Abstract
The early detection and intervention of oral squamous cell carcinoma (OSCC) using histopathological images are crucial for improving patient outcomes. The current literature for identifying OSCC predominantly relies on models pre-trained on ImageNet to minimize the need for manual data annotations in model fine-tuning. However, a significant data divergence exists between visual domains of natural images in ImageNet and histopathological images, potentially limiting the representation and transferability of these models. Inspired by recent self-supervised research, in this work, we propose HistoMoCo, an adaptation of Momentum Contrastive Learning (MoCo), designed to generate models with enhanced image representations and initializations for OSCC detection in histopathological images. Specifically, HistoMoCo aggregates 102,228 histopathological images and leverages the structure and features unique to histological data, allowing for more robust feature extraction and subsequent downstream fine-tuning. We perform OSCC detection tasks to evaluate HistoMoCo on two real-world histopathological image datasets, including NDB-UFES and Oral Histopathology datasets. Experimental results demonstrate that HistoMoCo consistently outperforms traditional ImageNet-based pre-training, yielding more stable and accurate performance in OSCC detection, achieving AUROC results up to 99.4% on the NDB-UFES dataset and 94.8% on the Oral Histopathology dataset. Furthermore, on the NDB-UFES dataset, the ImageNet-based pre-training solution achieves an AUROC of 89.32% using 40% of the training data, whereas HistoMoCo reaches an AUROC of 89.58% using only 10% of the training data. HistoMoCo addresses the issue of domain divergence between natural images and histopathological images, achieving state-of-the-art performance in two OSCC detection datasets. More importantly, HistoMoCo significantly reduces the reliance on manual annotations in the training dataset. We release our code and pre-trained parameters for further research in histopathology or OSCC detection tasks.
1. Introduction
The increasing incidence of cancer globally underscores the urgent need for effective screening and diagnostic strategies to enable timely detection and treatment. Among various types of cancer, oral squamous cell carcinoma (OSCC) has emerged as a significant health concern [1], particularly affecting regions with high prevalence rates, such as Asia. According to recent reports, OSCC ranks 16th in terms of global incidence and mortality, with Asia accounting for approximately 65.8% of cases and a striking death rate of 74.0% [1]. Early detection and intervention are crucial for improving outcomes, as they substantially enhance the chances of complete remission. Despite advancements in awareness programs and clinical protocols, the effective screening of OSCC remains challenging, emphasizing the importance of developing robust diagnostic tools. These tools are particularly needed to address the significant variations in clinical presentations and the impact of risk factors like tobacco use [2]. Therefore, enhancing OSCC detection methodologies is vital to reduce the disease burden and improve patient survival rates.
In recent years, deep learning-based solutions [3,4] have exhibited state-of-the-art performance in extracting valuable insights from histopathological images [5,6], offering promising avenues to address these challenges. An AI-assisted workflow could potentially serve as a “second reader”, pre-screening normal samples to reduce pathologists’ workload, automatically highlighting suspicious regions to guide attention, and providing remote preliminary screening for underserved areas. Such systems could significantly reduce analysis time while improving diagnostic consistency and accessibility. However, developing effective AI systems requires an extremely large number of images with fine annotations to train deep neural networks (DNNs) and deliver decent performance in a supervised learning manner [7,8,9]. Unfortunately, large-scale annotated OSCC histopathological datasets remain scarce due to multiple factors: (1) stringent patient privacy regulations and ethical constraints limiting data sharing, (2) the prohibitive cost of expert annotation, with each image requiring meticulous labeling by specialists, and (3) heterogeneity in sample processing and imaging protocols across institutions, resulting in technical variability that complicates dataset standardization.
Most state-of-the-art models [10,11,12] used for histological image analysis are pre-trained on ImageNet due to the lack of large-scale annotated histopathological datasets. Although this approach has proven effective, the domain divergence between natural images in ImageNet and histopathological images may hinder the model’s ability to generalize effectively to medical image tasks. As shown in Figure 1, there is significant distributional divergence in Contrast, Homogeneity, Energy, and Correlation, between natural and histopathological images, posing challenges for model fine-tuning. Contrast measures local variations in pixel intensity within an image, mathematically represented as intensity differences between adjacent pixels in the Gray Level Co-occurrence Matrix (GLCM). High contrast values indicate significant intensity variations in tissue structures, typically reflecting abnormal cell density and tissue structure disruption in OSCC. Homogeneity measures the uniformity of image texture by evaluating the similarity of pixel pairs. Normal tissues typically exhibit higher homogeneity, while tumor tissues show lower homogeneity due to cellular atypia and irregular arrangements. Energy measures the orderliness and uniformity of texture, calculated as the sum of squared GLCM elements. High energy values indicate uniform texture or numerous repetitive patterns, typically higher in normal tissues than in tumor tissues. Correlation is a measure of linear dependencies in the GLCM, representing the degree of correlation between adjacent pixels. Correlation can help distinguish between normal organized structures (high correlation) and the disordered arrangement of tumor tissues (low correlation). As a result, there is a growing demand for pre-training strategies that are specifically designed to address the unique characteristics of histopathological images.
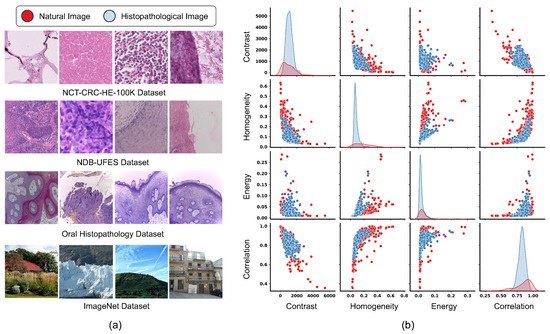
Figure 1.
(a) Eamples of natural images (from the ImageNet dataset) and histopathological images (from three real-world datasets). (b) The divergence between visual domains of natural images and histopathological images.
To reduce the required sample size and enable end-to-end adaptation to the target domain, recent approaches have proposed the self-supervised learning (SSL) paradigm, which enhances the performance of DNN models by directly learning visual features from target domain images, without the need for labels. Among the different self-supervised learning methods, contrastive learning algorithms [13,14,15] utilize a similarity-based metric to measure the distance between embeddings derived from different views of the same image. These views are generated through data augmentation techniques such as rotation, cropping, and shifting, while the embeddings are extracted from a DNN with learnable parameters. Specifically, in computer vision tasks, the contrastive loss is specifically computed using feature representations from the encoder network, which aids in clustering similar samples while separating dissimilar ones. Recent methods such as SwAV [16], SimCLR [13], MoCo [17], and PILR [18] have shown superior performance over traditional supervised learning methods on natural images.
Contrastive learning was initially proposed for natural images and has demonstrated promising results. In recent years, it has been gradually applied to various modalities of medical images, such as X-rays [19,20,21], MRI [22,23,24], and cellular images [25,26,27], proving its effectiveness.
In recent years, self-supervised learning methods have made significant progress in histopathological image analysis, with numerous innovative studies emerging [28,29,30,31,32,33]. These methods encompass various paradigms including contrastive learning, masked image modeling, and self-distillation, providing effective approaches to address the scarcity of annotated data. Our study aims to explore the applicability and optimization strategies of the MoCo framework in the specific domain of oral squamous cell carcinoma, complementing existing research.
In this work, we aim to extract more domain-specific features from histopathological images in an unsupervised manner, enhancing the model’s performance on downstream OSCC detection tasks. Our method involves pre-training a deep learning model using a self-supervised objective on a large set of histopathological images before fine-tuning on a smaller, labeled OSCC dataset.
The main contributions of this work are threefold:
- 1.
- To address the distributional divergence between histopathological images and natural images, we developed HistoMoCo, a custom pre-training framework tailored for histopathological images. HistoMoCo aims to generate enhanced image representations and initialize models for OSCC detection in histopathological images.
- 2.
- We provide comprehensive evaluations and analyses, highlighting the benefits of self-supervised learning for histopathological image analysis; demonstrating the proposed HistoMoCo provides high-quality representations and transferable initializations for histopathological images interpretation and OSCC detection.
- 3.
- We release our code and pre-trained parameters for further research in histopathology or OSCC detection tasks.
The remainder of this manuscript is organized as follows: Section 2 reviews related work on self-supervised learning and the application of MoCo in medical imaging. Section 3 provides the necessary background on MoCo. Section 4 presents detailed specifications of our method. Section 5 outlines our experimental methodology and detailed experimental results, while Section 6 concludes the work.
2. Related Work
2.1. Detection of Oral Squamous Cell Carcinoma
Oral squamous cell carcinoma (OSCC) detection increasingly utilizes deep learning, particularly for pre-trained models based on large datasets like ImageNet. Several studies have demonstrated the effectiveness of convolutional neural networks (CNNs) in analyzing histopathology images for OSCC diagnosis. Redie et al. [12] evaluated ten pre-trained CNN models, finding that VGG-19 achieved a classification accuracy of 96.26% when combined with data augmentation. Kavyashree et al. [11] compared DenseNet architectures, reporting that DenseNet-201 reached an accuracy of 85.00%, while DenseNet-169 achieved a training accuracy of 98.96%. Mohan et al. [1] proposed OralNet, which utilized a four-stage approach for OSCC detection, achieving over 99.50% accuracy with histopathology images. These findings highlight the potential of pre-trained models for feature extraction and classification. However, most existing work [6,11,12] relies on models pre-trained on ImageNet, which may not fully capture the unique characteristics of histopathological images. Thus, there is a need for specialized pre-training methods tailored to histopathology to improve OSCC detection accuracy and enhance clinical outcomes.
2.2. Application of MoCo in Medical Image Analysis
Contrastive learning is one of the mainstream paradigms of self-supervised learning (SSL), aiming to learn consistent representations through the comparison of positive and negative sample pairs without requiring additional annotations. Given the tremendous potential of label-efficient learning [17,34], SSL has garnered significant attention in the field of medical image analysis [19,20,21,22,23,25,26]. Specifically, Liao et al. [20,21] integrated multi-task learning into the self-supervised paradigm of MoCo to enhance the model’s representational ability on X-ray images. Chaitanya et al. [22] proposed a strategy to extend the contrastive learning framework for segmenting 3D MRI images in semi-supervised settings by leveraging domain-specific and problem-specific prompts. Cao et al. [35] applied MoCo to enhance the performance of deep learning models in detecting rib fractures on CT images.
Recent advances have seen the emergence of foundation models for computational pathology. Chen et al. [30] introduced UNI, a general-purpose self-supervised model trained on over 100 million images from 100,000+ whole-slide images across 20 tissue types, demonstrating superior performance on 34 computational pathology tasks. Vorontsov et al. [32] presented Virchow, a 632-million parameter vision transformer model trained using DINOv2 on 1.5 million H&E stained slides, achieving state-of-the-art performance on pan-cancer detection and various benchmarks. Lu et al. [31] developed CONCH, a visual-language foundation model trained on diverse histopathology images and over 1.17 million image-caption pairs, excelling in tasks including classification, segmentation, and retrieval. Xiang et al. [33] introduced MUSK, a vision-language foundation model pretrained on 50 million pathology images and one billion text tokens using unified masked modeling, demonstrating strong performance across 23 benchmarks and various outcome prediction tasks. Our study aims to explore the applicability and optimization strategies of the MoCo framework in the specific domain of oral squamous cell carcinoma, complementing existing research.
3. Preliminaries
3.1. Contrastive Learning
Contrastive learning enables the self-supervised learning of image representations. Given a dataset , the objective is to find a mapping function where , that satisfies the following condition:
where measures image similarity, while F is responsible for both representation learning and dimensionality reduction. In this context, positive and negative samples are denoted as and , respectively, where is similar to x and is dissimilar. The model learns representations by maximizing the agreement between different augmented views and of the same example x, utilizing a contrastive loss in the latent space. These augmented views and are generated through data augmentation techniques denoted as .
3.2. Momentum Contrastive (MoCo) Learning
Momentum Contrastive (MoCo) is a state-of-the-art contrastive learning method that surpasses supervised pre-training on various downstream tasks. Unlike other contrastive learning approaches, MoCo does not require a large batch size or a memory bank. Instead, it utilizes a memory queue to store a set of previously computed representations. Furthermore, MoCo introduces a momentum encoder, which will be elaborated on shortly.
3.2.1. Dictionary as a Queue
MoCo trains an encoder to perform a dictionary lookup task, where a query q and encoded samples act as the keys in the dictionary. A match occurs when the query q is similar to the positive sample , while there is no match for the negative samples . Specifically, MoCo utilizes two visual encoders, labeled and , to learn query representations and key representations . Here, denotes the query sample, and refers to the key sample. To allow the encoder to reuse previously encoded samples, MoCo employs the dictionary as a queue. The pre-trained model is trained using the following loss function:
where is a temperature hyperparameter, and and denote representations of positive and negative samples, respectively.
3.2.2. Momentum Update
MoCo employs a momentum update strategy for the parameters of the visual encoders. Let represent the parameters of and those of . The parameters are updated through back-propagation using the contrastive loss from Equation (2), and the parameters are updated using the following equation:
where is a momentum coefficient. This updating strategy ensures a smoother evolution of compared to [17].
4. Methods
4.1. Histopathological Datasets and OSCC Detection Tasks
In this study, we utilize four publicly available histopathological image datasets, including NCT-CRC-HE-100K [36], EBHI-Seg [37], NDB-UFES [38], and Oral Histopathology [39].
- 1.
- NCT-CRC-HE-100K [36] is a large pathology dataset comprising 100,000 H&E-stained histological images of human colorectal cancer and healthy tissues, extracted from 86 patients. It covers nine tissue types, with each image sized at 224 × 224 pixels.
- 2.
- EBHI-Seg [37] is a dataset for segmentation tasks, containing 2228 original H&E images and corresponding ground truth annotations, with each original H&E image also having a resolution of 224 × 224 pixels.
- 3.
- NDB-UFES [38] dataset (OSCC detection dataset) presents a total of 237 samples with histopathological images and sociodemographic and clinical data. Its subset comprises 3763 image patches for downstream classification tasks, with each patch classified as either OSCC or Normal, and sized at 512 × 512 pixels.
- 4.
- Oral Histopathology [39] (OSCC detection dataset) is composed of histopathological images of the normal epithelium of the oral cavity and images of OSCC. It includes two different magnifications, 100× and 400×, with a total of 290 samples classified as normal and 934 samples classified as OSCC, all with image sizes of 2048 × 1536.
4.2. HistoMoCo Pre-Training for Histopathological Interpretation
We applied MoCo pre-training to histopathological images. Figure 2a shows how data augmentation is used to generate views of a specific image, followed by contrastive learning to learn embeddings in an unsupervised manner.
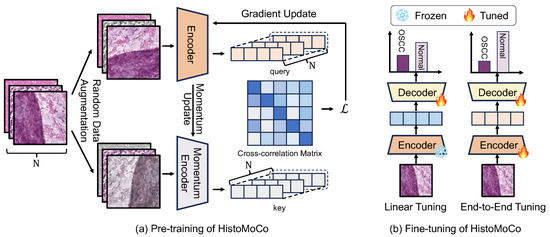
Figure 2.
The framework of HistoMoCo. (a) HistoMoCo maximizes agreement of embeddings generated by different augmentations of the same histological image. (b) Two fine-tuning solutions used in HistoMoCo. HistoMoCo employs ResNet-18 and ResNet-50 as encoders, using a linear classifier as the decoder.
We chose to use MoCo due to the limitations of large-scale computational resource costs in medical imaging AI. In comparison to other self-supervised frameworks such as SimCLR [13], MoCo requires significantly smaller batch sizes during pre-training [40]. The MoCo implementation used a batch size of 256 and achieved performance similar to SimCLR on ImageNet, while SimCLR struggled with a batch size of 256 [40]. MoCo mitigates the reliance on large batch sizes by utilizing a momentum-updated queue of previous samples to generate contrastive pair encodings.
We used 80% of the images from NCT-CRC-HE-100K [36] and the entire EBHI-Seg [37] datasets for MoCo pre-training. We chose to apply MoCo pre-training on models with ImageNet to leverage potential convergence advantages [41]. Due to the widespread availability of ImageNet pre-trained weights, initializing models with ImageNet weights before MoCo pre-training incurs no additional cost.
We update MoCo to be customized for histopathological images. Specifically, the data augmentation techniques commonly used in self-supervised learning for natural images may not be suitable for histopathological images. For example, color jittering and random grayscale can significantly alter the staining variations in histopathological images. Therefore, we disable these augmentations. Instead, we introduce multi-scale cropping to simulate tissue views at different magnification levels. In addition, the queue length was set to 216 (larger than the standard MoCo setting), with a momentum update coefficient of 0.999 to better maintain representation consistency across different batches, which is particularly important for subtle feature differences in histopathological images. The detailed experimental parameter settings for HistoMoCo include a batch size of 256, a learning rate of 0.03, a feature dimension of 128, a queue size of , and a MoCo momentum of 0.999.
We observed that using higher-resolution images significantly increases GPU memory consumption during model training. For example, when using ResNet-50 as the backbone with a batch size of 256, even when fine-tuning only the linear layer, images with a resolution of 512 × 512 require 6040 MB of memory, while those with a resolution of 1024 × 1024 demand 18,904 MB. This imposes a substantial computational burden [42]. Therefore, we downsampled the images to a resolution of 224 × 224 in both the pre-training and fine-tuning stages to account for computational resource constraints and ensure compatibility with the standard MoCo implementation.
Two encoder networks, ResNet-18 and ResNet-50 [43], were used to evaluate our consistency across model architectures. In downstream tasks, we evaluated the model performance in both Linear Tuning and End-to-End Tuning settings.
4.3. HistoMoCo Fine-Tuning
We perform fine-tuning on NCT-CRC-HE-100K [36] and two OSCC detection datasets, cluding NDB-UFES [38] and Oral Histopathology [39]. Specifically, we partition these three datasets, with the detailed division ratios presented in Figure 3. Notably, the 80% of NCT-CRC-HE-100K [36] used for fine-tuning is identical to the 80% previously used for pre-training. This setup ensures that the validation and test sets of NCT-CRC-HE-100K [36] remain unseen during the training process. Figure 3 shows the overall training process using HistoMoCo pre-training and subsequent fine-tuning.
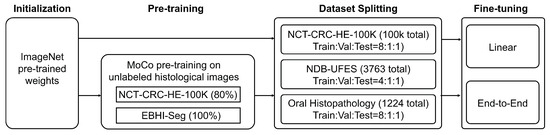
Figure 3.
HistoMoCo training pipeline. MoCo acts as a self-supervised training agent. The model is subsequently fine-tuned using the NCT dataset and two other external OSCC datasets.
We retained the model checkpoint with the best performance over 100 training epochs based on the AUROC metric (using macro-AUROC for NCT-CRC-HE-100K, trained for 10 epochs) on the validation set for evaluation on the test set. To evaluate the transferability of the representations, as shown in Figure 2b, we froze the encoder model and trained a linear classifier on top of it using labeled data (Linear Tuning). Additionally, we unfroze all layers and fine-tuned the entire model end-to-end using labeled data to assess the overall transferability of the performance (End-to-End Tuning). For Linear Tuning, a learning rate of 30 and a batch size of 256 were used, whereas for End-to-End Tuning, the learning rate was decreased to .
5. Experiments
5.1. Statistics of the Datasets
We use the NCT-CRC-HE-100K [36], EBHI-Seg [37], NDB-UFES [38], and Oral Histopathology [39] datasets for the pre-training, fine-tuning, and evaluation of HistoMoCo. The statistical information of these datasets is shown in Table 1, along with their classification/segmentation targets as follows:

Table 1.
Statistics of the histopathological datasets used in HistoMoCo.
- NCT-CRC-HE-100K [36]: Nine tissue types: adipose, background, debris, lymphocytes, mucus, smooth muscle, normal colon mucosa, cancer-associated stroma, colorectal adenocarcinoma epithelium.
- EBHI-Seg [37]: Six tumor differentiation stages: normal, polyp, low-grade intraepithelial neoplasia, high-grade intraepithelial neoplasia, serrated adenoma, and adenocarcinoma.
- NDB-UFES [38]: Two types: oral squamous cell carcinoma, normal epithelium.
- Oral Histopathology [39]: Two types: oral squamous cell carcinoma, normal epithelium.
5.2. Experimental Setups
5.2.1. Baseline Models
We propose the following baselines for comparison:
- Scratch: models are initialized using Kaiming’s random initialization [44] and then fine-tuned on the target datasets.
- ImageNet: models are initialized with the officially released weights pre-trained on the ImageNet dataset and fine-tuned on the target datasets.
- MoCo: models are initialized with the officially released weights (https://github.com/facebookresearch/moco (accessed on 12 March 2025)) from [17] and fine-tuned on the target datasets.
5.2.2. Evaluation Metrics
To evaluate these algorithms, we conduct a comparison of their performance by assessing and contrasting Macro-AUROC (MaROC.), Micro-AUROC (MiROC.), Macro-AUPRC (MaPRC.), Micro-AUPRC (MiPRC.), and Accuracy (Acc.) for multi-class classification task (colorectal tissue classification on NCT-CRC-HE-100K [36]), AUROC (ROC.), AUPRC (PRC.), Accuracy (Acc.), Precision (Pre.), Sensitivity (Sen.), Specificity (Spe.) and F1 Score (F1) for binary classification task (OSCC detection on NDB-UFES [38] and Oral Histopathology [39]).
5.3. Experimental Results
5.3.1. Transfer Performance of HistoMoCo Representations
We investigated whether representations acquired through HistoMoCo pre-training are of higher quality than those transferred from Scratch, ImageNet, and MoCo [40]. To evaluate these representations, we used the Linear Tuning protocol [45,46], where a linear classifier is trained on a frozen base model, and test performance is used as a proxy for representation quality. We also explored whether HistoMoCo pre-training translates to higher model performance by conducting End-to-End Tuning. The results are shown in Table 2. It can be observed that with ResNet-50 as the architecture, although the performance of ResNet-50 models on the NCT-CRC-HE-100K dataset is close to saturation, HistoMoCo still shows significant improvements compared to ImageNet and MoCo, especially in terms of AUPRC and accuracy metrics. This indirectly demonstrates the stronger robustness of HistoMoCo pre-training parameters for imbalanced positive and negative samples. ResNet-18 also reflects similar patterns.

Table 2.
Model performance on colorectal tissue classification task of NCT-CRC-HE-100K dataset. In each experimental setup, the best-performing results are shown in bold, and the second-best results are underlined with underline. When using ResNet-50 as the backbone with End-to-End Tuning, HistoMoCo achieves a 100% Macro-AUROC and Micro-AUROC. This is due to rounding to three decimal places. In fact, under this setting, HistoMoCo achieves a Macro-AUROC of 0.999746 and a Micro-AUROC of 0.999557.
5.3.2. Transfer Benefit of HistoMoCo on OSCC Detection Task
We conducted experiments to test whether the HistoMoCo pre-trained histopathological image representations obtained from the colorectal dataset could be transferred to two OSCC detection datasets (NDB-UFES and Oral Histopathology datasets). Consistent with the previous section, the experiments were based on both Linear Tuning protocol and End-to-End Tuning settings, with results shown in Table 3 and Table 4. It can be observed that HistoMoCo achieved the overall best performance on both external datasets. Notably, the Linear Tuning showed significant improvements in key metrics such as AUROC and AUPRC compared to ImageNet pre-training. Under End-to-End Tuning, even ImageNet pre-training can achieve over 99% ROC and PRC on the OSCC detection task of the NDB-UFES dataset when using ResNet-50. This suggests that the simplicity of this dataset makes it easy to attain high performance, making it challenging to HistoMoCo. However, HistoMoCo still outperformed other baselines overall. Given the small data volume, we believe that the occasional deviation in individual metrics is acceptable. The results on external datasets confirm the generalizability of HistoMoCo to out-of-domain data from different tissue sites, suggesting that it can serve as a more robust pre-training foundation for subsequent histopathological image research. Figure 4 presents the ROC curves of ResNet-50 using various pre-training algorithms on two OSCC detection tasks, 95% confidence intervals were estimated using bootstrapping with 100 independent trials on the testing sets.

Table 3.
Model performance on OSCC detection task of NDB-UFES dataset. In each experimental setup, the highest-performing results are highlighted in bold, while the second-best results are marked with an underline.

Table 4.
Model performance on OSCC detection task of oral histopathology dataset. In each experimental setup, the best-performing results are highlighted in bold, while the second-best results are underlined with an underline.
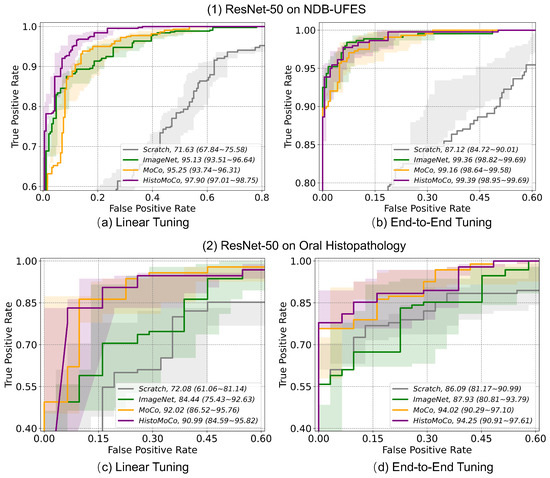
Figure 4.
Receiver operating characteristic (ROC) curves with AUC values (95% CI) for two OSCC detection tasks on the NDB-UFES and Oral Histopathology datasets.
Notably, initializing the backbone model from scratch and performing Linear Tuning resulted in highly biased outcomes, with both precision and sensitivity being 0. This indicates that, under this setting, the model exhibited an extreme tendency to classify all images as normal. The primary reason for this is that the scratch initialization did not provide the backbone model with any features relevant to histopathological images. Consequently, the model failed to generate meaningful feature representations for the subsequent linear classification layer, leading to such extreme results.
5.3.3. Performance Comparison with Similar Literature
We further introduce UNI [30], CONCH [31], DINO [47], and SwAV [16] as baseline models to evaluate the effectiveness of HistoMoCo. Specifically, we utilized UNI [30] and CONCH [31] as feature extractors and performed Linear Tuning experiments on the OSCC detection task of the NDB-UFES dataset (the larger dataset among the two OSCC detection datasets we used to ensure the reliability of the results). The experimental results are shown in Table 5. The experimental results demonstrate that both UNI2-h and CONCH achieved outstanding performance after extensive pre-training on large-scale histopathological image datasets. This further validates our hypothesis that pre-training should be conducted on data distributions consistent with the downstream task rather than on distributionally mismatched data, which could introduce biases. In comparison with UNI2-h, we openly acknowledge the performance limitations of HistoMoCo. UNI2-h achieved the best results in the majority of cases, including ROC, PRC, accuracy, and sensitivity. However, HistoMoCo remains noteworthy, as it achieved the best F1-score and maintained the second-best performance in most cases. More importantly, both UNI2-h and CONCH adopt the Vision Transformer [48] as model backbone, which, while effective, introduces a significant number of trainable parameters (630.76M for UNI2-h and 86.57M for CONCH). In contrast, HistoMoCo employs ResNet-50 [43] with only 25.56M parameters. Notably, when compared to CONCH, HistoMoCo achieved superior performance with fewer parameters. We believe that HistoMoCo strikes a balance between performance and computational efficiency, addressing an existing gap in current research.

Table 5.
Model performance on OSCC detection task of NDB-UFES dataset using HistoMoCo and SOTA models. In each experimental setup, the highest-performing results are highlighted in bold, while the second-best results are marked with an underline.
We implemented DINO [47] and SwAV [16] methods and compared them under the same experimental settings as MoCo, performing Linear Tuning and End-to-End Tuning experiments on the OSCC detection task of the NDB-UFES dataset. The experimental results are shown in Table 5. The experimental results indicate that HistoMoCo outperforms these state-of-the-art (SOTA) models, which is consistent with our hypothesis. Since both DINO and SwAV use natural images for pre-training, the divergence between natural images and histopathological images hinders the generalization of these models to downstream tasks.
5.3.4. Ablation Study
We note that the pre-training dataset, NCT-CRC-HE-100K, consists of histopathological images of colorectal cancer, whereas our target task focuses on OSCC detection. However, OSCC and colorectal cancer exhibit significant anatomical and morphological differences. Therefore, we conduct an ablation study on NCT-CRC-HE-100K to evaluate its effectiveness during the pretraining stage. The experimental results, as presented in Table 6, indicate that the removal of the NCT-CRC-HE-100K dataset leads to a significant performance drop in both Linear Tuning and End-to-End Tuning. We attribute this to HistoMoCo learning relevant histopathological representations from the NCT-CRC-HE-100K dataset, which are subsequently utilized for OSCC detection. Ideally, we believe that using only OSCC images for pre-training would yield the best results. However, due to the scarcity of such data, we resorted to colorectal cancer histopathological images as a related alternative to minimize the distribution gap between pre-training and fine-tuning datasets.

Table 6.
Ablation study of NCT-CRC-HE-100K in HistoMoCo. In each experimental setup, the better-performing results are highlighted in bold.
5.3.5. Sensitivity Analysis
We further analyze several hyperparameters in HistoMoCo, including the momentum update rate (moco-m), the queue size (moco-k), and projection head size (moco-dim), with the experimental results presented in Table 7, Table 8 and Table 9. In this experiment, we adhere to the controlled variable method, analyzing only a single hyperparameter in each experiment. When varying a specific hyperparameter, all other hyperparameters are set to their default values as specified in the methodology, i.e., momo-m = 0.999, moco-k = , and moco-dim = 128 (see Section 4.2). The results indicate that the hyperparameters queue size and projection head size have minimal impact on the final model performance, particularly in the case of End-to-End Tuning. However, the momentum update rate proves to be a crucial parameter. Our experiments suggest that selecting a relatively large momentum update rate (e.g., 0.999) is essential for maintaining a robust MoCo pre-training process.

Table 7.
Sensitivity analysis of the momentum update rate (moco-m) in HistoMoCo.

Table 8.
Sensitivity analysis of the queue size (moco-k) in HistoMoCo.

Table 9.
Sensitivity analysis of the projection head size (moco-dim) in HistoMoCo.
5.3.6. Robustness of HistoMoCo Against Data Insufficiency
We evaluate the robustness of HistoMoCo against insufficient training samples using the NDB-UFES Dataset with ResNet-18. To simulate data insufficiency, we reduce the image size of the training set from 80% to 50%/40%/30%/20%/10% of the entire dataset, while keeping the test set fixed for a fair comparison. Experiments are conducted across these different settings, and the performances of AUROC and AUPRC are plotted in Figure 5. As shown in Figure 5, HistoMoCo consistently outperforms the ImageNet pre-training solution in all settings. More importantly, with Linear Tuning, ImageNet achieved an AUROC of 89.32% using 40% of the training data, whereas HistoMoCo achieved an AUROC of 89.58% using only 10% of the training data. Similarly, with End-to-End Tuning, ImageNet reached an AUROC of 97.08% using 50% of the training data, while HistoMoCo attained an AUROC of 97.16% using only 40% of the training data. This further demonstrates that HistoMoCo enables deep learning models to achieve higher performance with less training data, reducing reliance on manually labeled data.
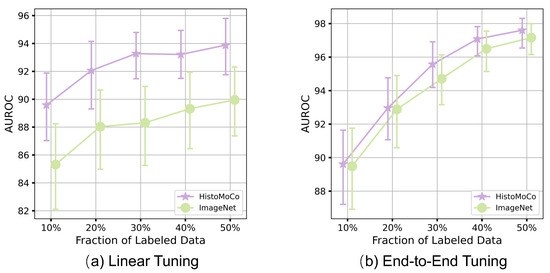
Figure 5.
AUROC on the NDB-UFES dataset for ResNet-18 with ImageNet and HistoMoCo pre-training using various sizes of fine-tuning datasets. HistoMoCo achieves higher performance than ImageNet using a smaller dataset.
6. Conclusions
We introduce HistoMoCo, a method that provides high-quality representations and transferable initializations for interpreting histopathological images, tackling the challenge of distributional divergence between histopathological and natural images. Despite the substantial differences in data and task properties between natural image classification and histopathological interpretation, HistoMoCo successfully adapts the MoCo pre-training approach to histopathological images. This highlights the potential for self-supervised methods to extend beyond natural image classification contexts.
We validated the effectiveness of HistoMoCo across various histopathological images, using the pre-training dataset NCT-CRC-HE-100K and two oral OSCC datasets: Oral Histopathology and NDB-UFES. Extensive experiments demonstrated the model’s generalization capability. Notably, the images in the external oral OSCC datasets were not included in the unsupervised pre-training of HistoMoCo and differed in anatomical sites, as the pre-training images were sourced from the rectum and colon.
Author Contributions
Methodology, W.L. and J.Z.; Formal analysis, Y.H. and B.J.; Writing—original draft, W.L.; Writing—review & editing, W.L.; Visualization, W.L.; Project administration, M.G. and X.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the National Natural Science Foundation of China (No. U23A20468).
Data Availability Statement
These data were derived from the following resources available in the public domain: [A histopathological image repository of normal epithelium of Oral Cavity and Oral Squamous Cell Carcinoma, https://data.mendeley.com/datasets/ftmp4cvtmb/1 (accessed on 12 March 2025); NDB-UFES: An oral cancer and leukoplakia dataset composed of histopathological images and patient data, https://data.mendeley.com/datasets/bbmmm4wgr8/4 (accessed on 12 March 2025); NCT-CRC-HE-100K, https://zenodo.org/records/1214456 (accessed on 12 March 2025); EBHI-Seg, https://figshare.com/articles/dataset/EBHI-SEG/21540159/1?file=38179080 (accessed on 12 March 2025)]. The complete datasets, code and pre-trained parameters are publicly accessible and documented publicly on Github: https://github.com/Heyffff/HistoMoCo (accessed on 12 March 2025).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Mohan, R.; Rama, A.; Raja, R.K.; Shaik, M.R.; Khan, M.; Shaik, B.; Rajinikanth, V. OralNet: Fused optimal deep features framework for oral squamous cell carcinoma detection. Biomolecules 2023, 13, 1090. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2020, 70, 313. [Google Scholar]
- Van der Laak, J.; Litjens, G.; Ciompi, F. Deep learning in histopathology: The path to the clinic. Nat. Med. 2021, 27, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef]
- Fu, Q.; Chen, Y.; Li, Z.; Jing, Q.; Hu, C.; Liu, H.; Bao, J.; Hong, Y.; Shi, T.; Li, K.; et al. A deep learning algorithm for detection of oral cavity squamous cell carcinoma from photographic images: A retrospective study. EClinicalMedicine 2020, 27, 100558. [Google Scholar] [CrossRef]
- Albalawi, E.; Thakur, A.; Ramakrishna, M.T.; Bhatia Khan, S.; SankaraNarayanan, S.; Almarri, B.; Hadi, T.H. Oral squamous cell carcinoma detection using EfficientNet on histopathological images. Front. Med. 2024, 10, 1349336. [Google Scholar] [CrossRef]
- Gao, J.; Zhu, Y.; Wang, W.; Wang, Z.; Dong, G.; Tang, W.; Wang, H.; Wang, Y.; Harrison, E.M.; Ma, L. A comprehensive benchmark for COVID-19 predictive modeling using electronic health records in intensive care. Patterns 2024, 5, 100951. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, C.; Gao, J.; Jiao, X.; Yu, Z.; Zhu, Y.; Wang, T.; Ma, X.; Wang, Y.; Tang, W.; et al. Mortality prediction with adaptive feature importance recalibration for peritoneal dialysis patients. Patterns 2023, 4, 100892. [Google Scholar] [CrossRef]
- Claudio Quiros, A.; Coudray, N.; Yeaton, A.; Yang, X.; Liu, B.; Le, H.; Chiriboga, L.; Karimkhan, A.; Narula, N.; Moore, D.A.; et al. Mapping the landscape of histomorphological cancer phenotypes using self-supervised learning on unannotated pathology slides. Nat. Commun. 2024, 15, 4596. [Google Scholar] [CrossRef]
- Ananthakrishnan, B.; Shaik, A.; Kumar, S.; Narendran, S.; Mattu, K.; Kavitha, M.S. Automated detection and classification of oral squamous cell carcinoma using deep neural networks. Diagnostics 2023, 13, 918. [Google Scholar] [CrossRef]
- Kavyashree, C.; Vimala, H.; Shreyas, J. Improving oral cancer detection using pretrained model. In Proceedings of the 2022 IEEE 6th Conference on Information and Communication Technology (CICT), Gwalior, India, 18–20 November 2022; pp. 1–5. [Google Scholar]
- Redie, D.K.; Bilgaiyan, S.; Sagnika, S. Oral cancer detection using transfer learning-based framework from histopathology images. J. Electron. Imaging 2023, 32, 053004. [Google Scholar] [CrossRef]
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A simple framework for contrastive learning of visual representations. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual, 13–18 July 2020; pp. 1597–1607. [Google Scholar]
- Li, Z.; Tang, H.; Peng, Z.; Qi, G.J.; Tang, J. Knowledge-guided semantic transfer network for few-shot image recognition. IEEE Trans. Neural Netw. Learn. Syst. 2023. [Google Scholar] [CrossRef]
- Liu, C.; Fu, Y.; Xu, C.; Yang, S.; Li, J.; Wang, C.; Zhang, L. Learning a few-shot embedding model with contrastive learning. In Proceedings of the AAAI Conference on Artificial Intelligence, Online, 2–9 February 2021; Volume 35, pp. 8635–8643. [Google Scholar]
- Caron, M.; Misra, I.; Mairal, J.; Goyal, P.; Bojanowski, P.; Joulin, A. Unsupervised learning of visual features by contrasting cluster assignments. Adv. Neural Inf. Process. Syst. 2020, 33, 9912–9924. [Google Scholar]
- He, K.; Fan, H.; Wu, Y.; Xie, S.; Girshick, R. Momentum contrast for unsupervised visual representation learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020; pp. 9729–9738. [Google Scholar]
- Misra, I.; Maaten, L.V.d. Self-supervised learning of pretext-invariant representations. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020; pp. 6707–6717. [Google Scholar]
- Sowrirajan, H.; Yang, J.; Ng, A.Y.; Rajpurkar, P. Moco pretraining improves representation and transferability of chest X-ray models. In Proceedings of the Medical Imaging with Deep Learning, Lübeck, Germany, 7–9 July 2021; pp. 728–744. [Google Scholar]
- Liao, W.; Xiong, H.; Wang, Q.; Mo, Y.; Li, X.; Liu, Y.; Chen, Z.; Huang, S.; Dou, D. Muscle: Multi-task self-supervised continual learning to pre-train deep models for X-ray images of multiple body parts. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; Springer: Berlin/Heidelberg, Germany; pp. 151–161. [Google Scholar]
- Liao, W.; Wang, Q.; Li, X.; Liu, Y.; Chen, Z.; Huang, S.; Dou, D.; Xu, Y.; Xiong, H. MTPret: Improving X-ray Image Analytics with Multi-Task Pre-training. IEEE Trans. Artif. Intell. 2024, 5, 4799–4812. [Google Scholar] [CrossRef]
- Chaitanya, K.; Erdil, E.; Karani, N.; Konukoglu, E. Contrastive learning of global and local features for medical image segmentation with limited annotations. Adv. Neural Inf. Process. Syst. 2020, 33, 12546–12558. [Google Scholar]
- Dufumier, B.; Gori, P.; Victor, J.; Grigis, A.; Wessa, M.; Brambilla, P.; Favre, P.; Polosan, M.; Mcdonald, C.; Piguet, C.M.; et al. Contrastive learning with continuous proxy meta-data for 3D MRI classification. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part II 24. Springer: Berlin/Heidelberg, Germany, 2021; pp. 58–68. [Google Scholar]
- Hollon, T.; Jiang, C.; Chowdury, A.; Nasir-Moin, M.; Kondepudi, A.; Aabedi, A.; Adapa, A.; Al-Holou, W.; Heth, J.; Sagher, O.; et al. Artificial-intelligence-based molecular classification of diffuse gliomas using rapid, label-free optical imaging. Nat. Med. 2023, 29, 828–832. [Google Scholar] [CrossRef]
- Nakhli, R.; Darbandsari, A.; Farahani, H.; Bashashati, A. Ccrl: Contrastive cell representation learning. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 397–407. [Google Scholar]
- Wu, H.; Wang, Z.; Song, Y.; Yang, L.; Qin, J. Cross-patch dense contrastive learning for semi-supervised segmentation of cellular nuclei in histopathologic images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 21–24 June 2022; pp. 11666–11675. [Google Scholar]
- Yang, M.; Yang, Y.; Xie, C.; Ni, M.; Liu, J.; Yang, H.; Mu, F.; Wang, J. Contrastive learning enables rapid mapping to multimodal single-cell atlas of multimillion scale. Nat. Mach. Intell. 2022, 4, 696–709. [Google Scholar] [CrossRef]
- Li, J.; Zheng, Y.; Wu, K.; Shi, J.; Xie, F.; Jiang, Z. Lesion-aware contrastive representation learning for histopathology whole slide images analysis. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 273–282. [Google Scholar]
- Wang, X.; Yang, S.; Zhang, J.; Wang, M.; Zhang, J.; Yang, W.; Huang, J.; Han, X. Transformer-based unsupervised contrastive learning for histopathological image classification. Med Image Anal. 2022, 81, 102559. [Google Scholar] [CrossRef]
- Chen, R.J.; Ding, T.; Lu, M.Y.; Williamson, D.F.; Jaume, G.; Song, A.H.; Chen, B.; Zhang, A.; Shao, D.; Shaban, M.; et al. Towards a general-purpose foundation model for computational pathology. Nat. Med. 2024, 30, 850–862. [Google Scholar] [CrossRef]
- Lu, M.Y.; Chen, B.; Williamson, D.F.; Chen, R.J.; Liang, I.; Ding, T.; Jaume, G.; Odintsov, I.; Le, L.P.; Gerber, G.; et al. A visual-language foundation model for computational pathology. Nat. Med. 2024, 30, 863–874. [Google Scholar] [CrossRef]
- Vorontsov, E.; Bozkurt, A.; Casson, A.; Shaikovski, G.; Zelechowski, M.; Liu, S.; Severson, K.; Zimmermann, E.; Hall, J.; Tenenholtz, N.; et al. Virchow: A million-slide digital pathology foundation model. arXiv 2023, arXiv:2309.07778. [Google Scholar]
- Xiang, J.; Wang, X.; Zhang, X.; Xi, Y.; Eweje, F.; Chen, Y.; Li, Y.; Bergstrom, C.; Gopaulchan, M.; Kim, T.; et al. A vision–language foundation model for precision oncology. Nature 2025, 638, 769–778. [Google Scholar] [CrossRef]
- Wu, L.; Fang, L.; He, X.; He, M.; Ma, J.; Zhong, Z. Querying labeled for unlabeled: Cross-image semantic consistency guided semi-supervised semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 8827–8844. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Xu, L.; Chen, D.Z.; Gao, H.; Wu, J. A robust shape-aware rib fracture detection and segmentation framework with contrastive learning. IEEE Trans. Multimed. 2023, 25, 1584–1591. [Google Scholar] [CrossRef]
- Kather, J.N.; Krisam, J.; Charoentong, P.; Luedde, T.; Herpel, E.; Weis, C.A.; Gaiser, T.; Marx, A.; Valous, N.A.; Ferber, D.; et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med. 2019, 16, e1002730. [Google Scholar] [CrossRef]
- Shi, L.; Li, X.; Hu, W.; Chen, H.; Chen, J.; Fan, Z.; Gao, M.; Jing, Y.; Lu, G.; Ma, D.; et al. EBHI-Seg: A Novel Enteroscope Biopsy Histopathological Haematoxylin and Eosin Image Dataset for Image Segmentation Tasks. arXiv 2022, arXiv:2212.00532. [Google Scholar] [CrossRef]
- Ribeiro-de Assis, M.C.F.; Soares, J.P.; de Lima, L.M.; de Barros, L.A.P.; Grão-Velloso, T.R.; Krohling, R.A.; Camisasca, D.R. NDB-UFES: An oral cancer and leukoplakia dataset composed of histopathological images and patient data. Data Brief 2023, 48, 109128. [Google Scholar] [CrossRef]
- Rahman, T.Y. A histopathological image repository of normal epithelium of oral cavity and oral squamous cell carcinoma. Mendeley Data 2019, 1. [Google Scholar] [CrossRef]
- Chen, X.; Fan, H.; Girshick, R.; He, K. Improved baselines with momentum contrastive learning. arXiv 2020, arXiv:2003.04297. [Google Scholar]
- Raghu, M.; Zhang, C.; Kleinberg, J.; Bengio, S. Transfusion: Understanding transfer learning for medical imaging. In Advances in Neural Information Processing Systems; Springer: Berlin/Heidelberg, Germany, 2019; Volume 32. [Google Scholar]
- Lu, S.; Chen, Y.; Chen, Y.; Li, P.; Sun, J.; Zheng, C.; Zou, Y.; Liang, B.; Li, M.; Jin, Q.; et al. General lightweight framework for vision foundation model supporting multi-task and multi-center medical image analysis. Nat. Commun. 2025, 16, 2097. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1026–1034. [Google Scholar]
- Bachman, P.; Hjelm, R.D.; Buchwalter, W. Learning representations by maximizing mutual information across views. Adv. Neural Inf. Process. Syst. 2019, 32. Available online: https://dl.acm.org/doi/abs/10.5555/3454287.3455679 (accessed on 12 March 2025).
- Kornblith, S.; Shlens, J.; Le, Q.V. Do better imagenet models transfer better? In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 2661–2671. [Google Scholar]
- Caron, M.; Touvron, H.; Misra, I.; Jégou, H.; Mairal, J.; Bojanowski, P.; Joulin, A. Emerging properties in self-supervised vision transformers. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 9650–9660. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. In Proceedings of the International Conference on Learning Representations, Addis Ababa, Ethiopia, 30 April 2020. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).