Multiple Differential Convolution and Local-Variation Attention UNet: Nucleus Semantic Segmentation Based on Multiple Differential Convolution and Local-Variation Attention
Abstract
1. Introduction
2. Materials and Methods
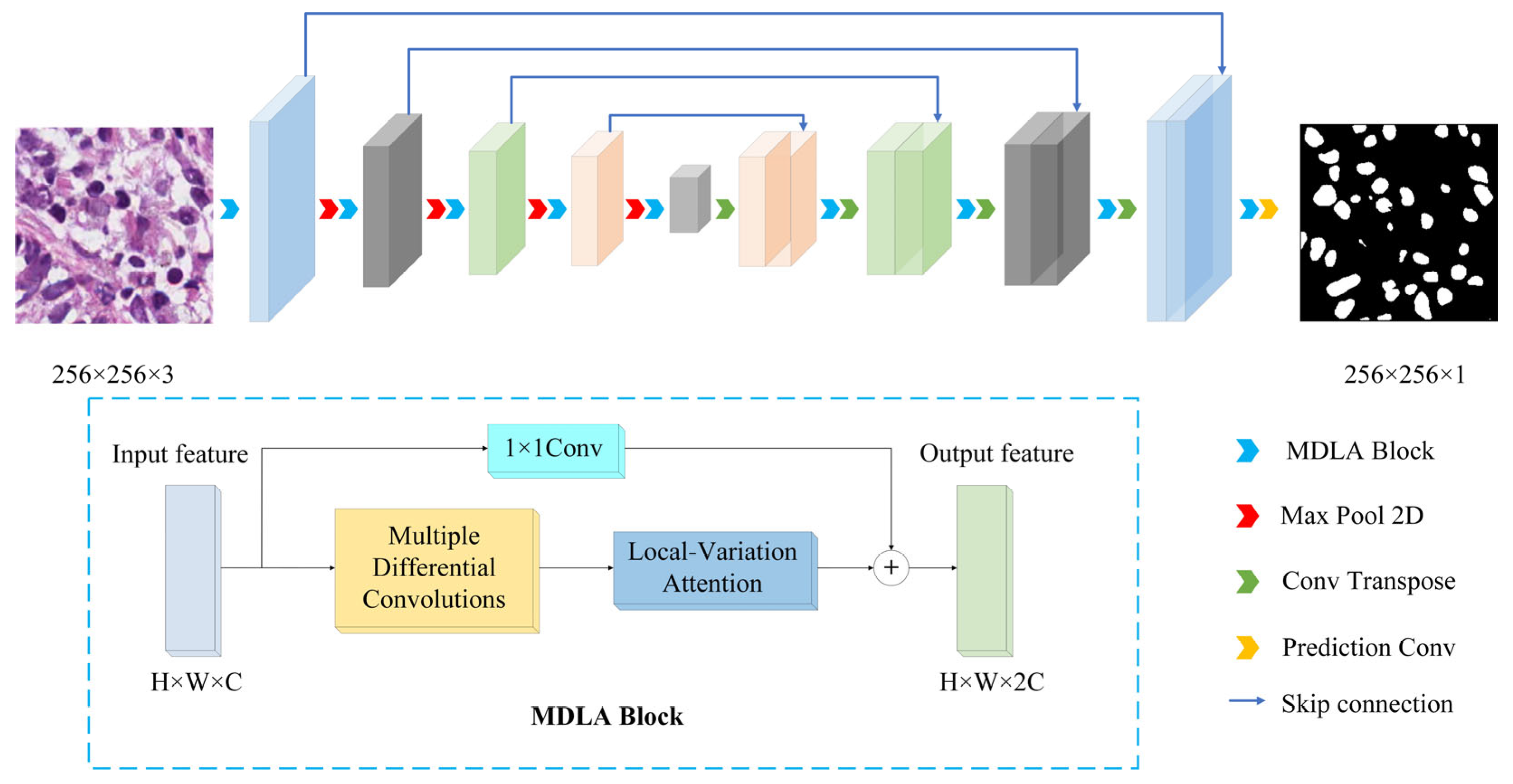
2.1. Network Structure
2.2. Multiple Differential Convolution Block (MDC)
2.3. Local-Variation Attention Block (LVA)
3. Experiment
3.1. Datasets and Preprocessing
3.2. Evaluation Metrics
3.3. Loss Function
3.4. Training Strategy and Parameter Settings
3.5. Detection Performance Comparative Experiment
4. Discussion
5. Conclusions and Future Work
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gheisari, M.; Ebrahimzadeh, F.; Rahimi, M.; Moazzamigodarzi, M.; Liu, Y.; Pramanik, P.K.D.; Heravi, M.A.; Mehbodniya, A.; Ghaderzadeh, M.; Feylizadeh, M.R.; et al. Deep Learning: Applications, Architectures, Models, Tools, and Frameworks: A Comprehensive Survey. CAAI Trans. Intell. Technol. 2023, 8, 581–606. [Google Scholar] [CrossRef]
- Najjar, R. Redefining Radiology: A Review of Artificial Intelligence Integration in Medical Imaging. Diagnostics 2023, 13, 2760. [Google Scholar] [CrossRef] [PubMed]
- Koetzier, L.R.; Mastrodicasa, D.; Szczykutowicz, T.P.; van der Werf, N.R.; Wang, A.S.; Sandfort, V.; van der Molen, A.J.; Fleischmann, D.; Willemink, M.J. Deep Learning Image Reconstruction for CT: Technical Principles and Clinical Prospects. Radiology 2023, 306, e221257. [Google Scholar] [CrossRef]
- Chakrabarty, N.; Mahajan, A. Imaging Analytics Using Arti Fi Cial Intelligence in Oncology: A Comprehensive Review. Clin. Oncol. 2024, 36, 498–513. [Google Scholar] [CrossRef]
- Gadermayr, M.; Tschuchnig, M. Multiple Instance Learning for Digital Pathology: A Review of the State-of-the-Art, Limitations & Future Potential. Comput. Med. Imaging Graph. 2024, 112, 102337. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, C.; Wei, X.; Yan, T.; Zhou, W.; Zhang, J.; Kang, H.; Yuan, Z.; Chen, X. Harnessing the Power of Optical Microscopy for Visualization and Analysis of Histopathological Images. Biomed. Opt. Express 2023, 14, 5451–5465. [Google Scholar] [CrossRef]
- Xing, F.; Xie, Y.; Su, H.; Liu, F.; Yang, L. Deep Learning in Microscopy Image Analysis: A Survey. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 4550–4568. [Google Scholar] [CrossRef]
- Basu, A.; Senapati, P.; Deb, M.; Rai, R.; Dhal, K.G. A Survey on Recent Trends in Deep Learning for Nucleus Segmentation from Histopathology Images. Evol. Syst. 2024, 15, 203–248. [Google Scholar] [CrossRef] [PubMed]
- Zinchuk, V.; Grossenbacher-Zinchuk, O. Machine Learning for Analysis of Microscopy Images: A Practical Guide and Latest Trends. Curr. Protoc. 2023, 3, e819. [Google Scholar] [CrossRef]
- Melanthota, S.K.; Gopal, D.; Chakrabarti, S.; Kashyap, A.A.; Radhakrishnan, R.; Mazumder, N. Deep Learning-Based Image Processing in Optical Microscopy. Biophys. Rev. 2022, 14, 463–481. [Google Scholar] [CrossRef]
- Shelhamer, E.; Long, J.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Falk, T.; Mai, D.; Bensch, R.; Cicek, O.; Abdulkadir, A.; Marrakchi, Y.; Boehm, A.; Deubner, J.; Jaeckel, Z.; Seiwald, K.; et al. U-Net: Deep Learning for Cell Counting, Detection, and Morphometry. Nat. Methods 2019, 16, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Stoyanov, D., Taylor, Z., Carneiro, G., Syeda-Mahmood, T., Martel, A., Maier-Hein, L., Tavares, J.M.R.S., Bradley, A., Papa, J.P., Belagiannis, V., et al., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 3–11. [Google Scholar]
- Long, F. Microscopy Cell Nuclei Segmentation with Enhanced U-Net. BMC Bioinf. 2020, 21, 8. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.-W.; Wu, J. UNet 3+: A Full-Scale Connected UNet for Medical Image Segmentation. In Proceedings of the ICASSP 2020—2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1055–1059. [Google Scholar] [CrossRef]
- Alom, M.Z.; Yakopcic, C.; Hasan, M.; Taha, T.M.; Asari, V.K. Recurrent Residual U-Net for Medical Image Segmentation. J. Med. Imaging 2019, 6, 014006. [Google Scholar] [CrossRef]
- Jafari, M.; Auer, D.; Francis, S.; Garibaldi, J.; Chen, X. DRU-Net: An Efficient Deep Convolutional Neural Network for Medical Image Segmentation. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1144–1148. [Google Scholar] [CrossRef]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-Like Pure Transformer for Medical Image Segmentation. In Proceedings of the Computer Vision—ECCV 2022 Workshops; Karlinsky, L., Michaeli, T., Nishino, K., Eds.; Springer Nature: Cham, Switzerland, 2023; pp. 205–218. [Google Scholar] [CrossRef]
- Chen, K.; Zhang, N.; Powers, L.; Roveda, J. Cell Nuclei Detection and Segmentation for Computational Pathology Using Deep Learning. In Proceedings of the 2019 Spring Simulation Conference (SpringSim), Tucson, AZ, USA, 29 April–2 May 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Kowal, M.; Żejmo, M.; Skobel, M.; Korbicz, J.; Monczak, R. Cell Nuclei Segmentation in Cytological Images Using Convolutional Neural Network and Seeded Watershed Algorithm. J. Digit. Imaging 2020, 33, 231–242. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.C.H.; Heinrich, M.P.; Misawa, K.; Mori, K.; McDonagh, S.G.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar] [CrossRef]
- Zeng, Z.; Xie, W.; Zhang, Y.; Lu, Y. RIC-Unet: An Improved Neural Network Based on Unet for Nuclei Segmentation in Histology Images. IEEE Access 2019, 7, 21420–21428. [Google Scholar] [CrossRef]
- Dogar, G.M.; Fraz, M.M.; Javed, S. Feature Attention Network for Simultaneous Nuclei Instance Segmentation and Classification in Histology Images. In Proceedings of the 2021 International Conference on Digital Futures and Transformative Technologies (ICoDT2), Islamabad, Pakistan, 20–21 May 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Ali, H.; ul Haq, I.; Cui, L.; Feng, J. MSAL-Net: Improve Accurate Segmentation of Nuclei in Histopathology Images by Multiscale Attention Learning Network. BMC Med. Inf. Decis. Making 2022, 22, 90. [Google Scholar] [CrossRef]
- Ghosh, S.; Das, S. Multi-Scale Morphology-Aided Deep Medical Image Segmentation. Eng. Appl. Artif. Intell. 2024, 137, 109047. [Google Scholar] [CrossRef]
- Wang, H.; Cao, P.; Yang, J.; Zaiane, O. Narrowing the Semantic Gaps in U-Net with Learnable Skip Connections: The Case of Medical Image Segmentation. Neural Netw. 2024, 178, 106546. [Google Scholar] [CrossRef]
- Tan, D.; Hao, R.; Zhou, X.; Xia, J.; Su, Y.; Zheng, C. A Novel Skip-Connection Strategy by Fusing Spatial and Channel Wise Features for Multi-Region Medical Image Segmentation. IEEE J. Biomed. Health Inf. 2024, 28, 5396–5409. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Zhang, D. Wavelet Transform. In Fundamentals of Image Data Mining. TCS; Springer: Cham, Switzerland, 2019; pp. 35–44. [Google Scholar] [CrossRef]
- Xu, G.; Liao, W.; Zhang, X.; Li, C.; He, X.; Wu, X. Haar Wavelet Downsampling: A Simple but Effective Downsampling Module for Semantic Segmentation. Pattern Recognit. 2023, 143, 109819. [Google Scholar] [CrossRef]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. CBAM: Convolutional Block Attention Module. In Proceedings of the Computer Vision—ECCV 2018; Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 3–19. [Google Scholar] [CrossRef]
- Kumar, N.; Verma, R.; Anand, D.; Zhou, Y.; Onder, O.F.; Tsougenis, E.; Chen, H.; Heng, P.-A.; Li, J.; Hu, Z.; et al. A Multi-Organ Nucleus Segmentation Challenge. IEEE Trans. Med. Imaging 2020, 39, 1380–1391. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Verma, R.; Sharma, S.; Bhargava, S.; Vahadane, A.; Sethi, A. A Dataset and a Technique for Generalized Nuclear Segmentation for Computational Pathology. IEEE Trans. Med. Imaging 2017, 36, 1550–1560. [Google Scholar] [CrossRef]
- Naylor, P.; Lae, M.; Reyal, F.; Walter, T. Segmentation of Nuclei in Histopathology Images by Deep Regression of the Distance Map. IEEE Trans. Med. Imaging 2019, 38, 448–459. [Google Scholar] [CrossRef]
- Mahbod, A.; Schaefer, G.; Bancher, B.; Löw, C.; Dorffner, G.; Ecker, R.; Ellinger, I. CryoNuSeg: A Dataset for Nuclei Instance Segmentation of Cryosectioned H&E-Stained Histological Images. Comput. Biol. Med. 2021, 132, 104349. [Google Scholar] [CrossRef]
- Lin, T.-L.; Lu, C.-T.; Karmakar, R.; Nampalley, K.; Mukundan, A.; Hsiao, Y.-P.; Hsieh, S.-C.; Wang, H.-C. Assessing the Efficacy of the Spectrum-Aided Vision Enhancer (SAVE) to Detect Acral Lentiginous Melanoma, Melanoma In Situ, Nodular Melanoma, and Superficial Spreading Melanoma. Diagnostics 2024, 14, 1672. [Google Scholar] [CrossRef]






| Datasets | Networks | ACC (%) | SE (%) | JS (%) | DC (%) | |
|---|---|---|---|---|---|---|
| MoNuSeg | U-Net [12] | 89.67 | 86.15 | 62.29 | 76.10 | |
| R2U-Net [18] | 89.04 | 84.02 | 60.57 | 75.27 | ||
| Attention U-Net [23] | 91.40 | 79.59 | 64.02 | 77.72 | ||
| UNet++ [14] | 90.91 | 84.99 | 64.62 | 77.98 | ||
| Swin-Unet [20] | w/o pretrain | 91.05 | 80.23 | 63.66 | 77.57 | |
| w/ pretrain | 90.36 | 82.68 | 62.46 | 76.44 | ||
| Morph-UNet- EfficinetNetB4 [27] | w/o pretrain | 91.29 | 79.35 | 64.00 | 77.92 | |
| w/ pretrain | 89.67 | 75.33 | 58.85 | 73.57 | ||
| UDTransNet [28] | w/o pretrain | 91.28 | 80.52 | 64.34 | 78.19 | |
| w/ pretrain | 91.86 | 81.04 | 66.01 | 79.41 | ||
| FSCA-Net [29] | 91.84 | 83.41 | 66.34 | 79.50 | ||
| MDLA-UNet (Ours) | 92.38 | 85.17 | 68.72 | 81.36 | ||
| TNBC | U-Net | 95.41 | 81.50 | 68.34 | 81.11 | |
| R2U-Net | 92.16 | 55.82 | 45.89 | 62.37 | ||
| Attention U-Net | 95.29 | 83.66 | 67.93 | 80.84 | ||
| UNet++ | 95.58 | 85.39 | 70.07 | 82.33 | ||
| Swin-Unet | w/o pretrain | 94.07 | 79.68 | 61.97 | 76.39 | |
| w/ pretrain | 94.78 | 81.34 | 65.19 | 78.83 | ||
| Morph-UNet- EfficinetNetB4 | w/o pretrain | 94.50 | 77.78 | 58.83 | 74.02 | |
| w/ pretrain | 91.50 | 72.35 | 51.77 | 67.89 | ||
| UDTransNet | w/o pretrain | 93.43 | 78.47 | 62.88 | 77.14 | |
| w/ pretrain | 95.47 | 81.60 | 68.21 | 81.04 | ||
| FSCA-Net | 95.55 | 81.24 | 68.81 | 81.40 | ||
| MDLA-UNet (Ours) | 95.92 | 84.43 | 71.73 | 83.46 | ||
| CryoNuSeg | U-Net | 90.15 | 81.13 | 65.71 | 79.11 | |
| R2U-Net | 87.49 | 75.32 | 58.62 | 73.61 | ||
| Attention U-Net | 89.83 | 83.93 | 65.82 | 79.15 | ||
| UNet++ | 90.42 | 84.36 | 67.18 | 80.17 | ||
| Swin-Unet | w/o pretrain | 89.54 | 83.01 | 64.83 | 78.42 | |
| w/ pretrain | 90.63 | 83.00 | 67.43 | 80.41 | ||
| Morph-UNet- EfficinetNetB4 | w/o pretrain | 87.82 | 78.38 | 60.05 | 74.86 | |
| w/ pretrain | 85.14 | 75.95 | 54.60 | 70.49 | ||
| UDTransNet | w/o pretrain | 88.35 | 76.49 | 60.61 | 75.32 | |
| w/ pretrain | 89.71 | 78.47 | 64.39 | 78.25 | ||
| FSCA-Net | 90.42 | 82.50 | 66.88 | 79.94 | ||
| MDLA-UNet (Ours) | 90.75 | 84.52 | 68.00 | 80.73 | ||
| Networks | Params (M) | Test Speed (img/s) |
|---|---|---|
| U-Net | 8.64 | 1115 |
| R2U-Net | 9.78 | 454 |
| Attention U-Net | 8.73 | 846 |
| UNet++ | 10.20 | 715 |
| Swin-Unet | 41.34 | 193 |
| Morph-UNet-EfficinetNetB4 | 0.42 | 237 |
| UDTransNet | 33.80 | 200 |
| FSCA-Net | 43.36 | 178 |
| MDLA-UNet (Ours) | 39.08 | 181 |
| Datasets | Network | ACC (%) | SE (%) | JS (%) | DC (%) | ||
|---|---|---|---|---|---|---|---|
| U-Net | MDC | LVA | |||||
| MoNuSeg | √ | 89.67 | 86.15 | 62.29 | 76.10 | ||
| √ | √ | 91.71 | 84.96 | 66.42 | 79.67 | ||
| √ | √ | 91.80 | 84.73 | 66.99 | 80.09 | ||
| √ | √ | √ | 92.38 | 85.17 | 68.72 | 81.36 | |
| TNBC | √ | 95.41 | 81.50 | 68.34 | 81.11 | ||
| √ | √ | 95.94 | 83.50 | 71.36 | 83.20 | ||
| √ | √ | 95.55 | 83.06 | 69.36 | 81.85 | ||
| √ | √ | √ | 95.92 | 84.43 | 71.73 | 83.46 | |
| CryoNuSeg | √ | 90.15 | 81.13 | 65.71 | 79.11 | ||
| √ | √ | 90.74 | 83.61 | 67.87 | 80.66 | ||
| √ | √ | 89.77 | 84.32 | 65.85 | 79.21 | ||
| √ | √ | √ | 90.75 | 84.52 | 68.00 | 80.73 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.; Li, S.; Chen, Y.; Chen, J.; Geng, H.; Sun, K.; Zhu, Y.; Su, B.; Zhang, H. Multiple Differential Convolution and Local-Variation Attention UNet: Nucleus Semantic Segmentation Based on Multiple Differential Convolution and Local-Variation Attention. Electronics 2025, 14, 1058. https://doi.org/10.3390/electronics14061058
Sun X, Li S, Chen Y, Chen J, Geng H, Sun K, Zhu Y, Su B, Zhang H. Multiple Differential Convolution and Local-Variation Attention UNet: Nucleus Semantic Segmentation Based on Multiple Differential Convolution and Local-Variation Attention. Electronics. 2025; 14(6):1058. https://doi.org/10.3390/electronics14061058
Chicago/Turabian StyleSun, Xiaoming, Shilin Li, Yongji Chen, Junxia Chen, Hao Geng, Kun Sun, Yuemin Zhu, Bochao Su, and Hu Zhang. 2025. "Multiple Differential Convolution and Local-Variation Attention UNet: Nucleus Semantic Segmentation Based on Multiple Differential Convolution and Local-Variation Attention" Electronics 14, no. 6: 1058. https://doi.org/10.3390/electronics14061058
APA StyleSun, X., Li, S., Chen, Y., Chen, J., Geng, H., Sun, K., Zhu, Y., Su, B., & Zhang, H. (2025). Multiple Differential Convolution and Local-Variation Attention UNet: Nucleus Semantic Segmentation Based on Multiple Differential Convolution and Local-Variation Attention. Electronics, 14(6), 1058. https://doi.org/10.3390/electronics14061058









