34 | 85.72 | 82.97 | 86.67 |
| FLAIR→T2 | 91.71 | 86.49 | 82.53 | 86.91 |
| Reuse Modality→Missing Modality | Dice (%) | |||
|---|---|---|---|---|
| WT | TC | ET | Avg | |
| Missing FLAIR | 86.65 | 81.38 | 79.59 | 82.54 |
| T1→FLAIR | 89.53 | 86.36 | 82.78 | 86.22 |
| T2→FLAIR | 89.47 | 86.53 | 81.89 | 85.96 |
| T1ce→FLAIR | 89.36 | 85.66 | 82.89 | 85.97 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, P.; Zhang, R.; Hu, C.; Guo, B. GMDNet: Grouped Encoder-Mixer-Decoder Architecture Based on the Role of Modalities for Brain Tumor MRI Image Segmentation. Electronics 2025, 14, 1658. https://doi.org/10.3390/electronics14081658
Yang P, Zhang R, Hu C, Guo B. GMDNet: Grouped Encoder-Mixer-Decoder Architecture Based on the Role of Modalities for Brain Tumor MRI Image Segmentation. Electronics. 2025; 14(8):1658. https://doi.org/10.3390/electronics14081658
Chicago/Turabian StyleYang, Peng, Ruihao Zhang, Can Hu, and Bin Guo. 2025. "GMDNet: Grouped Encoder-Mixer-Decoder Architecture Based on the Role of Modalities for Brain Tumor MRI Image Segmentation" Electronics 14, no. 8: 1658. https://doi.org/10.3390/electronics14081658
APA StyleYang, P., Zhang, R., Hu, C., & Guo, B. (2025). GMDNet: Grouped Encoder-Mixer-Decoder Architecture Based on the Role of Modalities for Brain Tumor MRI Image Segmentation. Electronics, 14(8), 1658. https://doi.org/10.3390/electronics14081658






