1. Introduction
Electronic health records (EHRs) are invaluable data sources for healthcare research and administration, yet they contain sensitive patient information that must be protected [
1]. Regulatory frameworks such as the USA’s Health Insurance Portability and Accountability Act (HIPAA) and the European Union’s General Data Protection Regulation (GDPR) mandate the protection of personally identifiable information in healthcare data [
2,
3]. During the COVID-19 pandemic, the importance of preserving privacy became even more apparent, as extensive data collection via contact tracing highlighted the necessity for robust deidentification techniques [
4]. Deidentification techniques play a crucial role in preserving patient privacy by removing or anonymizing sensitive information.
Figure 1 illustrates the deidentification of a piece of Dutch medical text. The original text contains Protected Health Information (PHI) instances (Patient, Persoon, Locatie, Instelling, Datum, Leeftijd, Patientnummer, Telefoonnummer, Url; for translations to English, see Table 1). The text goes through the deidentification process through different NER techniques, until all PHI instances have been removed.
Despite advancements in deidentification techniques, no system is infallible. For example, the leading named-entity-recognition (NER) system [
5] in the benchmark CoNLL 2003 (English) (
https://www.clips.uantwerpen.be/conll2003/ner/ (accessed on 12 April 2025)) has an
score of 94.6% (
https://paperswithcode.com/paper/automated-concatenation-of-embeddings-for-1 (accessed on 12 April 2025)). Deidentification errors can lead to data leakage. To mitigate this risk, a conservative approach could involve aggressively removing any information suspected of being PHI. However, an overly aggressive strategy may result in excessive data loss, rendering the deidentified texts unusable due to the removal of essential non-PHI elements, such as drug names, medical codes, or other relevant clinical information. For example, Berg et al. [
6] evaluated the impact of the quality of deidentification on downstream clinical NER used to extract body parts, disorders, drugs, or findings from texts, and found that poor deidentification led to poorer downstream clinical NER performance.
Deidentification is by no means a new task, and indeed impressive performance on English-language datasets has been achieved. The authors in Neamatullah et al. [
7] reported a recall score of 0.967 for their pattern matching method, which was trained and evaluated on a test corpus comprising 1836 nursing notes. These systems do not generalize out-of-the-box to the Dutch language. As an example, two reasons are mentioned here. First, Dutch names often have
tussenvoegsels, i.e., words that come between the first name and the last name (such as van der or de). Such a format is not common in English, which can lead to names not being recognized, and therefore leaked. Second, Dutch institutions often have names such as Mondrian (
https://www.mondriaan.eu/ (accessed on 12 April 2025)), which do not give an indication of their nature. Again, this can lead to data leakage.
This paper presents a comparative study of two prominent deidentification approaches–rule-based and neural network methods–focusing on Dutch medical texts. The
Rule-Based Approach uses pre-defined rules, language knowledge, and domain-specific dictionaries to identify or replace PHI in text. The authors in Douglass et al. [
8] used lexical lookup tables, regular expressions, and heuristic methods to deidentify medical care text data in US hospitals. The authors in Menger et al. [
9] applied these techniques to a test corpus of Dutch medical care notes and treatment plans. They created a pattern-matching method called Deduce to automatically deidentify sensitive information. The
Machine-Learning approach [
10] consists of training a machine-learning system from labelled data to learn complex textual patterns and contextual structures, a process that, unlike rule-based methods, does not require explicit programming. The authors in Trienes et al. [
11] conducted a comparative study on rule-based and machine learning methods for Dutch medical record deidentification. Their labeling process, however, was different from that of Deduce. Therefore, their results do not adequately demonstrate the superiority of state-of-the-art machine learning architectures over rule-based methods in the deidentification of Dutch medical texts. We aim to answer the following research question:
How do the rule-based and neural network approaches to deidentification compare in terms of effectiveness, adaptability, and generalization when applied to Dutch medical data, and how can the strengths of these methodologies be used to improve the deidentification process and enhance patient privacy protection?
To address this question, we conducted a replication-extension study aimed at comparing deidentification methodologies in Dutch medical data, building upon the research carried out by Trienes et al. [
11] that investigated rule-based Menger et al. [
9] and neural network approaches.
We address this question by conducting comparative experiments using correctly labeled datasets and exploring the feasibility of both rule-based methods and deep learning algorithms for deidentification in Dutch medical texts. We provide clear, accurate results and insights into effective deidentification techniques by employing an effective assessment methodology. Ultimately, this research aims to contribute to the body of knowledge enriching privacy protection in the medical records of the Netherlands. By exploring established and emerging deidentification methods, we hope to offer a comprehensive view of the field as it stands today and provide insights for future improvement and development.
2. Materials and Methods
2.1. Data
This experiment relied on two dataset sources, each generated by a different method, to study deidentification methods in Dutch medical texts.
The initial dataset format used for this experiment is a collection of files in JSON Lines text format from the UMC Utrecht EHR system. This format is a convenient choice for working with large datasets since each row is a complete and independent data object.
The data consists of Dutch text, metadata, and span tags, reflecting various key information about individuals. Each row in the data set corresponds to a unique record, formatted as a JSON object. The main fields in the JSON object are “text”, “meta”, and “spans”.
The “text” field contains a string of text in Dutch, usually representing a medical context. It includes patient information such as name, email address, phone number, patient number, facility location, appointment date, and name of the treating physician.
The “meta” field contains additional metadata about the record, such as the individual’s first name, last name, initials, data source, year, and unique identifier (uid).
The ”spans” field is an array of objects, each object containing information about a particular tag present in the text. Each object consists of the start and end indices of the tags in the text and the tags themselves. The tags represent various types of identifiable information, such as “patient”, “person”, “location”, “institution”, “date”, “age”, “patient number”, “phone number”, and “url”.
In this study, two deidentification methods, Deduce and Deidentify, are compared. Deduce and Deidentify were chosen because they are the leading out-of-the-box deidentification systems for Dutch texts [
12,
13], which makes them the tools most likely to be used in real practice. Deduce is currently used at the UMC Utrecht. Deduce’s rule-based method uses a clear anonymization strategy. Deidentify offers an alternative strategy based on machine learning, which should increase its power, but does not prove its superiority over Deduce. To fairly compare the two, we must use Deduce’s labeling strategy for data annotation, and subsequently train a Deidentify model with the annotated data.
2.1.1. Annotated Dataset
This dataset was created at the UMC Utrecht, using an annotation strategy inspired by Deduce [
9]. Prodigy was used for document annotation, with version 1.10.8 in token mode. The annotation categories align with those specified by Deduce. The strategy was honed over multiple rounds of document annotation, with a particular challenge being the decision to annotate complete or partial words. Prodigy’s design makes whole-word tagging easier, but partial-word tagging is more in line with Deduce’s design. We settled for whole-word tagging for simplicity.
2.1.2. Synthetic Dataset
The second dataset source was synthetically generated using OpenAI’s GPT-4 model [
14] and manual annotations. This two-step approach aims to create a comprehensive, consistent, and specific patient medical dataset in Dutch. First, GPT-4 generated English medical data containing various key elements such as patient name, contact information, location, appointment details, and age. Previous research has demonstrated the effectiveness of using generative language models, such as LSTM and GPT-2, to create synthetic EHR datasets annotated for named-entity recognition, highlighting their utility for downstream NLP tasks like deidentification [
15]. The prompt given to GPT-4 was as follows:
I want you to help me to generate medical text. Within this medical text, it should include eight categories. Here is the example data:
Intake interview with Jan Jansen (e: j.g.jsnen_1966@email.com, t: 0612345678, patnr: 1243567). It concerns a 51-year-old man who will be treated at the outpatient clinic of the UMCU from 14 March to 31 July due to a heart attack. gloom complaints. The patient lives at Voorstraat 45b in Utrecht and will be treated here by Peter de Visser. Please generate 10 sets of such data for me.
After generating the English dataset, the text was translated into Dutch using Google Translate. The translated data was then fed back into the GPT-4 model to generate corresponding JSON files, containing patient information, metadata, and indexed tags. A custom Python script was used to accurately identify and annotate the start and end index of each label, creating a synthetic Dutch healthcare dataset containing information on 100 patients. This method provides a useful tool for generating medical datasets in Dutch while protecting real individuals’ privacy. All our code is at
https://github.com/PabloMosUU/DeidCompare (commit 9f6a8a1) (accessed on 12 April 2025).
2.2. Methodology
2.2.1. Deduce
Deduce [
9] is an automated method for deidentifying Dutch medical texts by masking private information. This process protects patient privacy and complies with local laws and regulations, such as the General Data Protection Regulation (GDPR). Deduce focuses on several Protected Health Information (PHI) categories, including names, addresses, social security numbers, etc. The method uses a combination of lookup tables, decision rules, and fuzzy string matching to deidentify sensitive information. Deduce has been shown to be effective in test environments, and it is used at the UMC Utrecht.
2.2.2. Deidentify
Deidentify [
11] represents a significant advancement in the field of automatic deidentification of health records, offering a practical solution to balance patient confidentiality with the exploitation of information in unstructured data. We use the pre-trained Deidentify model
bistmcrf_fast-v0.2.0.
Deidentify uses a neural approach termed BiLSTM-CRF (Bidirectional Long Short-Term Memory–Conditional Random Field). This approach minimizes the reliance on hand-crafted features intrinsic to traditional CRF-based deidentification, making it a more versatile and generalizable solution. The BiLSTM-CRF architecture, paired with contextual string embeddings, delivers state-of-the-art results for sequence labeling tasks.
2.3. Evaluation Metrics
We employed precision, recall, and their harmonic mean, the
score, defined as follows:
where
(true positives) is the number of correctly identified sensitive entities,
(false positives) is the number of non-sensitive entities incorrectly identified as sensitive, and
(false negatives) is the number of sensitive entities that were not identified. We do not consider partial entity matches.
4. Discussion
4.1. Deduce
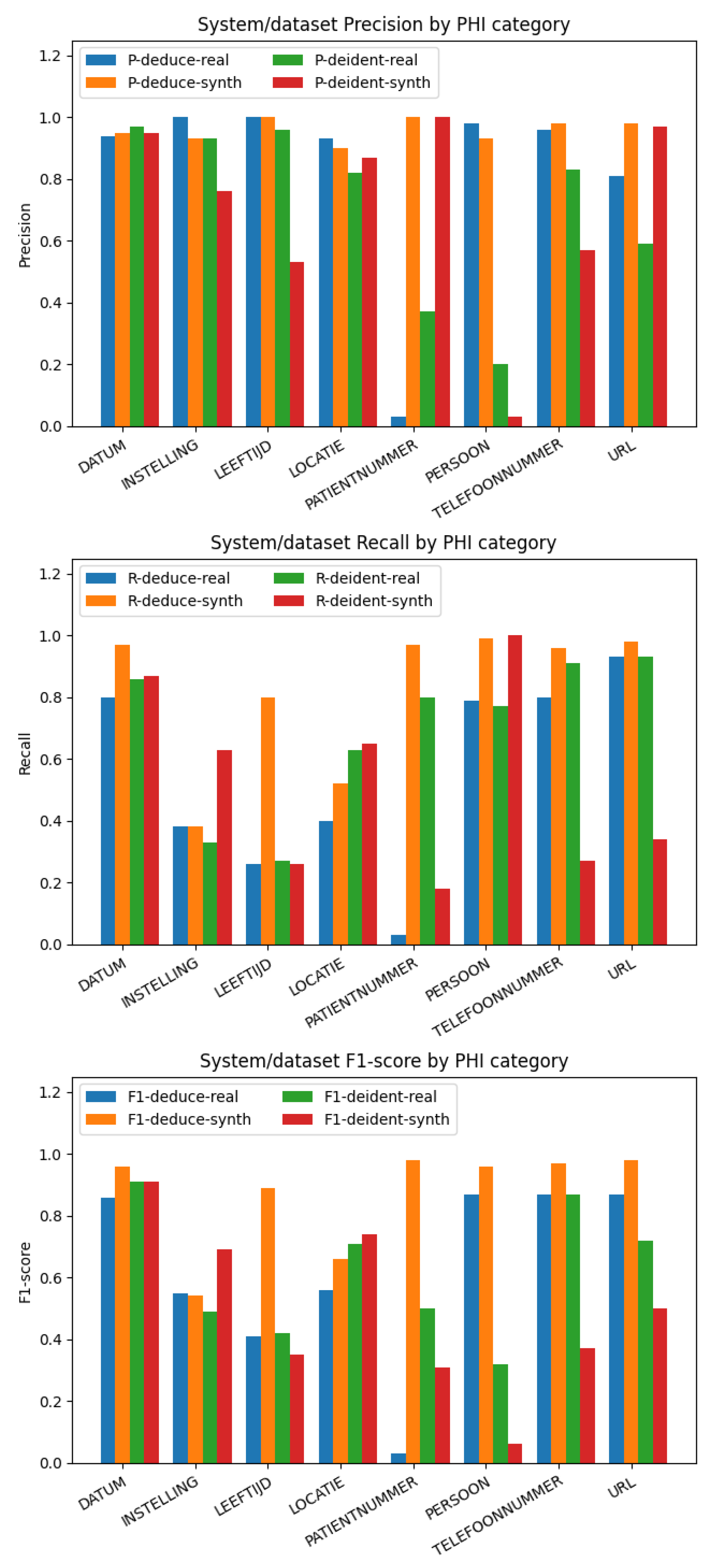
On the synthetic dataset, Deduce demonstrates a high score in tagging elements such as DATUM, LEEFTIJD, PATIENTNUMMER, PERSOON, TELEFOONNUMMER, and URL. The limitation of the detection of Dutch institution tags “INSTELLING” may be due to the fact that Deduce’s institution names are mainly from a list of psychiatric care institutions, and names made up by GPT-4 might not be in that list. In addition, there are various ways to express the name of the medical institution, such as different abbreviation formats or a reversed order of the text before and after. This variability can pose challenges for Deduce to identify them correctly. To overcome this problem, enriching Deduce’s data source of institution names is proposed. By integrating more diverse names of medical institutions across the Netherlands and employing flexible matching algorithms or natural language processing techniques, it should be possible to account for the different representation formats. Similarly, for the “LOCATIE” category, Deduce has predefined records for 2647 places of residence. In our Synthetic Dataset, geographical locations are complex and randomized, which means they might not always appear in the whitelist provided by Deduce. In some instances, the data might only contain the postcode and city without including the street and house number, further complicating the task. This complexity could lead to the observed lower recall rates.
On the real-world annotation-based dataset, Deduce performs well in DATUM, INSTELLING, LEEFTIJD, LOCATIE, PERSOON, and TELEFOONNUMMER, but PATIENTNUMMER has a surprisingly low performance, which means that Deduce does not recognize the correct patient number well in the real-world data set. In some instances, phone numbers are written with the prefix separate from the rest of the number, for example: 06 12345678. Deduce often makes an error in these cases, tagging “1234567” as a patient number instead of recognizing it as part of a phone number.
4.2. Deidentify
Deidentify shows mixed performance on the synthetic dataset. Interestingly, the precision of Deidentify was relatively high, except for the INSTELLING, LEEFTIJD, PERSOON, and TELEFOONNUMMER classes. However, Deduce outperformed Deidentify in recognizing INSTELLING, LEEFTIJD, LOCATIE, PERSOON, TELEFOONNUMMER, and URL. The recall of Deidentify was lower than that of Deduce for the DATUM, LEEFTIJD, PATIENTNUMMER, TELEFOONNUMMER, and URL classes. Despite Deidentify being a pre-trained neural network model, its performance on the same synthetic datasets used by Deduce was not outstanding. Only in the case of INSTELLING and LOCATIE were its generalization abilities superior to that of Deduce. Thus, the overall performance of Deidentify was lower than that of Deduce on synthetic datasets. In terms of overall performance, Deduce surpassed Deidentify. However, Deidentify demonstrated a better generalization ability in complex patterns such as INSTELLING and LEEFTIJD in unknown data.
Importantly, we note that we are using a pre-trained Deidentify model that was trained on data that was annotated using a different strategy from the one used by Deduce. Thus, like previous work that unfairly inflated Deidentify’s performance with respect to Deduce [
11], we acknowledge that we inflate the performance of Deduce with respect to Deidentify. This underscores the conclusion drawn in previous work that Dutch deidentification systems do not generalize well out of the box, and that further work on the data is required before using them [
9,
11]. This can be annotations, as well as the modification of rules of the re-training of machine-learning models. Future work should re-train the Deidentify model using data that was annotated in the Deduce format.
4.3. Generalizability
The observations regarding the performance of Deduce and Deidentify on our datasets encompass two phenomena. On the one hand, there are limitations to both systems regarding how well they can deidentify the texts they were designed to deidentify. On the other hand, there is the challenge of generalizing to a new dataset. Comparing our results in
Table 1 and
Table 2 to those of previous work [
9,
11], we note that Deduce performs much worse on institutions on our datasets than in the original paper. This is likely due to the fact that Deduce uses curated lists of institutions for deidentification, and our new and synthetic datasets include institutions that are probably not in the list in Deduce. This highlights the importance of tailoring a rule-based system such as Deduce before use, which can be a time-consuming process.
Interestingly, Deidentify showed a performance of 0.74 and 0.71 on locations on the synthetic and annotated datasets, respectively, while the original study had a performance of 0.84 on addresses. Although our observed performance is lower, this can be explained by the fact that we did not fine-tune the model on our dataset, as pointed out in
Section 4.2. It would be interesting to repeat this study on data from outside the Netherlands, where addresses have a different format, to see if the performance would drop dramatically in that case.
5. Conclusions
Our experiments revealed that both Deduce and Deidentify have strengths and limitations, demonstrating variable performance with Dutch medical data. In fixed-format synthetic data, Deduce surpasses Deidentify in terms of effectiveness, achieving a higher score for several categories like DATUM, LEEFTIJD, PATIENTNUMMER, PERSOON, TELEFOONNUMMER, and URL, and on average. Its rule-based approach, while requiring continuous rule modification and updating, provides a more precise and effective method for deidentifying structured data and numeric categories. On the other hand, Deidentify’s neural network approach demonstrates better adaptability and generalization capability when handling complex patterns like DATUM and LOCATIE on unknown or real-world data.
When facing real-world data, after verifying the true labels of the dataset with accurate format checks and ensuring no errors, for entity categories such as INSTELLING, PATIENTNUMMER, PERSOON, TELEFOONNUMMER, and URL, the Deduce method should be employed for deidentification. However, before utilizing Deduce, the lookup list, trie-based hashing, and regular expressions should be appropriately modified to enhance Deduce’s performance. For the other entity categories, the Deidentify method can be considered for deidentification. Importantly, before using Deidentify, it would be beneficial to fine-tune the pre-trained model of Deidentify on the required dataset. By utilizing the performance of Deduce in deidentifying structured and numeric categories with Deidentify’s adaptability and generalization ability for complex and unstructured data, the de- identification techniques’ effectiveness, adaptability, and generalization on Dutch medical text can be improved. Thus, we show a summary of the recommended deidentification methods for different PHI categories in
Table 3. In essence, this study sheds light on the distinct strengths of Deduce and Deidentify. However, the full spectrum of their comparative efficacies can only be deeply understood with broader access to the real-world datasets. Consequently, while the findings guide the current best practices in Dutch medical text deidentification, there remains a window of opportunity for further research, especially with access to richer and diverse real-world datasets.
6. Limitations and Future Work
As mentioned elsewhere in the paper, the current study does not fine-tune Deidentify on our datasets, nor does it curate the Deduce lists specifically for our purposes. Thus, both models are likely to underperform to some degree on our datasets. This is especially the case for Deidentify, which used a different annotation strategy at training time. In future work, we plan to fine-tune Deidentify and carefully evaluate the white lists used by Deduce before running the comparison.
The performance of deidentification methods is not only important for legal reasons, but it might also have implications on downstream tasks [
6]. For this reason, in future work, we plan to use our data for a downstream task such as violence risk assessment [
16], and use both deidentification systems, to see how the performance on the downstream task varies.
Finally, as mentioned in
Section 4.3, our study focused on Dutch clinical text. Some of the labels would perform significantly differently on non-clinical text or non-Dutch text. As an example, consider deidentifying addresses in countries with a different system for addresses. In future work, we will attempt to generate synthetic data from other countries to see how these deidentification systems perform.