Abstract
The increasing complexity and importance of medical data in improving patient care, advancing research, and optimizing healthcare systems led to the proposal of this study, which presents a novel methodology by evaluating the sensitivity of artificial intelligence (AI) algorithms when provided with real data, synthetic data, a mix of both, and synthetic features. Two medical datasets, the Pima Indians Diabetes Database (PIDD) and the Breast Cancer Wisconsin Dataset (BCWD), were used, employing the Gaussian Copula Synthesizer (GCS) and the Synthetic Minority Oversampling Technique (SMOTE) to generate synthetic data. We classified the new datasets using fourteen machine learning (ML) models incorporated into PyCaret AutoML (Automated Machine Learning) and two deep neural networks, evaluating performance using accuracy (ACC), F1-score, Area Under the Curve (AUC), Matthews Correlation Coefficient (MCC), and Kappa metrics. Local Interpretable Model-agnostic Explanations (LIME) provided the explanation and justification for classification results. The quality and content of the medical data are very important; thus, when the classification of original data is unsatisfactory, a good recommendation is to create synthetic data with the SMOTE technique, where an accuracy of 0.924 is obtained, and supply the AI algorithms with a combination of original and synthetic data.
1. Introduction
The dynamic of artificial intelligence brings many benefits in all activity domains, mainly in the medical field. The analysis of large datasets containing patient details informs public health decisions. Given the importance of healthcare, it is essential that AI tools generating medical datasets produce accurate and reliable results. These types of data can have undesired effects on the training and testing of AI classifiers.
Healthcare datasets contain extensive amounts of data about people’s health. These databases typically contain a wide range of patient data, including diagnostics, prescription usage, medical history, and the results of diagnostic tests. One of the many creative ways organizations can share datasets with more researchers is by using synthetic data. However, there are not many papers that examine the possibilities and uses in healthcare [1,2].
In data augmentation and data generation, creating synthetic data that resemble real data is essential. Computer simulations or algorithms artificially generate synthetic data to replace real-world data. It is used when real data are scarce, unavailable, or confidential and helps in training and testing machine learning models without privacy concerns. Synthetic data find their applications in a multitude of contexts. In machine learning, adequate, high-quality data continue to be necessary. Sometimes, privacy issues prevent access to real data, and other times it seems like there are not enough data to train the machine learning model.
Data or metadata, whether real or synthetic, supply the ML and AutoML. Synthetic data are typically obtained from real data. A synthetic data generator can produce a synthetic dataset. The most effective synthetic data generators (SDGs) were made in the Python environment. These include data profiling, which offers advanced exploratory data analysis that helps users understand the data consistently and visually, letting them quickly analyze them [3]; the Synthetic Data Vault (SDV), which creates generative models of relational databases [4]; and the Gretel-synthetics package, which lets you make a lot of changes to the data you create [5]. The proposed study employs the Gaussian Copula Synthesizer, a statistical method that trains a model to generate synthetic data [6] and SMOTE.
AutoML is an innovative approach that facilitates the exploration of multiple options to identify the optimal models by automating the entire ML pipeline. AutoML aims to decrease the need for data scientists by allowing domain experts to automatically create machine learning applications with little to no statistical or machine learning expertise [7]. We proposed PyCaret AutoML, a completely automated, supervised learning system that is extremely scalable and streamlines the training process of numerous candidate models and stacked ensembles into a single function [8].
In practice, classification or regression tasks use various machine learning models, each containing a wide range of algorithms, but not all of them are sensitive to synthetic data. Starting with this idea, we bring a novel approach, testing the sensibility of synthetic data with PyCaret AutoML. A justification of classification and important features by an XAI tool is necessary.
Due to the lack of transparency in the outcomes provided by AI algorithms, resulting from the data quality that powers them, testing with engineering methods such as synthetic data and feature creation is necessary. Considering medical data, it is essential that these datasets are clean, as they can significantly influence the binary classification of diagnostics. In this context, to achieve confident outcomes, the sensitivity of AI algorithms applied in the medical field can be evaluated using three approaches: original data, a combination of original and synthetic data, and original data enhanced with synthetic features.
A set of processes and approaches termed “explainable AI” aims to provide a clear and comprehensible rationale for the decisions made by artificial intelligence and machine learning models. LIME is a prevalent explainable artificial intelligence (XAI) method that aims to elucidate the local structure of a certain model instance [9].
In this sense, we brought the following contributions:
- (i)
- Proposing the BCWD and PIDD publicly available datasets and building from them the new synthetic dataset (SD) using GCS and SMOTE techniques;
- (ii)
- Synthetic feature creation for the PIDD by applying equal-width and equal-frequency discretization techniques and transforming numerical features into categorical. Combining these synthetic features with the original features improved the classification results.
- (iii)
- PyCaret AutoML and two DNNs were supplied by both PIDD and BCWD datasets, including the original, synthetic, and mixed versions.
- (iv)
- From the confusion matrix, the ACC, AUC, F1-score, MCC, and Kappa metrics were computed.
- (v)
- Explaining the classification with the LIME XAI tool was achieved.
2. Related Works
Recent advances in combining synthetic data generation with explainable AI (XAI) methods have opened new frontiers in machine learning, particularly in addressing privacy concerns, improving model transparency, and enabling more robust testing of model explanations. These advances offer several promising directions for research and practical applications [10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44].
In the medical domain, the research in synthetic data is growing in two key areas: (i) developing new methods for generating synthetic data and (ii) testing how well these methods work in real-world applications. Modern synthetic data generators (SDG) [20,21] include Generative Adversarial Networks (GANs) [22,23], Variational Autoencoders (VAEs) [24], Gaussian Copula Synthesizer [25], and Differentially Private Synthetic Data Generation (DPSDG) [26]. However, despite growing interest, the field of synthetic data still lacks a clear, unified definition [20]. This leads to inconsistent interpretations and uses across different contexts, which affects transparency and reproducibility in research involving synthetic data.
On the other hand, Explainable AI (XAI) methods are crucial for interpreting and understanding the decisions made by machine learning models, particularly in complex or “black box” algorithms. Notable XAI methods include LIME (Local Interpretable Model-agnostic Explanations), which provides local approximations; SHapley Additive exPlanations (SHAP), which offers consistent feature attribution; Saliency Maps, which highlight influential areas in image-based models; and Counterfactual Explanations (CE), which demonstrate how slight input changes affect predictions. These methods enhance transparency and trust in AI systems.
A growing number of studies are actively exploring how machine learning (ML), synthetic data (SD), and explainable AI (XAI) methods can be applied to enhance medical technologies and improve patient care. For instance, a study [27] proposes a novel XAI method called Feature Selection with Explainable Random Forest (FSXRF) to improve COVID-19 diagnosis from blood test samples. This approach integrates feature selection techniques with a random forest (RF) classifier to improve diagnostic accuracy. The machine learning-based method achieved several key performance metrics: accuracy was 89.98%, the F1 score was 78.12%, sensitivity reached 71.69%, specificity was 92.96%, and the Area Under the Receiver Operating Characteristic Curve (AUROC) was 92.88%. The study shows the use of explainable AI techniques, specifically Local Interpretable Model-Agnostic Explanations (LIME), to provide transparency by highlighting the most influential features in the diagnosis process. In another study [28], the researchers used ten machine learning (ML) algorithms to predict heart disease risk, with a particular focus on support vector machines (SVM), XGBoost, bagging, Decision Trees (DT), and Random Forests (RF). Among these methods, the XGBoost algorithm achieved the highest performance, with an accuracy of 97.57%, a sensitivity of 96.61%, a specificity of 90.48%, a precision of 95.00%, an F1 score of 92.68%, and an AUC of 98%. The study also implemented an explainable AI method using SHAP to clarify the model’s predictions. In [29], the use of machine learning (ML) models for heart disease prediction using medical data is explored. The study assesses various classification algorithms, including Logistic Regression (LR), K-Nearest Neighbors (KNN), Decision Trees (DT), XGBoost, and CatBoost. Among these, XGBoost and CatBoost outperformed the others, particularly in terms of both accuracy (97%) and interpretability. To further enhance the transparency of these predictions, the study incorporates Explainable AI (XAI) methods such as Conformalized Quantile Regression (CQR) and the Explainable Boosting Classifier (EBC). Another study [30] presents an automatic system for predicting diabetes using machine learning techniques, developed with a private dataset of female patients from Bangladesh, supplemented by the Pima Indian diabetes dataset. To improve the model’s accuracy, a feature selection algorithm based on mutual information was used, and a semi-supervised model with extreme gradient boosting (XGBoost) was applied to predict insulin-related features. The study addresses the issue of class imbalance by using SMOTE and Adaptive Synthetic (ADASYN) methods. Several ML models, including DT, SVM, RF, LR, KNN, and various ensemble techniques, were tested. Among these, the XGBoost classifier with the ADASYN approach performed best, achieving 81% accuracy, an F1 score of 0.81, and an AUC of 0.84. To enhance the system’s transparency, explainable AI techniques using LIME and SHAP were integrated, helping to clarify the model’s predictions. Similarly, the study [31] evaluated several ML models, including Artificial Neural Networks (ANN), Random Forest, Support Vector Machines, Logistic Regression, AdaBoost, and XGBoost. The researchers employed an ensemble model that used a soft voting mechanism to combine predictions from these diverse algorithms, leading to significant improvements in both accuracy and interpretability. To enhance transparency, explainable AI techniques, specifically SHAP, were used to provide insights into how each model arrived at its predictions. The proposed method achieved a balanced accuracy of 90% and an F1 score of 89%, underscoring its effectiveness in identifying diabetes risk while making the decision-making process comprehensible.
However, a crucial aspect often overlooked in practice is how these algorithms handle corrupted or noisy data. While some algorithms can maintain high performance in the presence of corrupted data, others can be significantly impacted, leading to poor generalization, reduced accuracy, and faulty predictions. In this direction, various methods [32,33] were proposed to address the presence of noisy or corrupted data (such as noisy labels, missing values, and outliers) that can affect machine learning models in medical applications, where high-quality labels are critical. Furthermore, assessing the impact of blending real and synthetic data to evaluate the performance of machine learning algorithms is also important. A proper evaluation can confirm if blending improves model reliability while maintaining performance standards.
In this context, our study introduces a novel approach by testing the sensitivity of machine learning models when they are supplied with both real and synthetic data. This approach allows us to explore how well the models can adapt to diverse data sources, helping to identify potential weaknesses or strengths in their generalization capabilities. Understanding this sensitivity is crucial, particularly in applications such as healthcare, where data quality and diversity are critical to model performance. Additionally, we use LIME to gain deeper insights into how these models make predictions when trained on both real and synthetic data.
3. Methodology
The proposed methodology expressed by Figure 1 comprises four main steps. (i) The first step involves presenting the public medical dataset and dividing it into a 70:30 training/testing ratio. (ii) The “new dataset” block outlines all new datasets and the methods for preprocessing the initial data; (iii) The PyCaret AutoML tools, equipped with fourteen ML classifiers and two DNNs, receive the datasets from the previous block. (iv) The final block includes the quantization and explanation of the classification.
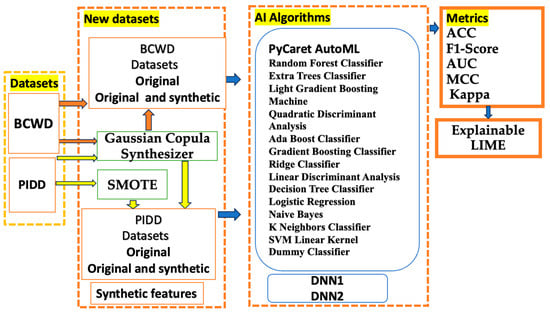
Figure 1.
The methodology of the proposed study.
The PIDD and BCWD datasets are detailed in the following section, including a description of their features and the number of records. We aimed to supply AutoML with four types of datasets: the original, synthetic, original and synthetic, and original and synthetic features datasets, with synthetic data generated by the GCS and SMOTE techniques.
After training the machine learning models, the quality of classification on the test set was evaluated using several performance metrics, including accuracy (ACC), F1-score, Area Under the Curve (AUC), Matthews Correlation Coefficient (MCC), and Kappa statistics. These metrics provide a comprehensive assessment of the model’s classification performance.
For each input dataset, the relevant metrics are computed. Also, an explainable tool, LIME, is proposed, and the interpretation of the proposed methodology is presented in the Section 8. Table 1 presents the optimized classifiers for each data category, while Table 2 and Table 3 display the metrics computed for the 14 classifiers used in the binary classification of diabetic and non-diabetic classes.

Table 1.
The optimal classifiers selected by the PyCaret and DNN architectures.

Table 2.
The metrics computed with DNN1 and DNN2 for original, original and synthetic, and synthetic and extracted from two databases.

Table 3.
The metrics computed with PyCaret for original, original and synthetic, and synthetic, and synthetic features extracted from two databases.
4. AI Algorithms
4.1. Gaussian Copula Synthesizer (CGS)
The Gaussian Copula Synthesizer [34] is a statistical technique designed to generate synthetic data while preserving the relationships between variables. It achieves this by modeling the joint distribution of the data using a Gaussian copula, which effectively maintains the correlations among variables as it transforms the data to conform to specified distributions. To create synthetic data, GCS first estimates the marginal distributions of the individual variables. Then, it fits a Gaussian copula to capture the dependencies between these variables. Finally, synthetic samples are generated by sampling from the copula and adjusting the results to align with the original distributions.
This method is particularly beneficial in fields such as healthcare, where maintaining variable dependencies is crucial. For instance, GCS can help resolve issues like class imbalance in machine learning datasets by generating high-quality synthetic data that mirror real-world relationships while protecting individual privacy. Implementing the Gaussian Copula Synthesizer in Python involves using libraries such as NumPy, SciPy, and Copulas.
4.2. SMOTE
SMOTE (Synthetic Minority Oversampling Technique, version 0.12.4), introduced in the paper [39], is a technique used to generate synthetic data for the class that have a smaller number of samples in case the dataset is not balanced. It is improving the performance of classifiers trained on imbalanced datasets. The SMOTE technique generates synthetic data by first identifying the class with fewer samples using KNN and then generating new samples by interpolating between the current sample and one of the KNN’s neighbors. Each feature undergoes the interpolation process.
4.3. Synthetic Features
The PIDD dataset’s synthetic feature creation used two different discretization methods on the numerical columns, but not on the outcome target column. The first method used equal-width interval discretization, which took the values in each column and split them into five equal-width intervals based on the lowest and highest values. Each interval was given a number label from 0 to 4 to show its size. Range-based classification is always conducted this way. Equal-frequency interval discretization was the second way to make new features. In this method, each column was split into five intervals, and each interval had about the same number of data points (same frequency), which showed how the data were spread out. From 0 to 4, each number was given a label based on its quantile range. Equal-width intervals focused on range, and equal-frequency intervals focused on density. These two methods together created new category features and gave different views on the data. The study used the changed dataset, which had both real and synthetic features.
4.4. AutoML PyCaret
PyCaret [35] is an open-source, low-code library in Python that helps automate the machine learning process from start to finish. It makes it easier for users, whether they are new to data science or have experience, to build and deploy machine learning models. PyCaret rapidly evaluates a range of models, giving the data scientist a clear image of which models perform successfully and reliably across regression and classification tasks.
One of the standout features of PyCaret is its AutoML capabilities. This allows users to try out different machine learning algorithms and adjust their settings with very little coding required. PyCaret can handle various tasks, such as classification, regression, clustering, anomaly detection, and natural language processing. The proposed AutoML contains the following classifiers: Random Forest Classifier (RF), Extra Trees Classifier (ET), Light Gradient Boosting Machine (LIGHTGBM), Quadratic Discriminant Analysis (QDA), Ada Boost Classifier (ADA), Gradient Boosting Classifier (GBC), Ridge Classifier (RIDGE), Linear Discriminant Analysis (LDA), Decision Tree Classifier (DT), Logistic Regression (LR), Naive Bayes (NB), K Neighbors Classifier (KNN), Support Vector Machine (SVM), and Dummy Classifier (DUMMY). The PyCaret library contains the following types of ML algorithms: decision three (RF, ET, DT), boosting (GBC, ADA, LIGHTGBM), discriminant analysis (QDA, LR, LDA), rules (NB, DUMMY, RIDGE), and distance-based (KNN, SVM).
4.5. DNN
Deep Neural Networks (DNNs) have been applied in various domains in the last decade with success for image, video, audio, and natural text data. In the medical domain, DNNs were used. A DNN is a network of artificial neurons placed in multiple layers that are connected and learn the relationship between input features and output. Usage of DNNs in the medical field was found in [40,41,42,43].
In this study, two DNN architectures were used. The first DNN architecture is a lightweight model that has four layers: 32 neurons on the first layer, 16 neurons on the second, and 8 neurons on the third layer, and uses the ReLU (Rectified Linear Unit) as the activation function. On the last layer, there is a single neuron with the sigmoid activation function being a classification problem.
The second DNN architecture is a more complex model than the first one: 5 layers with larger numbers of neurons (256/128/63/32/1 neurons) and 3 layers for regularization that are present after each of the first three layers of neurons (dropout layers that deactivate 30% of the neurons from the previous layer and try to reduce the overfitting problem).
In order to establish the final architecture for the two DNNs, the ablation study was performed. The number of epochs varies between 5 and 50, the number of neurons between 8 and 526, and the number of dense layers between 1 and 5; also, the percentage of test sets varied between 10% and 30%. The optimum architectures that provide the best accuracy are described in the Section 8.
4.6. LIME Explainable AI Tool
LIME (Local Interpretable Model-agnostic Explanations) [36] is a tool that helps explain how machine learning models make predictions. It focuses on explaining specific predictions rather than the whole model. When a model gives a result, LIME slightly changes the input data and sees how these changes affect the output. This process helps identify which features are most important for that prediction, making it easier to understand why the model made its decision.
LIME can be used with any machine learning model, making it flexible. By showing how decisions are made, LIME helps build trust in AI systems, especially in important areas like healthcare, where understanding the reasoning behind predictions is crucial.
5. Databases
5.1. Pima Indians Diabetes Database (PIDD)
The PIDD is publicly available at the web address https://www.kaggle.com/datasets/mathchi/diabetes-data-set/data, the last access was 8 September 2024. The proposed dataset has the following features: pregnancies, glucose, blood pressure, skin thickness, insulin, body mass index (BMI), diabetes pedigree function, age, and outcome as target features. It belongs to the National Institute of Diabetes and Digestive and Kidney Diseases and contains 769 records collected from female patients of at least 21 years of age. We have chosen the SMOTE method to generate synthetic data for the PIDD dataset because the PIDD dataset has a class imbalance (33% diabetic and 67% non-diabetic).
5.2. Breast Cancer Wisconsin Dataset (BCWD)
The BCWD is publicly available at the web address https://archive.ics.uci.edu/dataset/17/breast+cancer+wisconsin+diagnostic, the last access was 12 September 2024. The BCWD corresponds to 569 patients, with benign (357 cases) and malignant (212 cases) tumor diagnoses [44]. The database was developed at the University of Wisconsin Hospital, Madison, USA. There are 32 quantitative features and 699 instances described in the database.
5.3. Confidence Interval
A confidence interval (CI) analysis to determine an appropriate sample size for our datasets was incorporated. This ensures that our results are statistically reliable and generalizable. In this sense, we computed the minimum sample size (n) required for our study using the formula for the confidence interval of a proportion:
where
Z is the Z-score for the desired confidence level (1.96 for 95% confidence), p is the estimated proportion of the outcome of interest (classification success rate), and E is the margin of error (desired precision) [45].
We have assumed an expected classification accuracy (p) of 0.80 (from preliminary experiments), a margin of error (E) of 5%, and a confidence level of 95%; the required sample size is n = 246. A minimum of 246 samples was required to ensure that our classification accuracy estimates were within +5% and −5% of the true value with 95% confidence.
Pima Indian Diabetes Dataset (PIDD): the dataset contains 768 samples, exceeding the minimum requirement and ensuring statistical robustness.
Breast Cancer Wisconsin Dataset (BCWD): the dataset contains 569 samples; it is slightly above the required minimum.
5.4. Violin Graph
Comparable to box plots, violin plots further display the data’s probability density at various values, typically with a kernel density estimator applied for smoothness. The same information that is shown in a box plot can also be seen in a violin plot: the data’s median, interquartile range, and, if the sample size is small enough, all of the points in the data. The median value is symbolized with a white dot situated on the vertical axis. The graphs were drawn for the original, synthetic, and original and synthetic from both the BCWD and PIDD datasets. Also, for all features, the clustering value around the median was monitored. Figure 2 shows violin graphs for each feature. The BCWD dataset has 32 features that encapsulate various aspects of cell nuclei features; Figure 3 shows ten features for the mean. In the shown graph, the synthetic data were created with GCS.
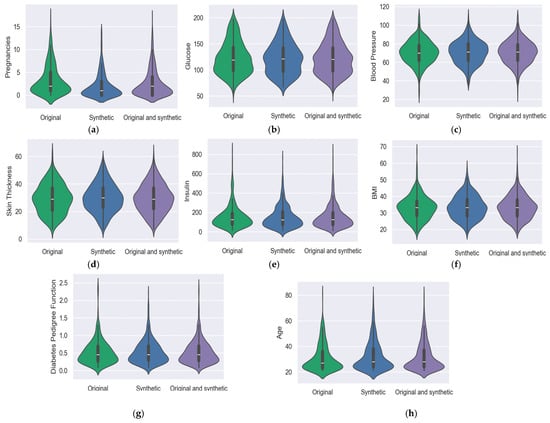
Figure 2.
PIDD data analysis with a violin graph: (a) Pregnancies; (b) Glucose; (c) Blood pressure; (d) Skin thickness; (e) Insulin; (f) Body mass index; (g) Diabetes pedigree function; (h) Age.
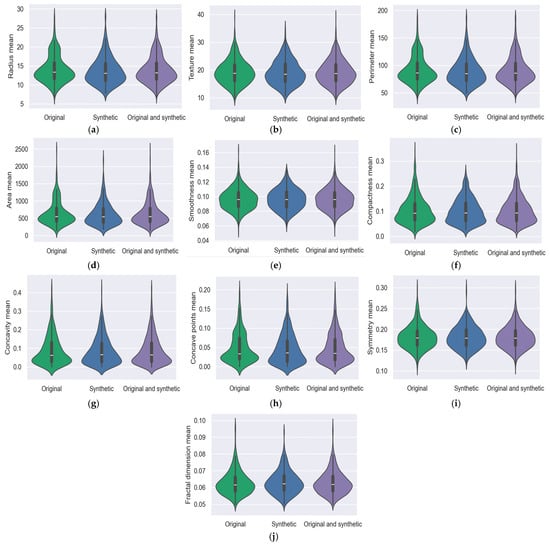
Figure 3.
BCWD data analysis with a violin graph: (a) radius mean; (b) texture mean; (c) perimeter mean; (d) area mean; (e) smoothness mean; (f) compactness mean; (g) concavity mean; (h) concave points mean; (i) symmetry mean; and (j) fractal dimension mean.
To illustrate the data’s distribution form, kernel density estimates are displayed on either side of the black line. The likelihood that some members of the population will adopt the specified value increases as the violin plot’s width decreases.
6. Hardware and Software
A PC with the following architecture—Apple M1 Pro, 16 GB unified RAM, Chip Apple, and 20-Core GPU—performed the experiments. The Python software environment, version 3.12.8 (https://www.python.org/downloads/release/python-3128/ (accessed on 22 December 2024)), was utilized, along with the following libraries and their respective versions:
- NumPy 2.2.1 (https://pypi.org/project/numpy/ (accessed on 22 December 2024));
- Pandas 2.2.3 (https://pandas.pydata.org/docs/whatsnew/index.html (accessed on 22 December 2024));
- TensorFlow 2.17.0 (https://blog.tensorflow.org/2024/07/whats-new-in-tensorflow-217.html (accessed on 22 December 2024));
- Scikit-learn 1.6.1 (https://pypi.org/project/scikit-learn/ (accessed on 11 January 2025));
- PyCaret 3.0.4 (https://pycaret.readthedocs.io/en/stable/installation.html (accessed on 22 December 2024));
- Seaborn 0.13.2 (https://seaborn.pydata.org/installing.html (accessed on 22 December 2024))
- LIME 8.2.2. (https://lib.haxe.org/p/lime/ (accessed on 22 December 2024)).
7. Metrics
The main objective of this research is to evaluate binary classification models using a confusion matrix. Evaluating the outcomes of a binary classification is an ongoing difficulty in computational statistics and machine learning. A contingency table known as a two-class confusion matrix can be created each time an algorithm is used to distinguish between elements of a dataset with two conditions (such as positive and negative). This table shows the proportion of elements that were correctly predicted and incorrectly classified [10,11,12]. An effective method for assessing how well a classifier can identify pairs of distinct classes is the confusion matrix, which is typically used to quantify the accuracy of a classifier or predictor. An alternative machine learning evaluation metric called the F1-score elaborates on a model’s performance within a class, as opposed to evaluating the model’s overall performance based on accuracy. Besides these, the Area Under the Curve (AUC) for measuring the degree of separability of studied classes, the Matthews Correlation Coefficient (MCC) for summarizing the classifier performance, and Kappa for model agreement metrics were used, and PRE is for chance agreement [11,12,13].
where TP (True Positive) is the number of instances where the model correctly predicted the positive class; TN (True Negative) is the number of instances where the model correctly predicted the negative class; FP (False Positive) is the number of instances where the model incorrectly predicted the positive class; and FN (False Negative) is the number of instances where the model incorrectly predicted the negative class.
8. Results and Discussions
In this section, we present a detailed description of the results in concordance with the proposed methodology.
We conducted a thorough investigation with the AutoML PyCaret tool and two DNNs. Table 1 displays the optimal classifier selected for original, synthetic, original and synthetic, and original and synthetic features. The model’s objective was to distinguish between diabetic and non-diabetic cases (PIDD dataset), as well as the benign and malignant classes of breast cancer (BCWD dataset), by dividing the data into 70% for training and 30% for testing. Additionally, Figure 4 showcases the architectures of DNN1 with three dense layers and DNN2 with five dense layers and three dropout layers, each designed with varying numbers of layers and categories, and also the layer parameters.
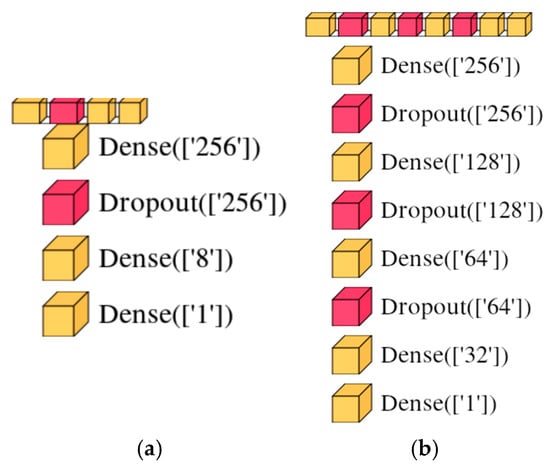
Figure 4.
The architectures of the proposed DNNs: (a) DNN1; (b) DNN2.
From BCWD were derived the synthetic, original, and synthetic data when the synthetic data were created with the GCS techniques. From the PIDD dataset were obtained synthetic, original, and synthetic data. When the synthetic data were created with the GCS and SMOTE techniques, additional synthetic features were created for the same dataset. The PIDD dataset’s poor classification results led to the creation of supplementary features. Supplementary Materials section contains the download link for original data, synthetic data and synthetic features that were used in this study.
The DNN1 was used for the classification of the BCWD dataset for original, synthetic, and original and synthetic when the synthetic data were created with the GCS. For the PIDD dataset, besides the datasets and the DNN1 used previously, the DNN2 was proposed along with the original, synthetic, and original and synthetic data when the synthetic data were created with SMOTE. Figure 5 displays the classification results for DNNs, while Table 3 displays the results for PyCaret AutoML.
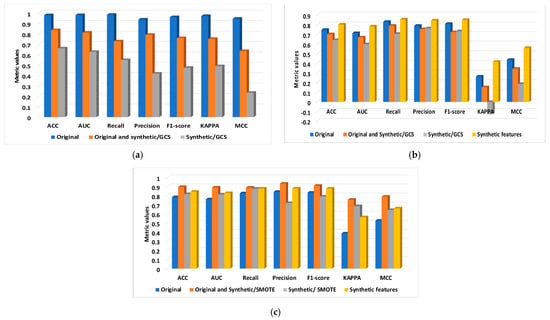
Figure 5.
The metrics computed with DNN1 and DNN2 for original, original and synthetic, and synthetic and extracted from two databases: (a) BCWD/DNN1; (b) PIDD/DNN1; and (c) PIDD/DNN2.
In choosing the number of layers, the number of neurons on each layer, and the dropout rate for networks DNN1 and DNN2, we have applied neural networks and deep learning best practices: (i) a pyramid-like structure—a higher number of neurons on the first layers to extract hierarchical features and a reduced number of neurons on the next layers; (ii) a common practice is to choose the number of neurons in powers of two (256, 128, 8) to optimize memory allocation and various matrix operations; and (iii) by making multiple tests and tuning hyperparameters.
For DNN1 we have chosen 256/8/1—on the first layer 256 neurons capture complex patterns, 8 neurons on the next layer help reduce overfitting while maintaining expressiveness, and one neuron in the output layer with a sigmoid activation for binary classification.
For the DNN2 network, we have chosen 256/128/64/32/1—the first layers with 256 and 128 neurons capture high-dimensional data relationships, the next layers with 64 and 32 neurons induce a balance in the feature complexity and regularization, and the last layer with one neuron is used for binary classification with sigmoid activation.
Dropout is used in DNN1 and DNN2 because it is a regularization technique that helps prevent overfitting by randomly deactivating neurons during training.
In the literature, it is recommended to have dropout values for deep learning tasks between 0.2 and 0.5, but this depends on the dataset size and model complexity. In our case in DNN1, we have introduced a dropout of 0.3 after the first layer with 256 neurons that helps to reduce the overfitting while preserving enough information for classification to be accurate. In DNN2 we have used a dropout rate of 0.3 after the first layers with 256, 128, and 64 neurons. Having multiple dropout layers provides robust regularization and is recommended in models with moderate complexity.
ML and DNNs have specific parameters, and their descriptions are provided above. Criteria compute the probability that a specific variable is incorrectly classified when chosen randomly; it is optimized with each iteration of the algorithm. The value of the learning rate is very important because a large learning rate could prevent the loss value from converging, leading to a failure in the learning process, whereas a very small learning rate could cause the learning process to proceed too slowly [43]. The number of estimators controls the number of trees used in the classification process. Also, max features help to determine the number of attributes to consider for the optimal partitioning of the dataset. The rectified linear activation function, abbreviated as ReLU, is a standard for neural network architectures due to its facilitation of model training. Its output is either ‘0’ or ‘1’, depending on its argument. The sigmoid function is among the most frequently used activation functions, and its output is a very small value if its argument is close to 0 [44].
The results stored in Table 2 are obtained with the two DNNs and six types of datasets (original, original and synthetic/GCS, synthetic/GCS, original and synthetic/SMOTE, synthetic/SMOTE, and original and synthetic features). The obtained results for DNN1 supplied with original data belong to BCWD, for these three data categories denote that the classification accuracy for original data is 0.979. When the GCS mixes the original data with synthetic data, the results drop to 0.853 for both the original and synthetic data and 0.660 for the synthetic data. This indicates that the BCWD contains accurate data, negating the need for ablation.
For PIDD, DNN1 obtained a poor accuracy of 0.746 for the original data; in this case, the same dataset category as previously mentioned was used, and the classification accuracy also decreased. Therefore, the DNN2 was built, and the synthetic data with the SMOTE technique was performed. In this case, the classification accuracy increased from 0.785 for original data to 0.897 for original and synthetic and 0.819 for synthetic data.
Table 3 shows the best metrics that PyCaret AutoML could find, the relevant classifier that was chosen from a tuning process of fourteen classifiers, the datasets, the type of data used in the study, and the GCS and SMOTE techniques for the datasets that are not balanced. For BCWD, the RF classifier yields the best results for original data, with a classification accuracy of 0.972. A special concern occupies the results obtained for the PPID dataset because the initial results obtained for the original data (the GBC classifier; accuracy of 0.799) found further solutions, and the AutoML was supplied with the same dataset as the DNN. Significant results are obtained for the combination of the original and synthetic/SMOTE and the ET classifier (accuracy of 0.942).
Time-consuming factors include the hardware, the software settings, and how well the AI tools work. The testing and training time consumed by DNN1 and DNN2, as well as the testing time consumed by PyCaret AutoML, are stored in Table 4 and Table 5 for each dataset and data type. The training accuracy for the DNNs is shown in Table 5. However, for AutoML, it is not possible to compute training accuracy directly due to its automated model selection and optimization process.

Table 4.
The time consumption and training accuracy of DNN1 and DNN2 across original, original and synthetic, and synthetic data, computed for two databases.

Table 5.
The time consumption of PyCaret AutoML across original, original and synthetic, and synthetic data, computed for two databases.
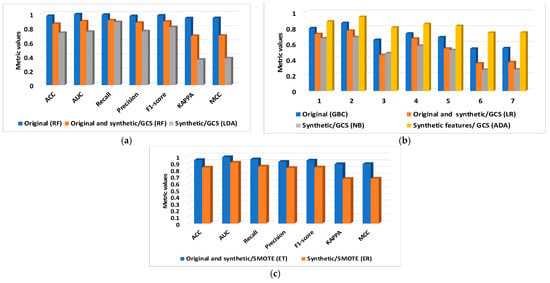
Figure 6.
The metrics computed with PyCaret for original, original and synthetic, and synthetic, and the synthetic features extracted from two databases: (a) BCWD and GCS; (b) PIDD and GCS; and (c) PIDD and SMOTE.
This research evaluates the consistency of the content of the dataset with an AI classifier and different data types and a justification of the results with the LIME XAI framework, a widely used model-agnostic approach in the literature, with particular emphasis on its ability to enhance the interpretability of tabular models, thus highlighting the significance of training ML on synthetic or real data and the advantages of using strong ML in the classification process.
For the binary classification task, two classes are proposed: class “0” (non-diabetic) and class “1” (diabetic), as shown in Figure 7, Figure 8, Figure 9, Figure 10, Figure 11 and Figure 12. The left figure displays the prediction probabilities of the “0” and “1” classes. The figures from the middle return the most important features with their scores. The color coding is consistent across sections. On the right side, the figures display the top selected features. Because the results obtained for BCWD are satisfactory for the original data, only the results obtained for the PIDD dataset are interpreted.
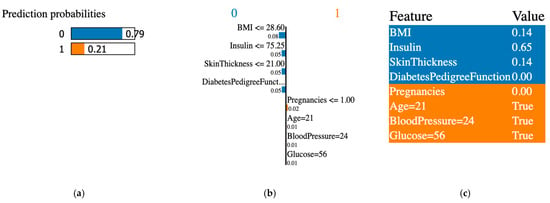
Figure 7.
XAI LIME explanation for original data from the PIDD dataset. (a) prediction probabilities for class 0 (non-diabetic) and class 1 (diabetic); (b) influential features that pushed the prediction toward class 0 or class 1; (c) features values, features contributing to class 0 are shown in blue and features contributing to class 1 are shown in orange.
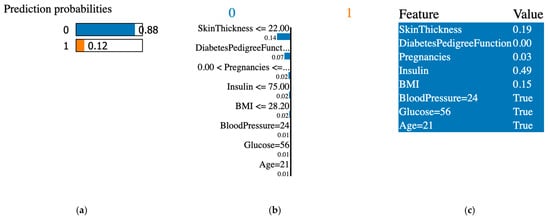
Figure 8.
XAI LIME for synthetic and original, where synthetic data were generated with the GCS. (a) prediction probabilities for class 0 (non-diabetic) and class 1 (diabetic); (b) influential features that pushed the prediction toward class 0 or class 1; (c) features values, features contributing to class 0 are shown in blue and features contributing to class 1 are shown in orange.

Figure 9.
XAI LIME for synthetic data generated with the GCS. (a) prediction probabilities for class 0 (non-diabetic) and class 1 (diabetic); (b) influential features that pushed the prediction toward class 0 or class 1; (c) features values, features contributing to class 0 are shown in blue and features contributing to class 1 are shown in orange.

Figure 10.
XAI LIME for synthetic features generated with the GCS. (a) prediction probabilities for class 0 (non-diabetic) and class 1 (diabetic); (b) influential features that pushed the prediction toward class 0 or class 1; (c) features values, features contributing to class 0 are shown in blue and features contributing to class 1 are shown in orange.

Figure 11.
Original and synthetic data created with the SMOTE technique. (a) prediction probabilities for class 0 (non-diabetic) and class 1 (diabetic); (b) influential features that pushed the prediction toward class 0 or class 1; (c) features values, features contributing to class 0 are shown in blue and features contributing to class 1 are shown in orange.
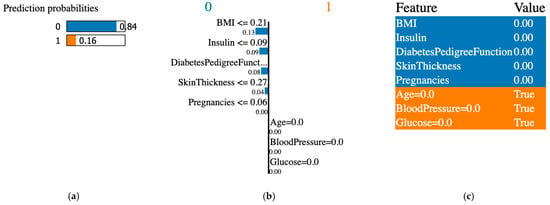
Figure 12.
XAI LIME synthetic data created with the SMOTE technique. (a) prediction probabilities for class 0 (non-diabetic) and class 1 (diabetic); (b) influential features that pushed the prediction toward class 0 or class 1; (c) features values, features contributing to class 0 are shown in blue and features contributing to class 1 are shown in orange.
The XAI interpreted the original data from the PIDD datasets; Figure 7, Figure 8, Figure 9, Figure 10, Figure 11 and Figure 12 display the results of applying the LIME tool to both original and synthetic data and their mixture: Figure 7 for original data; Figure 8 for both original and synthetic data; Figure 9 for synthetic data; and Figure 10 for synthetic features. The interpretation continues with Figure 11 and Figure 12 when the synthetic data were created with the SMOTE technique. The last column shows the value associated with each feature, indicating the contribution to the classification of diabetes (the PIDD dataset).
The association of colors in Figure 7, Figure 8, Figure 9, Figure 10, Figure 11 and Figure 12 is the following: class “0” (non-diabetic) corresponds to blue, and the orange color is for class “1” (diabetic). In LIME XAI explanations, Boolean values represent the model’s outcome or decision, and the true result indicates a positive result in agreement with the model’s classification.
XAI LIME explanations: the subfigures shown on the first figure are dedicated to original data, and Figure 8, Figure 9, Figure 10, Figure 11 and Figure 12 to prediction probability; the middle column of each figure shows the important features with their scores, and the last column shows the top features. In Figure 7, Figure 8 and Figure 9, the results for the original, synthetic and original, and synthetic data, respectively, were displayed, and the GCS pre-processing technique produced the synthetic data. Figure 10 shows the interpretation of the dataset with synthetic features. Figure 11 and Figure 12 illustrate the interpretation of both original and synthetic and synthetic data, created using the SMOTE technique.
The DNN, AutoML, and the LIME tool corroborate the results and offer a clear interpretation of the possible classification outcomes.
The LIME tool selects important features for the PIDD dataset, providing a first direction of interpretation. In this context, the metrics score increases when BMI and insulin are selected as the first two important features. The ET classifier achieves an accuracy of 0.942 for both original and synthetic/SMOTE data, with a BMI score of 0.14 and an insulin score of 0.08. The DNN2 classification accuracy of 0.897 confirmed the same results as the derived dataset from the PIDD.
For synthetic data created with the SMOTE, the accuracy classification is higher compared with the data created with the GCS; initially, the accuracy of 0.669 (skin thickness score of 0.19) obtained for the Synthetic/GCS and NB classifier increased to 0.832 for the Synthetic/SMOTE and FR classifier (BMI of 0.13).
A synthetic feature is a solution for improving the classification accuracy: DNN2 provides an accuracy of 0.844 and the DNA classifier an accuracy of 0.884, respectively.
A comparison between our study and the scientific literature on synthetic data generation, machine learning, explainable AI methods, and various databases is provided in Table 6. This reveals that most studies are concentrated on achieving high performance metrics, with less interest in the sensitivity of machine learning models to the integration of synthetic data—an important aspect that our work explores. By examining this sensitivity, we aim to understand how synthetic data influence model behavior, robustness, and generalization. Our approach not only measures the models’ performance but also investigates potential biases, errors, or instabilities introduced by synthetic data, ensuring a more thorough and reliable assessment of model capabilities in real-world applications.

Table 6.
Summary of results comparison with recent relevant studies.
9. Conclusions
The proposed study focused on verifying ML and DNN behaviors when fed with original, synthetic, and original and synthetic data. By changing the input with synthetic data, the study determines the strongest machine learning that yields a favorable classification. The study aimed to investigate the potential LIME tool to explain the ML classification. The experimental results demonstrated that the ET algorithm is the strongest with synthetic data, with an ACC of 0.942 for the combination of original and synthetic/SMOTE. In this case, the LIME XAI selected the first two important features: BMI with a score of 0.14 and insulin of 0.08. Although the accuracy is low when the synthetic data are created with the GCS technique. Remarkably, the ET classifier confirms the results obtained by DNN2.
Specifically, if an ML model’s performance decreases when exposed to synthetic data, it indicates that the model is sensitive and does not qualify as a strong ML model.
Future work could explore advanced AutoML tools, explain results using other XAI tools, and uncover additional vulnerabilities of machine learning.
Supplementary Materials
The following supporting information can be downloaded at https://github.com/simonamoldovanu/synthetic_data_features (accessed on 22 December 2024).
Author Contributions
Conceptualization, S.M., M.M. and D.M.; methodology, D.M.; software, M.M.; validation, S.M., M.M. and D.M.; formal analysis, D.M.; investigation, S.M. and D.M.; resources, S.M.; data curation, D.M.; writing—original draft preparation, S.M.; writing—review and editing, M.M.; visualization D.M.; supervision, S.M.; and M.M. for the term explanation. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially supported under contract no. RF 2461/31.05.2024, “Software application dedicated to improve diagnostic in medical imaging” at the University Dunărea de Jos of Galați. This work was supported by scientific research contract 825/30.09.2024 “Research Study on the Use of AI Techniques for Sensitivity Testing with Synthetic Data” from Dunărea de Jos University of Galati. This work was supported by scientific research contract 826/30.09.2024 “Research Study on the Applicability of Explainable Artificial Intelligence and Synthetic Data in the Fields of Medicine or Agriculture” from Dunărea de Jos University of Galati.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- An, Q.; Rahman, S.; Zhou, J.; Kang, J.J. A Comprehensive Review on Machine Learning in Healthcare Industry: Classification, Restrictions, Opportunities and Challenges. Sensors 2023, 23, 4178. [Google Scholar] [CrossRef]
- Gonzales, A.; Guruswamy, G.; Smith, S.R. Synthetic Data in Health Care: A Narrative Review. PLoS Digit. Health 2023, 2, e0000082. [Google Scholar] [CrossRef] [PubMed]
- Clemente, F.; Ribeiro, G.M.; Quemy, A.; Santos, M.S.; Pereira, R.C.; Barros, A. ydata-profiling: Accelerating Data-Centric AI with High-Quality Data. Neurocomputing 2023, 554, 126585. [Google Scholar] [CrossRef]
- Patki, N.; Wedge, R.; Veeramachaneni, K. The Synthetic Data Vault. In Proceedings of the 2016 IEEE International Conference on Data Science and Advanced Analytics (DSAA), Montreal, QC, Canada, 17–19 October 2016; pp. 399–410. [Google Scholar] [CrossRef]
- Juneja, T.; Bajaj, S.B.; Sethi, N. Synthetic Time Series Data Generation Using Time GAN with Synthetic and Real-Time Data Analysis. In Proceedings of the Lecture Notes in Electrical Engineering; Springer: Berlin/Heidelberg, Germany, 2023; Volume 1011, pp. 657–667. [Google Scholar] [CrossRef]
- Sei, Y.; Onesimu, J.A.; Ohsuga, A. Machine Learning Model Generation with Copula-Based Synthetic Dataset for Local Differentially Private Numerical Data. IEEE Access 2022, 10, 101656–101671. [Google Scholar] [CrossRef]
- He, X.; Zhao, K.; Chu, X. AutoML: A Survey of the State-of-the-Art. Knowl.-Based Syst. 2021, 212, 106622. [Google Scholar] [CrossRef]
- LeDell, E.; Poirier, S. H2O AutoML: Scalable Automatic Machine Learning. In Proceedings of the 7th ICML AutoML Workshop, Online Event, 18 July 2020; pp. 1–16. Available online: https://www.automl.org/wp-content/uploads/2020/07/AutoML_2020_paper_61.pdf (accessed on 23 October 2024).
- Salih, A.M.; Raisi-Estabragh, Z.; Galazzo, I.B.; Radeva, P.; Petersen, S.E.; Lekadir, K.; Menegaz, G. A Perspective on Explainable Artificial Intelligence Methods: SHAP and LIME. Adv. Intell. Syst. 2024, 7, 2400304. [Google Scholar] [CrossRef]
- Chicco, D.; Tötsch, N.; Jurman, G. The Matthews Correlation Coefficient (MCC) Is More Reliable Than Balanced Accuracy, Bookmaker Informedness, and Markedness in Two-Class Confusion Matrix Evaluation. BioData Min. 2021, 14, 13. [Google Scholar] [CrossRef]
- Tăbăcaru, G.; Moldovanu, S.; Răducan, E.; Barbu, M. A Robust Machine Learning Model for Diabetic Retinopathy Classification. J. Imaging 2023, 10, 8. [Google Scholar] [CrossRef]
- Rujas, M.; Herranz, R.M.G.; Fico, G.; Merino-Barbancho, B. Synthetic Data Generation in Healthcare: A Scoping Review of Reviews on Domains, Motivations, and Future Applications. Int. J. Med. Inform. 2024, 195, 105763. [Google Scholar] [CrossRef]
- Aziz, N.A.; Manzoor, A.; Qureshi, M.D.M.; Qureshi, M.A.; Rashwan, W. Explainable AI in Healthcare: Systematic Review of Clinical Decision Support Systems. medRxiv 2024. [Google Scholar] [CrossRef]
- Hernandez, M.; Epelde, G.; Beristain, A.; Álvarez, R.; Molina, C.; Larrea, X.; Alberdi, A.; Timoleon, M.; Bamidis, P.; Konstantinidis, E. Incorporation of Synthetic Data Generation Techniques within a Controlled Data Processing Workflow in the Health and Wellbeing Domain. Electronics 2022, 11, 812. [Google Scholar] [CrossRef]
- Ibrahim, M.; Al Khalil, Y.; Amirrajab, S.; Suna, C.; Breeuwer, M.; Pluim, J.; Elen, B.; Ertaylan, G.; Dumontier, M. Generative AI for Synthetic Data Across Multiple Medical Modalities: A Systematic Review of Recent Developments and Challenges. Comput. Biol. Med. 2025, 189, 109834. [Google Scholar] [CrossRef] [PubMed]
- Giuffrè, M.; Shung, D.L. Harnessing the Power of Synthetic Data in Healthcare: Innovation, Application, and Privacy. Npj Digit. Med. 2023, 6, 186. [Google Scholar] [CrossRef]
- Chen, R.J.; Lu, M.Y.; Chen, T.Y.; Williamson, D.F.; Mahmood, F. Synthetic Data in Machine Learning for Medicine and Healthcare. Nat. Biomed. Eng. 2021, 5, 493–497. [Google Scholar] [CrossRef]
- Pezoulas, V.C.; Zaridis, D.I.; Mylona, E.; Androutsos, C.; Apostolidis, K.; Tachos, N.S.; Fotiadis, D.I. Synthetic Data Generation Methods in Healthcare: A Review on Open-Source Tools and Methods. Comput. Struct. Biotechnol. J. 2024, 23, 2892–2910. [Google Scholar] [CrossRef]
- Dankar, F.K.; Ibrahim, M. Fake It Till You Make It: Guidelines for Effective Synthetic Data Generation. Appl. Sci. 2021, 11, 2158. [Google Scholar] [CrossRef]
- Wan, C.; Jones, D.T. Protein Function Prediction Is Improved by Creating Synthetic Feature Samples with Generative Adversarial Networks. Nat. Mach. Intell. 2020, 2, 540–550. [Google Scholar] [CrossRef]
- Mahmood, F.; Borders, D.; Chen, R.J.; McKay, G.N.; Saliman, K.; Baras, A. Deep Adversarial Training for Multi-Organ Nuclei Segmentation in Histopathology Images. IEEE Trans. Med. Imaging 2020, 39, 3257–3267. [Google Scholar] [CrossRef]
- Kingma, D.P.; Welling, M. Auto-Encoding Variational Bayes. arXiv 2013, arXiv:1312.6114. [Google Scholar] [CrossRef]
- Khosravi, H.; Das, S.; Al-Mamun, A.; Ahmed, I. Binary Gaussian Copula Synthesis: A Novel Data Augmentation Technique to Advance ML-Based Clinical Decision Support Systems for Early Prediction of Dialysis Among CKD Patients. arXiv 2023, arXiv:2403.00965. [Google Scholar] [CrossRef]
- Torfi, A.; Fox, E.A.; Reddy, C.K. Differentially Private Synthetic Medical Data Generation Using Convolutional GANs. Inf. Sci. 2022, 586, 485–500. [Google Scholar] [CrossRef]
- Rostami, M.; Oussalah, M. A Novel Explainable COVID-19 Diagnosis Method by Integration of Feature Selection with Random Forest. Inform. Med. Unlocked 2022, 30, 100941. [Google Scholar] [CrossRef]
- El-Sofany, H.; Bouallegue, B.; El-Latif, Y.M.A. A Proposed Technique for Predicting Heart Disease Using Machine Learning Algorithms and an Explainable AI Method. Sci. Rep. 2024, 14, 23277. [Google Scholar] [CrossRef]
- Titti, R.R.; Pukkella, S.; Radhika, T.S.L. Augmenting Heart Disease Prediction with Explainable AI: A Study of Classification Models. Comput. Math. Biophys. 2024, 12, 20240004. [Google Scholar] [CrossRef]
- Tasin, I.; Nabil, T.U.; Islam, S.; Khan, R. Diabetes Prediction Using Machine Learning and Explainable AI Techniques. Healthc. Technol. Lett. 2022, 10, 1–10. [Google Scholar] [CrossRef]
- Kibria, H.B.; Nahiduzzaman, M.; Goni, M.O.F.; Ahsan, M.; Haider, J. An Ensemble Approach for the Prediction of Diabetes Mellitus Using a Soft Voting Classifier with an Explainable AI. Sensors 2022, 22, 7268. [Google Scholar] [CrossRef]
- Karimi, D.; Dou, H.; Warfield, S.K.; Gholipour, A. Deep Learning with Noisy Labels: Exploring Techniques and Remedies in Medical Image Analysis. Med. Image Anal. 2020, 65, 101759. [Google Scholar] [CrossRef]
- Chuah, J.; Kruger, U.; Wang, G.; Yan, P.; Hahn, J. Framework for Testing Robustness of Machine Learning-Based Classifiers. J. Pers. Med. 2022, 12, 1314. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://docs.sdv.dev/sdv/single-table-data/modeling/synthesizers/gaussiancopulasynthesizer (accessed on 22 December 2024).
- Available online: https://pycaret.readthedocs.io/en/latest/ (accessed on 22 December 2024).
- Available online: https://lime.readthedocs.io/en/latest/ (accessed on 22 December 2024).
- Hospital Israelita Albert Einstein. Diagnosis of COVID-19 and Its Clinical Spectrum—Ai and Data Science Supporting Clinical Decisions (From 28th Mar to 3st Apr). Available online: https://www.kaggle.com/einsteindata4u/covid19 (accessed on 30 September 2024).
- Available online: https://www.kaggle.com/code/akhiljethwa/heart-failure-classification-knn-decision-tree (accessed on 30 September 2024).
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. (JAIR) 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Kaur, G.; Reddy, S.; Singh, D.; Sharma, N. A Deep Neural Architecture for Harmonizing 3-D Input Data Analysis and Decision Making in Medical Imaging. arXiv 2023, arXiv:2303.00175. [Google Scholar] [CrossRef]
- Nazir, S.; Kaleem, M. Federated Learning for Medical Image Analysis with Deep Neural Networks. Diagnostics 2023, 13, 1532. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Gutiérrez, M.E.; González-Pérez, P.P. Multi-Class Classification of Medical Data Based on Neural Network Pruning and Information-Entropy Measures. Entropy 2022, 24, 196. [Google Scholar] [CrossRef] [PubMed]
- Gasmi, A. Deep learning and health informatics for smart monitoring and diagnosis. arXiv 2022, arXiv:2208.03143. [Google Scholar] [CrossRef]
- Street, W.N.; Wolberg, W.H.; Mangasarian, O.L. Nuclear feature extraction for breast tumor diagnosis. In Proceedings of the IS&T/SPIE 1993 International Symposium on Electronic Imaging: Science and Technology, San Jose, CA, USA, 31 January–5 February 1993; Volume 1905, pp. 861–870. [Google Scholar] [CrossRef]
- Sellat, Q.; Bisoy, S.K.; Priyadarshini, R. Chapter 10—Semantic Segmentation for Self-Driving Cars Using Deep Learning: A Survey. In Cognitive Big Data Intelligence with a Metaheuristic Approach; Mishra, S., Tripathy, H.K., Mallick, P.K., Sangaiah, A.K., Chae, G.-S., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 211–238. [Google Scholar] [CrossRef]
- Morabito, F.C.; Kozma, R.; Alippi, C.; Choe, Y. Advances in AI, neural networks, and brain computing: An introduction. In Artificial Intelligence in the Age of Neural Networks and Brain Computing; Academic Press: Cambridge, MA, USA, 2024; pp. 1–8. [Google Scholar] [CrossRef]
- Sadiq, I.Z.; Usman, A.; Muhammad, A.; Ahmad, K.H. Sample size calculation in biomedical, clinical and biological sciences research. J. Umm Al-Qura Univ. Appl. Sci. 2024, 11, 133–141. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).