Fast Multi-View Subspace Clustering Based on Flexible Anchor Fusion
Abstract
1. Introduction
- (1)
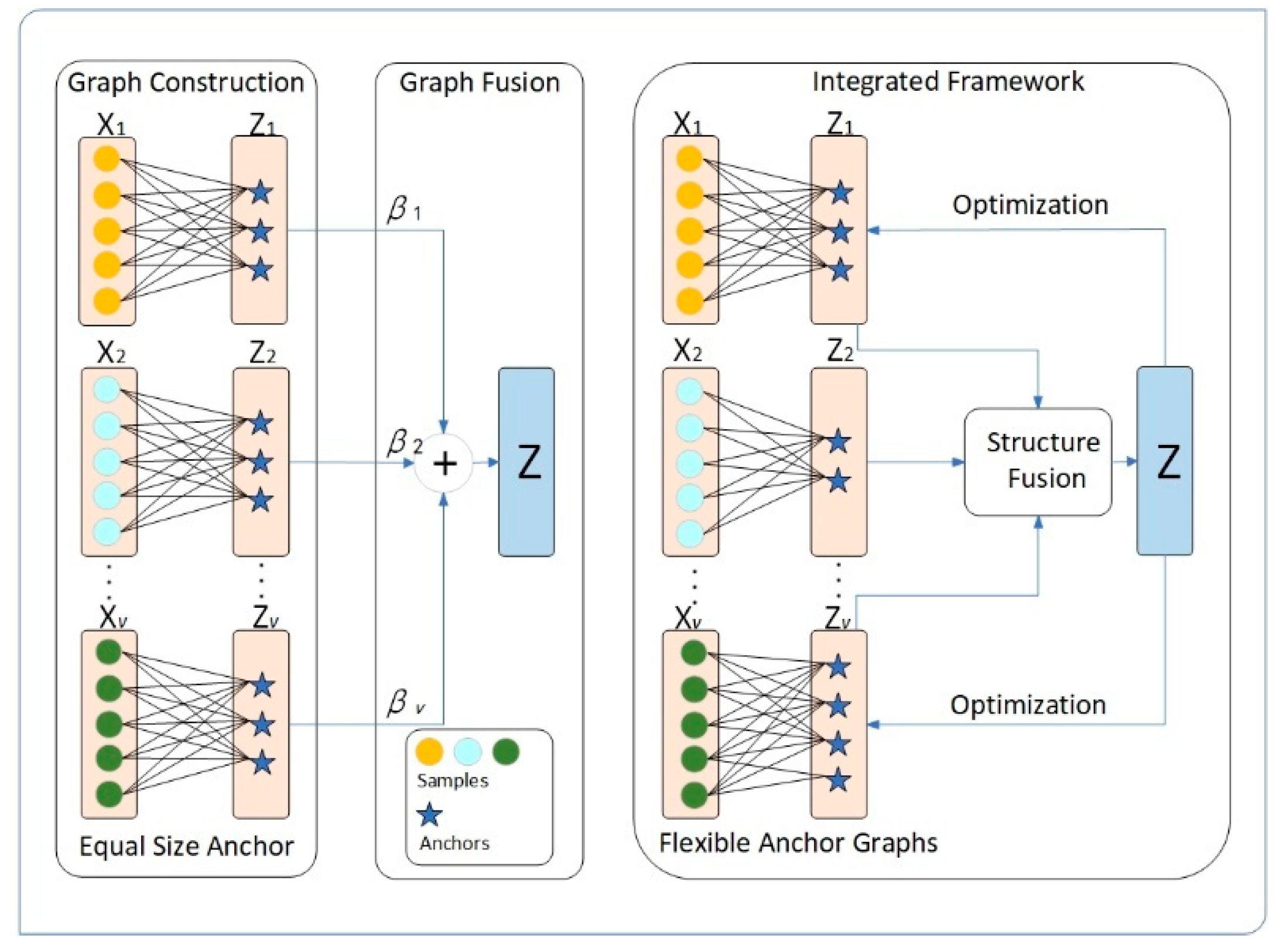
- Unlike existing frameworks, our proposed framework can automatically learn and update anchor points. We integrate the construction and optimization of the anchor matrix and anchor graph into a unified framework. By iteratively updating these components using consensus and complementary information, they can more accurately capture the underlying data structure.
- (2)
- We focus not only on capturing the global structure of the data but also exploring its local structure. Integrating both global and local structures helps to enhance the final clustering performance. Based on the global and local structures, we propose an alternative optimization method with linear complexity relative to the number of examples, and the experimental results demonstrate the effectiveness and efficiency of FMVSC.
- (3)
- Instead of using the same number of anchors, we believe that each view has its own unique underlying data space, and the number of anchors should be selected to match the characteristics of each view. Therefore, we propose a new framework which can learn anchor graphs that align with the data space of individual views and fuse anchor graphs of different sizes.
2. Related Work and Proposed Approach
2.1. Single-View Subspace Clustering
2.2. Multi-View Subspace Clustering
2.3. Anchor-Based Multi-View Subspace Clustering
2.4. Spectral Clustering
2.5. Proposed Approach
3. Optimization
| Algorithm 1 FMVSC |
| Input: Multi-view dataset , cluster number , parameter Initialization: Initialize 1: Repeat 2: Update in Equation (16) 3: Update in Equation (23) 4: Update in Equation (25) 5: Update in Equation (26) 6: Update in Equation (28) 7: Until converge 8: Obtain the spectral embedding matrix Output: Perform k-means clustering on to find the final results. |
4. Experiments
4.1. Dataset Descriptions
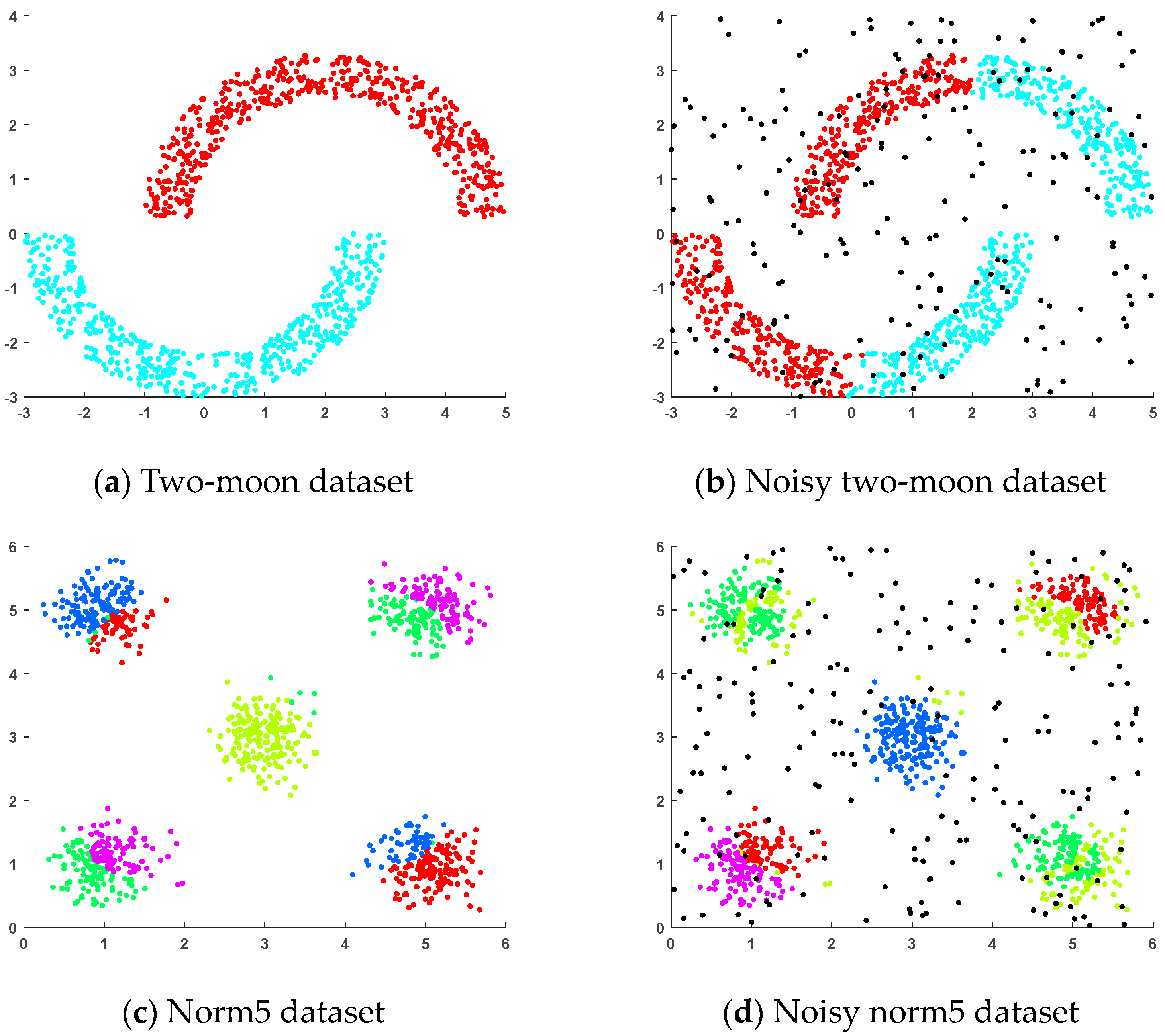
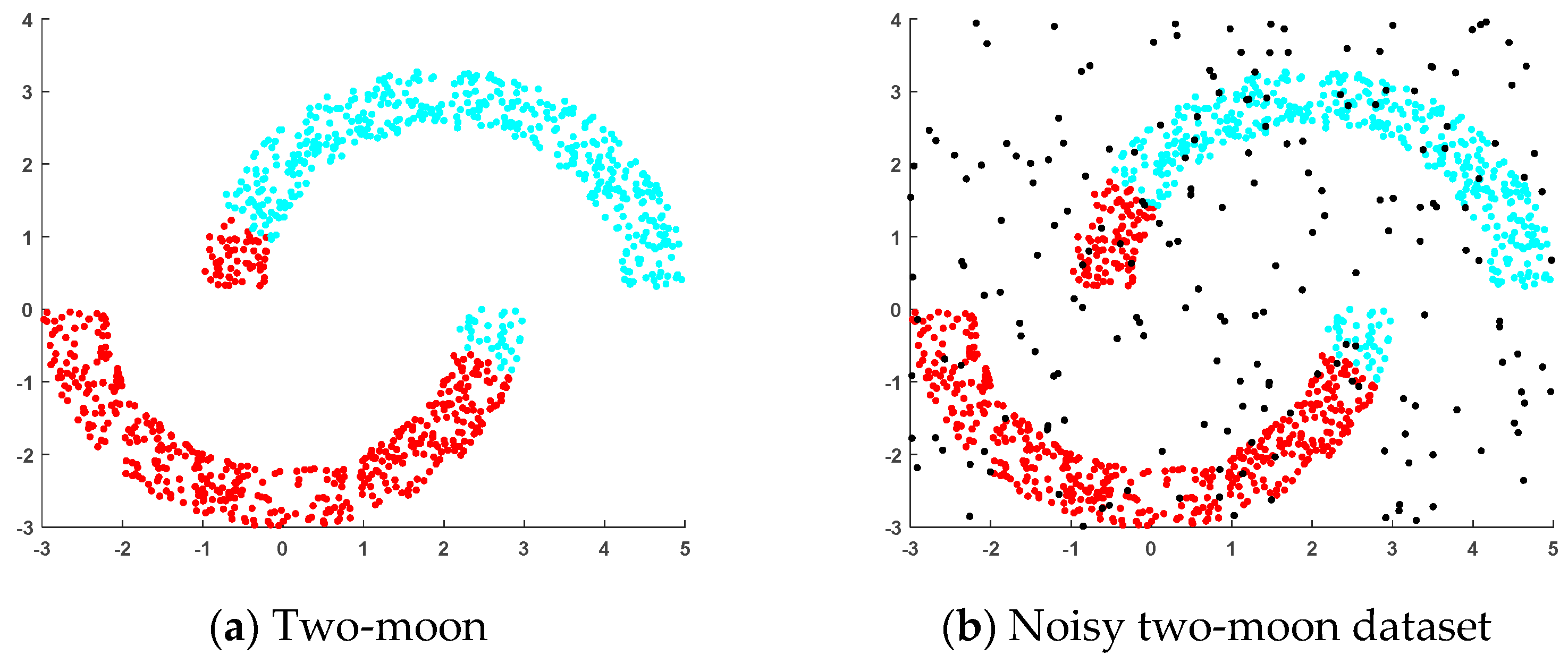
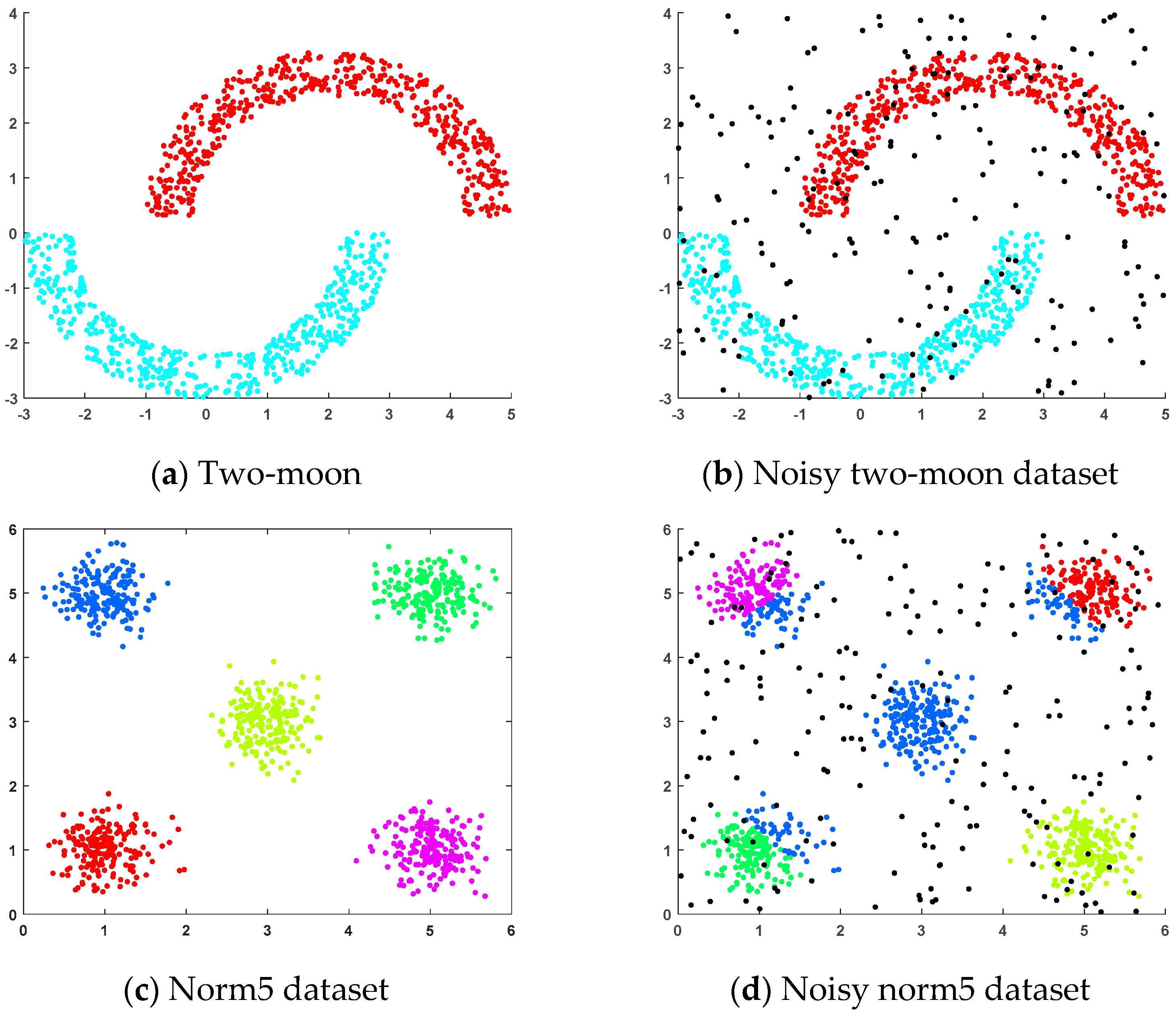
4.1.1. Artificial Datasets
- (a)
- Two-moon dataset: This dataset was generated randomly, where two sets of points were positioned next to each other in a moon shape and each set of points contained 500 random points.
- (b)
- Noisy two-moon dataset: This dataset was generated by adding 200 randomly generated noise points to the two-moon dataset.
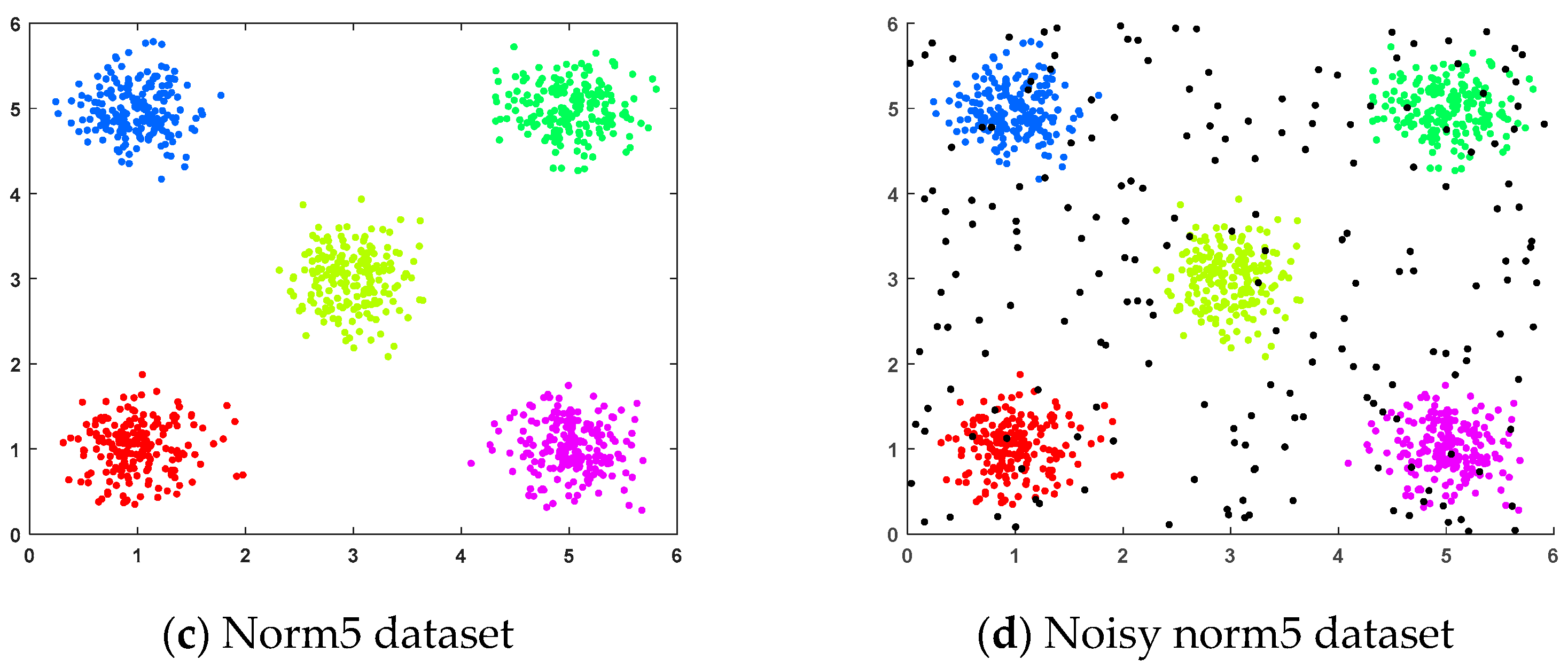
- (c)
- Norm5 dataset: This randomly generated dataset consisted of five clusters, each containing 200 samples, distributed across five spherical regions.
- (d)
- Noisy norm5 dataset: This dataset was generated by adding 200 randomly generated noise points to the norm5 dataset.
4.1.2. Real-World Datasets
4.2. Compared Methods
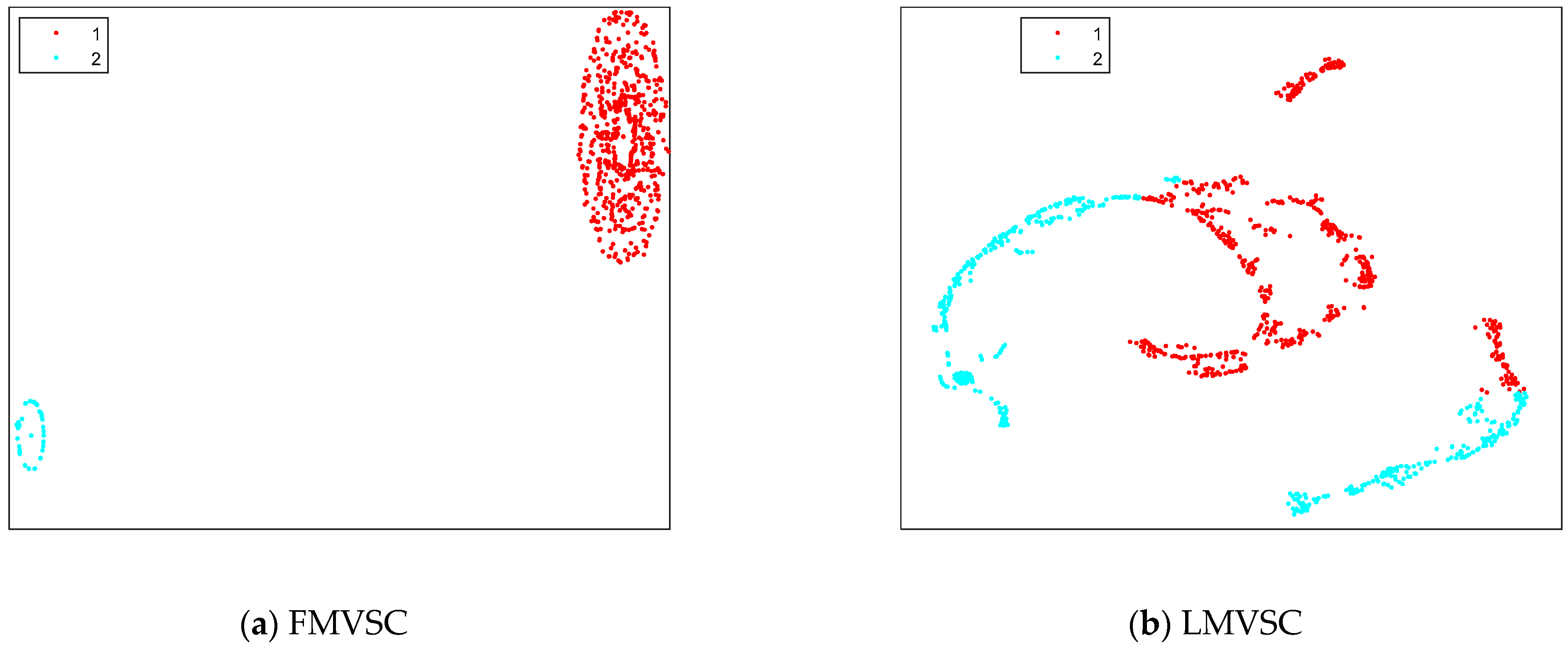
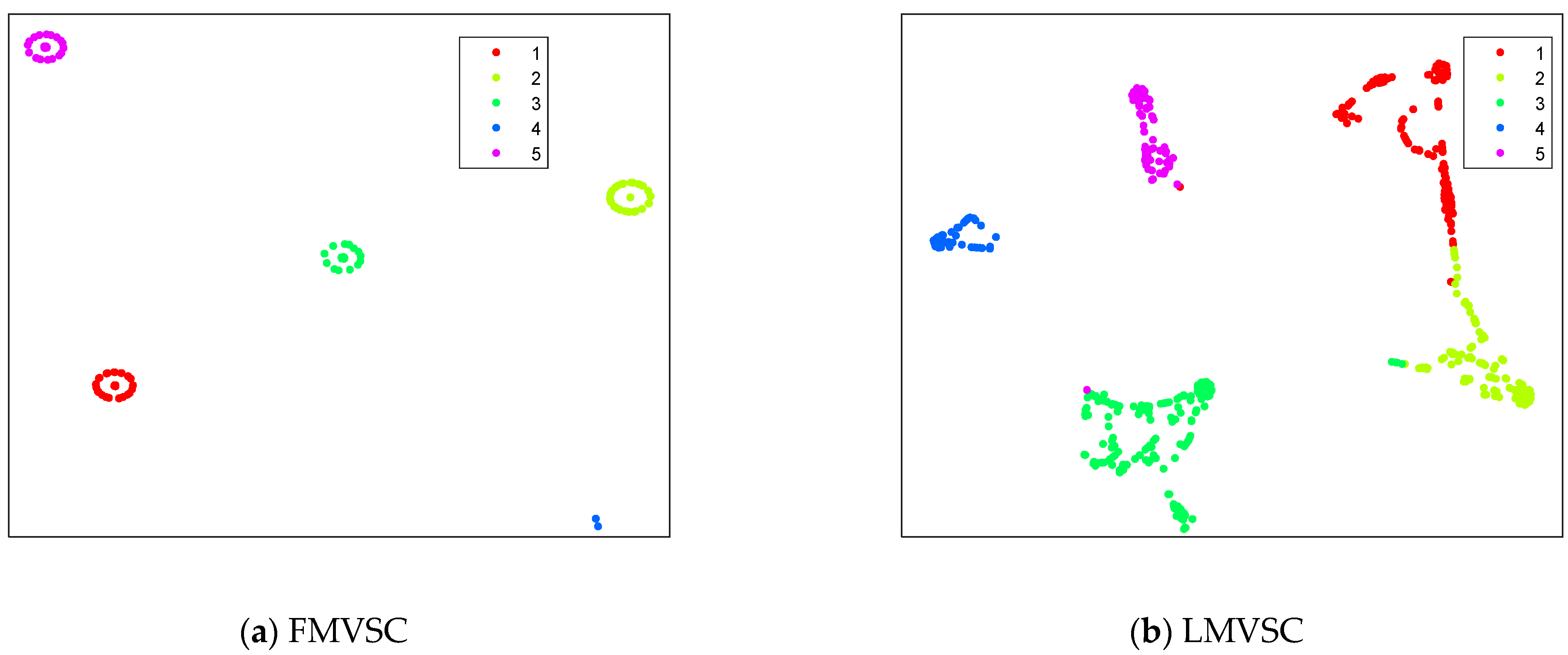
4.3. Results on Artificial Datasets
4.4. Experimental Results
- (a)
- Compared with other baseline algorithms, FMVSC demonstrated significant superiority in performance. Especially in terms of the ACC metric, FMVSC achieved the optimal value across all cases. Compared with the second-best algorithm, FMVSC improved the ACC scores by 0.44%, 1.4%, 4.9%, 3.1%, 2.5%, 2.1%, 2.0%, and 3.8%, respectively. In the other three metrics, FMVSC also achieved either the optimal or second-best results. This highlights the excellent clustering performance of FMVSC.
- (b)
- Compared with classic MVSC algorithms such as MLRSSC and MSC_IAS, FMVSC reduced both the time and space complexity. By leveraging a flexible anchor point sampling strategy, it is better suited for large-scale datasets.
- (c)
- FMVSC also achieved impressive results on smaller-scale datasets. When compared with fast MVSC methods such as LMVSC, MSGL, SMVSC, RAMCSF, and OMVCDR, FMVSC demonstrated clear performance advantages. This indicates that the flexible anchor graphs constructed for each view effectively captured the data space.
- (d)
- Compared with fast MVSC algorithms such as LMVSC, MSGL, SMVSC, and RAMCSF, the smaller standard deviation of FMVSC’s experimental results indicates that the variation in the results was low, suggesting that FMVSC is stable. Additionally, SFMC and OMVCDR do not rely on randomized methods like k-means clustering during the clustering process, which is why their experimental results had a standard deviation of zero.
| Dataset | MLRSSC | MSC_IAS | LMVSC | MSGL | SMVSC | SFMC | RAMCSF | OMVCDR | Ours |
|---|---|---|---|---|---|---|---|---|---|
| Year | 2018 | 2019 | 2020 | 2022 | 2022 | 2022 | 2023 | 2024 | 2024 |
| Notting-Hill | 80.61 ± 4.31 | 77.24 ± 5.46 | 77.82 ± 6.48 | 73.41 ± 5.77 | 75.45 ± 6.33 | 84.91 ± 0.00 | 86.64 ± 7.18 | 83.57 ± 0.00 | 88.38 ± 4.29 |
| WebKB | 90.77 ± 0.22 | 77.07 ± 1.48 | 68.34 ± 0.13 | 93.03 ± 0.00 | 90.87 ± 0.00 | 94.76 ± 0.00 | 94.19 ± 0.00 | 95.37 ± 0.00 | 95.81 ± 0.00 |
| Wiki | 15.67 ± 0.45 | 23.97 ± 1.26 | 49.83 ± 2.33 | 51.16 ± 1.97 | 50.03 ± 4.17 | 33.27 ± 0.00 | 43.94 ± 2.94 | 46.71 ± 0.00 | 56.04 ± 2.14 |
| CCV | 15.87 ± 0.31 | 14.10 ± 0.29 | 20.12 ± 0.58 | 15.44 ± 0.41 | 19.78 ± 0.46 | 11.13 ± 0.00 | 14.16 ± 0.87 | 19.74 ± 0.00 | 22.87 ± 0.34 |
| ALOI | OM | OM | 42.18 ± 1.71 | 16.93 ± 0.74 | 52.36 ± 2.24 | OM | 55.75 ± 2.37 | 63.84 ± 0.00 | 66.36 ± 2.11 |
| Animal | OM | OM | 10.63 ± 0.19 | 11.09 ± 0.21 | 13.16 ± 0.22 | OM | 15.67 ± 0.42 | 14.23 ± 0.00 | 17.77 ± 0.33 |
| NUSWIDE | OM | OM | 12.27 ± 0.27 | 13.60 ± 0.67 | 12.13 ± 0.37 | OM | 14.08 ± 0.53 | 13.53 ± 0.00 | 16.02 ± 0.39 |
| YoutubeFace | OM | OM | 14.25 ± 0.53 | 16.71 ± 0.57 | OM | OM | 17.64 ± 0.85 | OM | 21.39 ± 0.61 |
| Dataset | MLRSSC | MSC_IAS | LMVSC | MSGL | SMVSC | SFMC | RAMCSF | OMVCDR | Ours |
|---|---|---|---|---|---|---|---|---|---|
| Year | 2018 | 2019 | 2020 | 2022 | 2022 | 2022 | 2023 | 2024 | 2024 |
| Notting-Hill | 67.33 ± 4.09 | 71.93 ± 3.52 | 66.33 ± 4.04 | 59.14 ± 5.81 | 72.04 ± 4.85 | 81.62 ± 0.00 | 77.23 ± 6.47 | 76.15 ± 0.00 | 79.86 ± 3.61 |
| WebKB | 47.27 ± 0.31 | 19.09 ± 0.12 | 13.24 ± 0.08 | 58.14 ± 0.00 | 55.19 ± 0.00 | 62.74 ± 0.00 | 60.81 ± 0.00 | 64.29 ± 0.00 | 73.25 ± 0.00 |
| Wiki | 12.31 ± 0.68 | 18.65 ± 1.03 | 45.39 ± 2.07 | 48.63 ± 1.83 | 46.38 ± 3.25 | 31.29 ± 0.00 | 37.62 ± 2.03 | 41.94 ± 0.00 | 52.40 ± 1.50 |
| CCV | 11.98 ± 0.40 | 9.50 ± 0.35 | 16.48 ± 0.47 | 11.89 ± 0.53 | 14.82 ± 0.61 | 3.13 ± 0.00 | 7.62 ± 0.95 | 17.58 ± 0.00 | 19.27 ± 0.41 |
| ALOI | OM | OM | 51.23 ± 1.93 | 22.54 ± 0.81 | 61.47 ± 1.89 | OM | 64.73 ± 2.39 | 72.70 ± 0.00 | 80.65 ± 1.76 |
| Animal | OM | OM | 6.95 ± 0.17 | 6.47 ± 0.27 | 12.63 ± 0.24 | OM | 14.21 ± 0.32 | 13.29 ± 0.00 | 14.07 ± 0.24 |
| NUSWIDE | OM | OM | 8.13 ± 0.33 | 1.86 ± 0.60 | 6.39 ± 0.29 | OM | 9.27 ± 0.38 | 13.01 ± 0.00 | 15.10 ± 0.28 |
| YoutubeFace | OM | OM | 5.20 ± 0.37 | 3.24 ± 0.29 | OM | OM | 15.19 ± 0.41 | OM | 17.3 ± 0.43 |
| Dataset | MLRSSC | MSC_IAS | LMVSC | MSGL | SMVSC | SFMC | RAMCSF | OMVCDR | Ours |
|---|---|---|---|---|---|---|---|---|---|
| Year | 2018 | 2019 | 2020 | 2022 | 2022 | 2022 | 2023 | 2024 | 2024 |
| Notting-Hill | 81.86+3.84 | 83.30 ± 4.09 | 80.59 ± 6.29 | 79.82 ± 5.33 | 83.41 ± 5.61 | 85.45 ± 0.00 | 86.91 ± 6.63 | 89.64 ± 0.00 | 89.14 ± 4.17 |
| WebKB | 90.77 ± 0.22 | 78.12 ± 0.19 | 68.34 ± 0.13 | 93.03 ± 0.00 | 90.87 ± 0.00 | 94.77 ± 0.00 | 94.20 ± 0.00 | 95.37 ± 0.00 | 95.81 ± 0.00 |
| Wiki | 15.37 ± 0.83 | 23.68 ± 1.27 | 54.31 ± 2.40 | 58.33 ± 2.04 | 51.79 ± 4.59 | 34.18 ± 0.00 | 42.36 ± 2.58 | 47.34 ± 0.00 | 61.36 ± 2.55 |
| CCV | 20.10 ± 0.32 | 18.25 ± 0.26 | 23.74 ± 0.43 | 20.04 ± 0.37 | 22.25 ± 0.49 | 11.68 ± 0.00 | 16.31 ± 0.89 | 21.36 ± 0.00 | 24.87 ± 0.33 |
| ALOI | OM | OM | 42.32 ± 1.64 | 22.15 ± 0.77 | 49.45 ± 2.31 | OM | 59.86 ± 1.92 | 64.77 ± 0.00 | 68.63 ± 1.81 |
| Animal | OM | OM | 18.57 ± 0.21 | 8.84 ± 0.26 | 14.21 ± 0.21 | OM | 15.17 ± 0.44 | 22.06 ± 0.00 | 21.76 ± 0.30 |
| NUSWIDE | OM | OM | 24.32 ± 0.31 | 25.18 ± 0.51 | 23.31 ± 0.44 | OM | 12.53 ± 0.32 | 23.57 ± 0.00 | 27.75 ± 0.34 |
| YoutubeFace | OM | OM | 27.32 ± 0.25 | 32.79 ± 0.33 | OM | OM | 28.64 ± 0.55 | OM | 31.60 ± 0.38 |
| Dataset | MLRSSC | MSC_IAS | LMVSC | MSGL | SMVSC | SFMC | RAMCSF | OMVCDR | Ours |
|---|---|---|---|---|---|---|---|---|---|
| Year | 2018 | 2019 | 2020 | 2022 | 2022 | 2022 | 2023 | 2024 | 2024 |
| Notting-Hill | 71.89 ± 3.77 | 75.61 ± 4.34 | 73.24 ± 5.57 | 77.82 ± 4.14 | 65.41 ± 5.59 | 72.45 ± 0.00 | 78.91 ± 7.09 | 82.64 ± 0.00 | 83.74 ± 3.95 |
| WebKB | 86.94 ± 0.17 | 78.39 ± 0.15 | 62.80 ± 0.11 | 89.89 ± 0.00 | 86.58 ± 0.00 | 92.52 ± 0.00 | 91.95 ± 0.00 | 93.56 ± 0.00 | 93.70 ± 0.00 |
| Wiki | 17.46 ± 0.47 | 25.74 ± 0.74 | 43.75 ± 2.08 | 39.77 ± 1.91 | 37.64 ± 3.41 | 21.38 ± 0.00 | 29.72 ± 2.24 | 31.54 ± 0.00 | 49.87 ± 2.24 |
| CCV | 9.85 ± 0.42 | 9.22 ± 0.12 | 11.70 ± 0.31 | 9.76 ± 0.46 | 11.93 ± 0.53 | 10.85 ± 0.00 | 9.51 ± 0.93 | 11.78 ± 0.00 | 13.28 ± 0.27 |
| ALOI | OM | OM | 38.43 ± 1.52 | 10.95 ± 0.74 | 41.67 ± 1.73 | OM | 47.82 ± 1.97 | 53.76 ± 0.00 | 57.43 ± 1.58 |
| Animal | OM | OM | 9.54 ± 0.08 | 11.18 ± 0.07 | 10.37 ± 0.15 | OM | 10.97 ± 0.24 | 9.27 ± 0.00 | 10.83 ± 0.16 |
| NUSWIDE | OM | OM | 8.84 ± 0.07 | 10.47 ± 0.43 | 11.39 ± 0.39 | OM | 9.33 ± 0.46 | 10.52 ± 0.00 | 11.67 ± 0.22 |
| YoutubeFace | OM | OM | 10.72 ± 0.19 | 13.81 ± 0.21 | OM | OM | 11.31 ± 0.87 | OM | 14.13 ± 0.35 |
4.5. Runtime Comparison
- (a)
- FMVSC achieved optimal or near-optimal running efficiency on five datasets, indicating that FMVSC inherits the linear time complexity of the anchor point strategy, significantly reducing the runtime. This is because FMVSC effectively leverages the consensus and complementary information between views, allowing it to converge in fewer iterations. At the same time, our algorithm only requires constructing smaller-sized anchor graphs for each view to represent the current view space.
- (b)
- Unlike the anchor-based methods, MLRSSC, MSC_IAS, and SFMC construct a representation matrix of for each view during the clustering process and then combine them into a consistent matrix of . This leads to for the space complexity and at least for the time complexity. Therefore, MLRSSC, MSC_IAS, and SFMC may encounter out-of-memory (OM) issues when dealing with large datasets.
- (c)
- Although LMVSC runs faster on Animal, NUSWIDE, and YoutubeFace, its simple heuristic process does not take full advantage of the consensus and complementary information between views, resulting in poor clustering performance.
| Dataset | MLRSSC | MSC_IAS | LMVSC | MSGL | SMVSC | SFMC | RAMCSF | OMVCDR | Ours |
|---|---|---|---|---|---|---|---|---|---|
| Year | 2018 | 2019 | 2020 | 2022 | 2022 | 2022 | 2023 | 2024 | 2024 |
| Notting-Hill | 25.02 | 18.78 | 5.02 | 18.84 | 9.04 | 11.65 | 2.91 | 5.82 | 4.13 |
| WebKB | 367.35 | 120.55 | 8.62 | 5.43 | 7.86 | 12.58 | 2.17 | 4.67 | 5.36 |
| Wiki | 294.33 | 93.08 | 7.89 | 6.20 | 19.63 | 35.06 | 4.21 | 6.12 | 3.74 |
| CCV | 2352.44 | 839.46 | 62.77 | 35.76 | 65.86 | 595.69 | 23.53 | 46.87 | 16.84 |
| ALOI | OM | OM | 140.61 | 203.54 | 712.91 | OM | 120.06 | 241.15 | 92.42 |
| Animal | OM | OM | 230.11 | 1417.36 | 903.93 | OM | 311.27 | 814.73 | 444.65 |
| NUSWIDE | OM | OM | 367.74 | 687.33 | 1684.32 | OM | 538.09 | 1725.61 | 589.64 |
| YoutubeFace | OM | OM | 353.93 | 2134.65 | OM | OM | 612.30 | OM | 537.95 |
4.6. Ablation Experiments
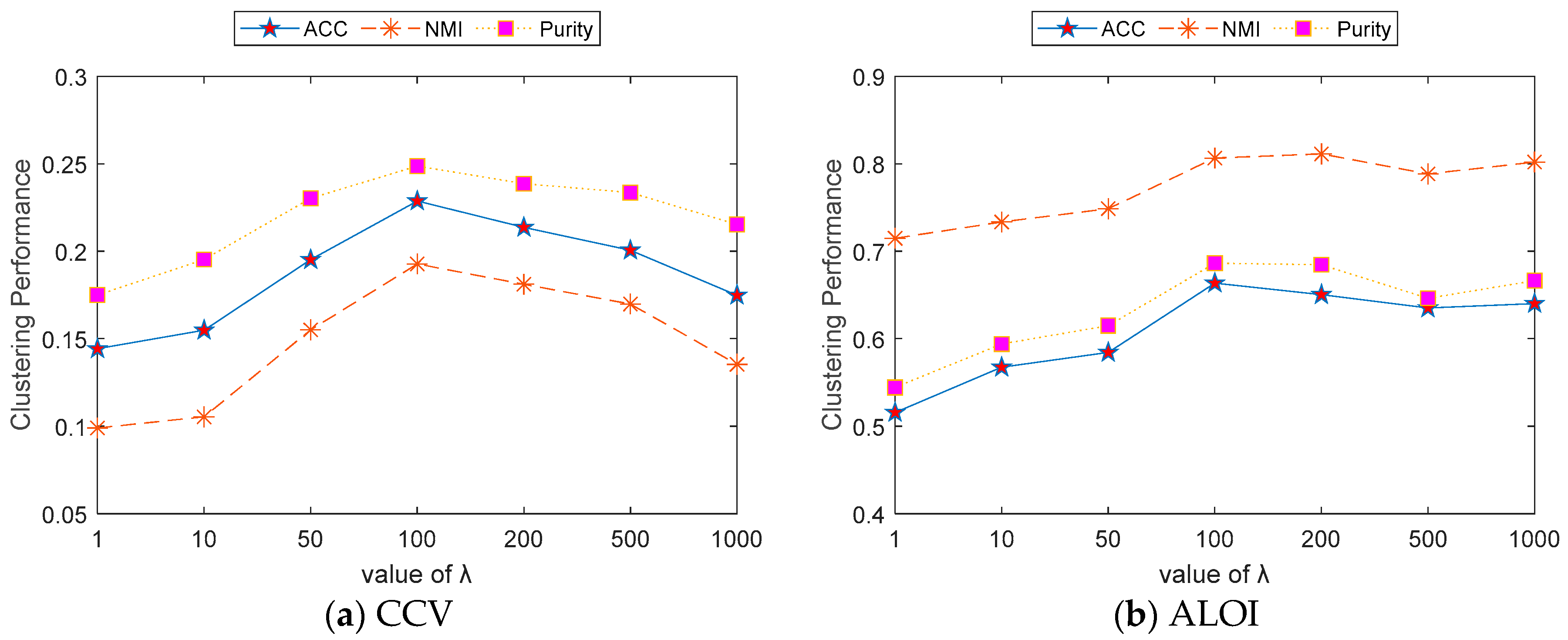
4.7. Parameter Analysis
- (a)
- When had a small value, such as 1–50, the performance of the ACC, NMI, and Purity datasets was relatively poor. This indicates that when is too low, the model may lack sufficient regularization or optimization strength, making it difficult to capture the underlying structure of the data.
- (b)
- As increased to a moderate range, such as 100–200, all indicators reached their peak performance. This suggests that at this range, the value of balanced the optimization of model performance with the prevention of overfitting.
- (c)
- When became excessively large, such as 500–1000, the performance of all indicators began to slightly decline. This indicates that excessive regularization may limit the model’s clustering capability, resulting in reduced performance.

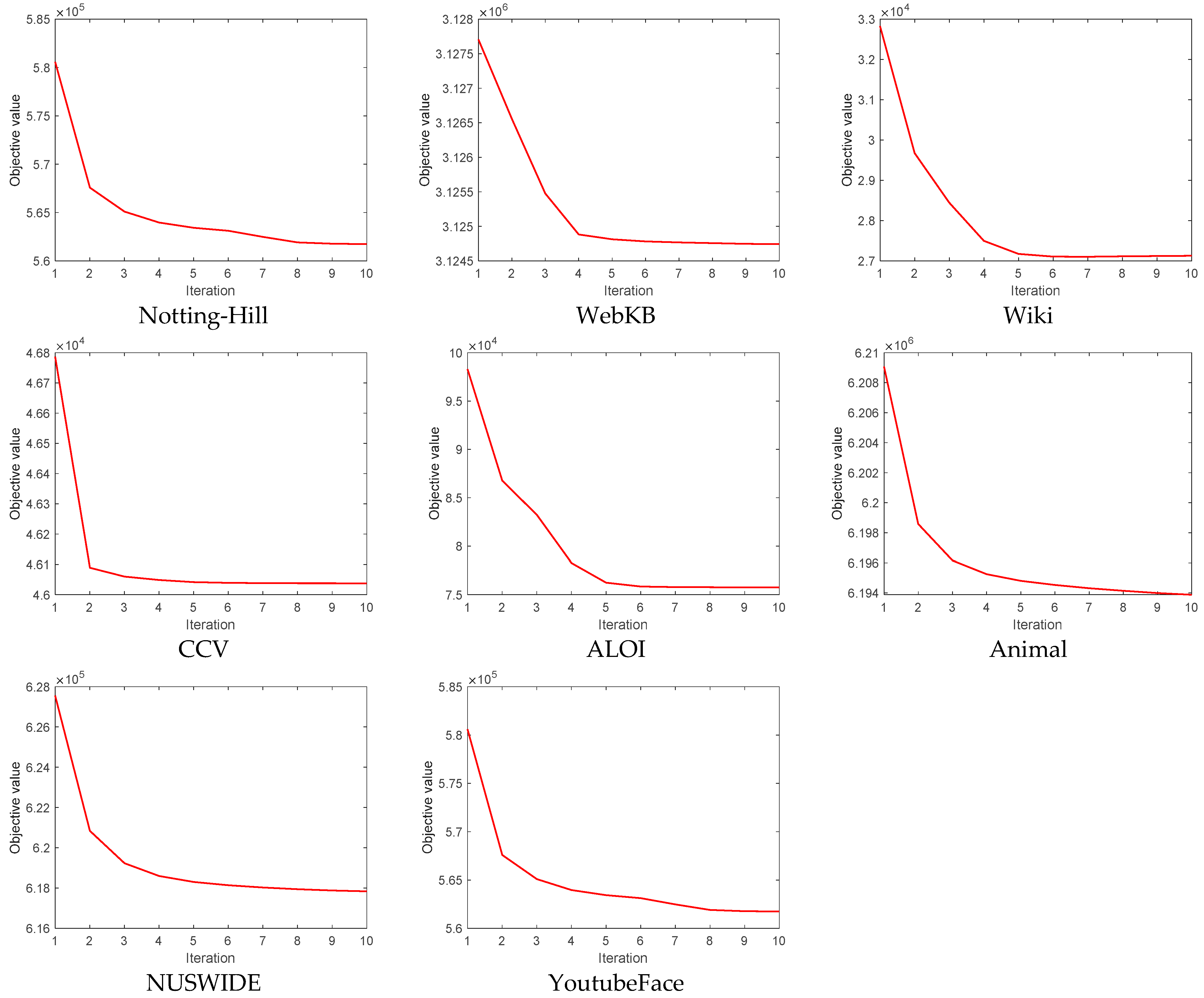
4.8. Convergence Analysis
- (a)
- There were differences in the convergence speeds across different datasets. The objective values of some datasets, like WebKB, Wiki, CCV, and ALOI, nearly reached convergence within the first five iterations, indicating that FMVSC can quickly find a good solution on these datasets. On the other hand, the objective values of datasets like Notting-Hill, Animal, NUSWIDE, and YoutubeFace required more iterations to reach convergence, indicating that the convergence process was more complex for these datasets, and more computation time was needed.
- (b)
- The objective values for all datasets stabilized after 10 iterations, indicating that our algorithm can reach a convergent state after only a few iterations across different datasets. This makes FMVSC particularly suitable for large-scale datasets.
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhou, S.; Yang, M.; Wang, X.; Song, W. Anchor-based scalable multi-view subspace clustering. Inf. Sci. 2024, 666, 120374. [Google Scholar] [CrossRef]
- Wang, S.; Liu, X.; Zhu, X.; Zhang, P.; Zhang, Y.; Gao, F.; Zhu, E. Fast parameter-free multi-view subspace clustering with consensus anchor guidance. IEEE Trans. Image Process. 2021, 31, 556–568. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, X.; Liu, S.; Tu, W.; Zhu, E. Scalable and structural multi-view graph clustering with adaptive anchor fusion. IEEE Trans. Image Process. 2024, 33, 4627–4639. [Google Scholar] [CrossRef]
- Peng, X.; Zhu, H.; Feng, J.; Shen, C.; Zhang, H.; Zhou, J.T. Deep clustering with sample-assignment invariance prior. IEEE Trans. Neural Netw. Learn. Syst. 2019, 31, 4857–4868. [Google Scholar] [CrossRef]
- Wang, Q.; Qin, Z.; Nie, F.; Li, X. Spectral embedded adaptive neighbors clustering. IEEE Trans. Neural Netw. Learn. Syst. 2018, 30, 1265–1271. [Google Scholar] [CrossRef]
- Yang, G.; Li, Q.; Yun, Y.; Lei, Y.; You, J. Hypergraph Learning-Based Semi-Supervised Multi-View Spectral Clustering. Electronics 2023, 12, 4083. [Google Scholar] [CrossRef]
- Xie, D.; Li, Z.; Sun, Y.; Song, W. Robust Tensor Learning for Multi-View Spectral Clustering. Electronics 2024, 13, 2181. [Google Scholar] [CrossRef]
- Chen, J.; Yi, Z. Sparse representation for face recognition by discriminative low-rank matrix recovery. J. Vis. Commun. Image Represent. 2014, 25, 763–773. [Google Scholar] [CrossRef]
- Liu, M.; Liang, K.; Zhao, Y.; Tu, W.; Zhou, S.; Gan, X.; Liu, X.; He, K. Self-supervised temporal graph learning with temporal and structural intensity alignment. IEEE Trans. Neural Netw. Learn. Syst. 2024, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Liu, J.; Liang, W.; Liu, X.; Wen, Y.; Zhu, E. Continual multi-view clustering. In Proceedings of the 30th ACM International Conference on Multimedia, Lisboa, Portugal, 10–14 October 2022; pp. 3676–3684. [Google Scholar]
- Gao, H.; Nie, F.; Li, X.; Huang, H. Multi-view subspace clustering. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 13–16 December 2015; pp. 4238–4246. [Google Scholar]
- Lan, M.; Meng, M.; Yu, J.; Wu, J. Generalized multi-view collaborative subspace clustering. IEEE Trans. Circuits Syst. Video Technol. 2021, 32, 3561–3574. [Google Scholar] [CrossRef]
- Chen, Y.; Xiao, X.; Peng, C.; Lu, G.; Zhou, Y. Low-rank tensor graph learning for multi-view subspace clustering. IEEE Trans. Circuits Syst. Video Technol. 2021, 32, 92–104. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Y.; Lu, G.; Le, W. Mixed structure low-rank representation for multi-view subspace clustering. Appl. Intell. 2023, 53, 18470–18487. [Google Scholar] [CrossRef]
- Lan, W.; Yang, T.; Chen, Q.; Zhang, S.; Dong, Y.; Zhou, H.; Pan, Y. Multiview subspace clustering via low-rank symmetric affinity graph. IEEE Trans. Neural Netw. Learn. Syst. 2023, 35, 11382–11395. [Google Scholar] [CrossRef]
- Ma, H.; Wang, S.; Zhang, J.; Yu, S.; Liu, S.; Liu, X.; He, K. Symmetric Multi-view Subspace Clustering with Automatic Neighbor Discovery. IEEE Trans. Circuits Syst. Video Technol. 2024, 34, 8766–8778. [Google Scholar] [CrossRef]
- Sun, M.; Zhang, P.; Wang, S.; Zhou, S.; Tu, W.; Liu, X.; Zhu, E.; Wang, C. Scalable multi-view subspace clustering with unified anchors. In Proceedings of the 29th ACM International Conference on Multimedia, Virtual Event, China, 20–24 October 2021; pp. 3528–3536. [Google Scholar]
- Brbić, M.; Kopriva, I. Multi-view low-rank sparse subspace clustering. Pattern Recognit. 2018, 73, 247–258. [Google Scholar] [CrossRef]
- Lu, C.; Feng, J.; Lin, Z.; Mei, T.; Yan, S. Subspace clustering by block diagonal representation. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 41, 487–501. [Google Scholar] [CrossRef]
- Chen, X.; Cai, D. Large scale spectral clustering with landmark-based representation. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 7–11 August 2011; pp. 313–318. [Google Scholar]
- Kang, Z.; Zhou, W.; Zhao, Z.; Shao, J.; Han, M.; Xu, Z. Large-scale multi-view subspace clustering in linear time. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; pp. 4412–4419. [Google Scholar]
- Li, Y.; Nie, F.; Huang, H.; Huang, J. Large-scale multi-view spectral clustering via bipartite graph. In Proceedings of the AAAI Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015. [Google Scholar]
- Liu, S.; Wang, S.; Zhang, P.; Xu, K.; Liu, X.; Zhang, C.; Gao, F. Efficient one-pass multi-view subspace clustering with consensus anchors. In Proceedings of the AAAI Conference on Artificial Intelligence, Vancouver, BC, Canada, 27 February–2 March 2022; pp. 7576–7584. [Google Scholar]
- Chen, J.; Yang, J. Robust subspace segmentation via low-rank representation. IEEE Trans. Cybern. 2013, 44, 1432–1445. [Google Scholar] [CrossRef]
- Elhamifar, E.; Vidal, R. Sparse subspace clustering: Algorithm, theory, and applications. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 2765–2781. [Google Scholar] [CrossRef]
- Nie, F.; Chang, W.; Wang, R.; Li, X. Learning an optimal bipartite graph for subspace clustering via constrained Laplacian rank. IEEE Trans. Cybern. 2021, 53, 1235–1247. [Google Scholar] [CrossRef]
- Parsons, L.; Haque, E.; Liu, H. Subspace clustering for high dimensional data: A review. ACM Sigkdd Explor. Newsl. 2004, 6, 90–105. [Google Scholar] [CrossRef]
- Zhou, S.; Wang, X.; Yang, M.; Song, W. Multi-view clustering with adaptive anchor and bipartite graph learning. Neurocomputing 2025, 611, 128627. [Google Scholar] [CrossRef]
- Shi, L.; Cao, L.; Wang, J.; Chen, B. Enhanced latent multi-view subspace clustering. IEEE Trans. Circuits Syst. Video Technol. 2024, 34, 12480–12495. [Google Scholar] [CrossRef]
- Kang, Z.; Lin, Z.; Zhu, X.; Xu, W. Structured graph learning for scalable subspace clustering: From single view to multiview. IEEE Trans. Cybern. 2021, 52, 8976–8986. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, H.; Wang, R.; Nie, F. Multiview clustering: A scalable and parameter-free bipartite graph fusion method. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 44, 330–344. [Google Scholar] [CrossRef]
- Yang, B.; Wu, J.; Zhang, X.; Lin, Z.; Nie, F.; Chen, B. Robust anchor-based multi-view clustering via spectral embedded concept factorization. Neurocomputing 2023, 528, 136–147. [Google Scholar] [CrossRef]
- Lao, J.; Huang, D.; Wang, C.-D.; Lai, J.-H. Towards scalable multi-view clustering via joint learning of many bipartite graphs. IEEE Trans. Big Data 2023, 10, 77–91. [Google Scholar] [CrossRef]
- Chan, P.K.; Schlag, M.D.; Zien, J.Y. Spectral k-way ratio-cut partitioning and clustering. In Proceedings of the 30th international Design Automation Conference, Dallas, TX, USA, 14–18 June 1993; pp. 749–754. [Google Scholar]
- Li, L.; He, H. Bipartite graph based multi-view clustering. IEEE Trans. Knowl. Data Eng. 2020, 34, 3111–3125. [Google Scholar] [CrossRef]
- Jiang, T.; Gao, Q.; Gao, X. Multiple graph learning for scalable multi-view clustering. arXiv 2021, arXiv:2106.15382. [Google Scholar]
- Wang, W.; Carreira-Perpinán, M.A. Projection onto the probability simplex: An efficient algorithm with a simple proof, and an application. arXiv 2013, arXiv:1309.1541. [Google Scholar]
- Wang, S.; Liu, X.; Zhu, E.; Tang, C.; Liu, J.; Hu, J.; Xia, J.; Yin, J. Multi-view clustering via late fusion alignment maximization. In Proceedings of the IJCAI, Macao, China, 10–16 August 2019; pp. 3778–3784. [Google Scholar]
- Bezdek, J.C.; Hathaway, R.J. Convergence of alternating optimization. Neural Parallel Sci. Comput. 2003, 11, 351–368. [Google Scholar]
- Zhang, Y.-F.; Xu, C.; Lu, H.; Huang, Y.-M. Character identification in feature-length films using global face-name matching. IEEE Trans. Multimed. 2009, 11, 1276–1288. [Google Scholar] [CrossRef]
- Cai, D.; He, X.; Wu, X.; Han, J. Non-negative matrix factorization on manifold. In Proceedings of the 2008 Eighth IEEE International Conference on Data Mining, Pisa, Italy, 15–19 December 2008; pp. 63–72. [Google Scholar]
- Wang, X.; Lei, Z.; Guo, X.; Zhang, C.; Shi, H.; Li, S.Z. Multi-view subspace clustering with intactness-aware similarity. Pattern Recognit. 2019, 88, 50–63. [Google Scholar] [CrossRef]
- Wan, X.; Liu, J.; Gan, X.; Liu, X.; Wang, S.; Wen, Y.; Wan, T.; Zhu, E. One-step multi-view clustering with diverse representation. IEEE Trans. Neural Netw. Learn. Syst. 2024, 1–13. [Google Scholar] [CrossRef] [PubMed]


| Symbol | Definition |
|---|---|
| Number of samples | |
| Number of classes | |
| Number of views | |
| Number of anchors in the pth view | |
| Dimensions of the pth view | |
| View coefficient vector | |
| Balance parameter | |
| The pth view data matrix | |
| The pth anchor points matrix | |
| The pth anchor graph | |
| The pth rotation matrix | |
| The fused spectral embedding matrix |
| Dataset | Samples | View | Class | Feature |
|---|---|---|---|---|
| Notting-Hill | 550 | 3 | 5 | 2000, 3304, 6750 |
| WebKB | 1051 | 2 | 2 | 1840, 3000 |
| Wiki | 2866 | 2 | 10 | 128, 10 |
| CCV | 6773 | 3 | 20 | 20, 20, 20 |
| ALOI | 10,800 | 4 | 100 | 77, 13, 64, 125 |
| Animal | 11,673 | 4 | 50 | 2689, 2000, 2001, 2000 |
| NUSWIDE | 30,000 | 5 | 31 | 65, 226, 145, 74, 129 |
| YoutubeFace | 101,499 | 5 | 31 | 64, 512, 64, 647, 838 |
| Metrics | Anchor Numbers | Datasets | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Notting-Hill | WebKB | Wiki | CCV | ALOI | Animal | NUSWIDE | YoutubeFace | ||
| ACC | equal | 82.31 ± 3.16 | 93.26 ± 0.00 | 51.99 ± 3.06 | 19.05 ± 0.44 | 61.09 ± 2.52 | 14.62 ± 0.36 | 11.96 ± 0.30 | 16.11 ± 0.65 |
| flexible | 88.38 ± 4.29 | 95.81 ± 0.00 | 56.04 ± 2.14 | 22.87 ± 0.34 | 66.36 ± 2.11 | 17.77 ± 0.33 | 16.02 ± 0.39 | 21.39 ± 0.61 | |
| NMI | equal | 71.64 ± 3.40 | 69.54 ± 0.00 | 50.10 ± 1.62 | 15.11 ± 0.38 | 76.99 ± 1.94 | 12.21 ± 0.55 | 11.83 ± 0.16 | 12.39 ± 0.29 |
| flexible | 79.86 ± 3.61 | 73.25 ± 0.00 | 52.40 ± 1.50 | 19.27 ± 0.41 | 80.65 ± 1.76 | 14.07 ± 0.24 | 15.10 ± 0.28 | 18.73 ± 0.43 | |
| Purity | equal | 83.02 ± 3.18 | 93.26 ± 0.00 | 56.74 ± 3.69 | 22.33 ± 0.39 | 63.44 ± 2.03 | 19.15 ± 0.37 | 23.22 ± 0.23 | 23.84 ± 0.52 |
| flexible | 89.14 ± 4.17 | 95.81 ± 0.00 | 61.36 ± 2.55 | 24.87 ± 0.33 | 68.63 ± 1.81 | 21.76 ± 0.30 | 27.75 ± 0.34 | 31.60 ± 0.38 | |
| F score | equal | 77.64 ± 2.23 | 90.87 ± 0.00 | 45.26 ± 3.43 | 11.13 ± 0.31 | 51.33 ± 1.47 | 09.53 ± 0.09 | 07.92 ± 0.17 | 10.37 ± 0.24 |
| flexible | 83.74 ± 3.95 | 93.70 ± 0.00 | 49.87 ± 2.24 | 13.28 ± 0.27 | 57.43 ± 1.58 | 10.83 ± 0.16 | 11.67 ± 0.22 | 14.13 ± 0.35 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Zhou, S.; Jin, G. Fast Multi-View Subspace Clustering Based on Flexible Anchor Fusion. Electronics 2025, 14, 737. https://doi.org/10.3390/electronics14040737
Zhu Y, Zhou S, Jin G. Fast Multi-View Subspace Clustering Based on Flexible Anchor Fusion. Electronics. 2025; 14(4):737. https://doi.org/10.3390/electronics14040737
Chicago/Turabian StyleZhu, Yihao, Shibing Zhou, and Guoqing Jin. 2025. "Fast Multi-View Subspace Clustering Based on Flexible Anchor Fusion" Electronics 14, no. 4: 737. https://doi.org/10.3390/electronics14040737
APA StyleZhu, Y., Zhou, S., & Jin, G. (2025). Fast Multi-View Subspace Clustering Based on Flexible Anchor Fusion. Electronics, 14(4), 737. https://doi.org/10.3390/electronics14040737





