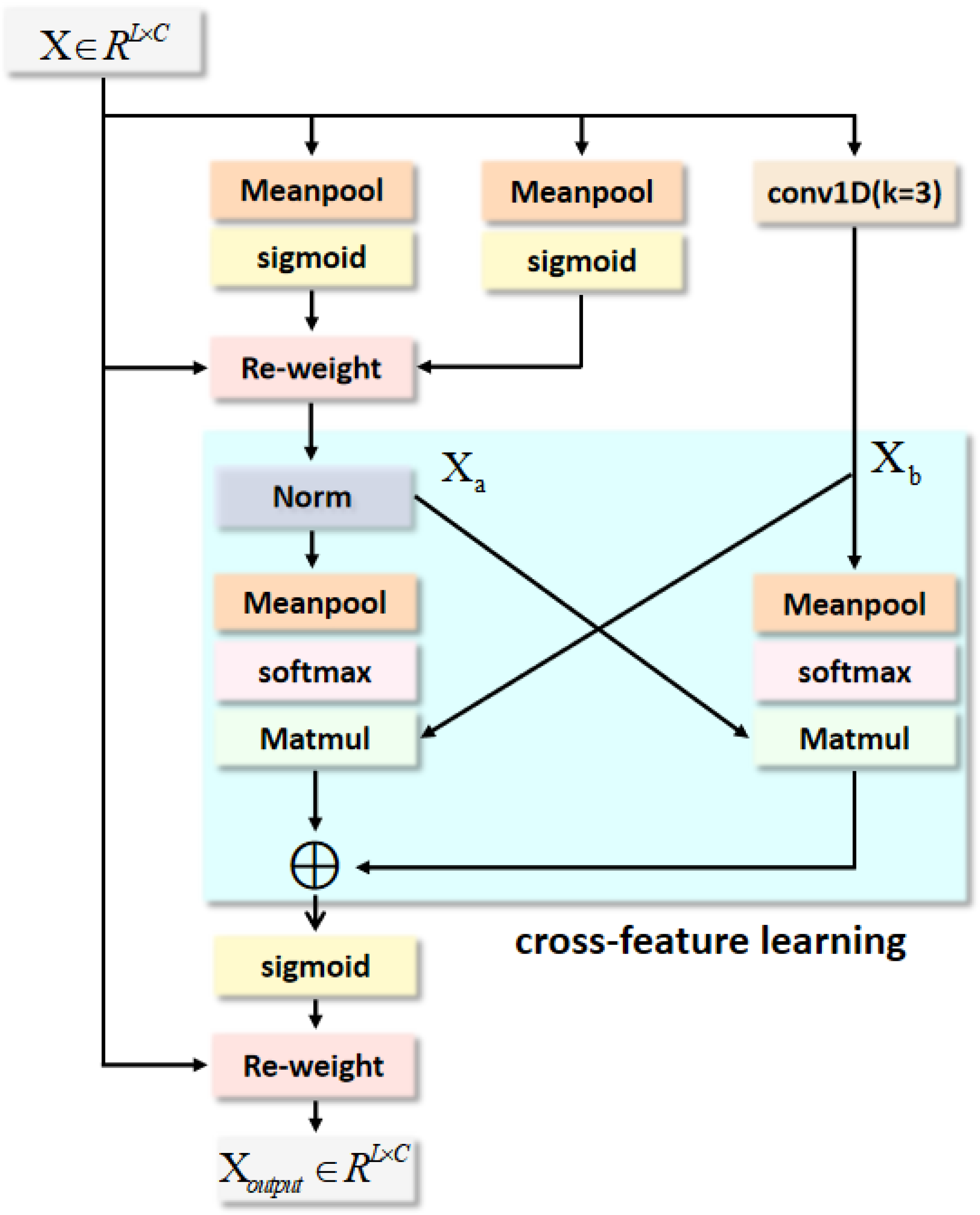
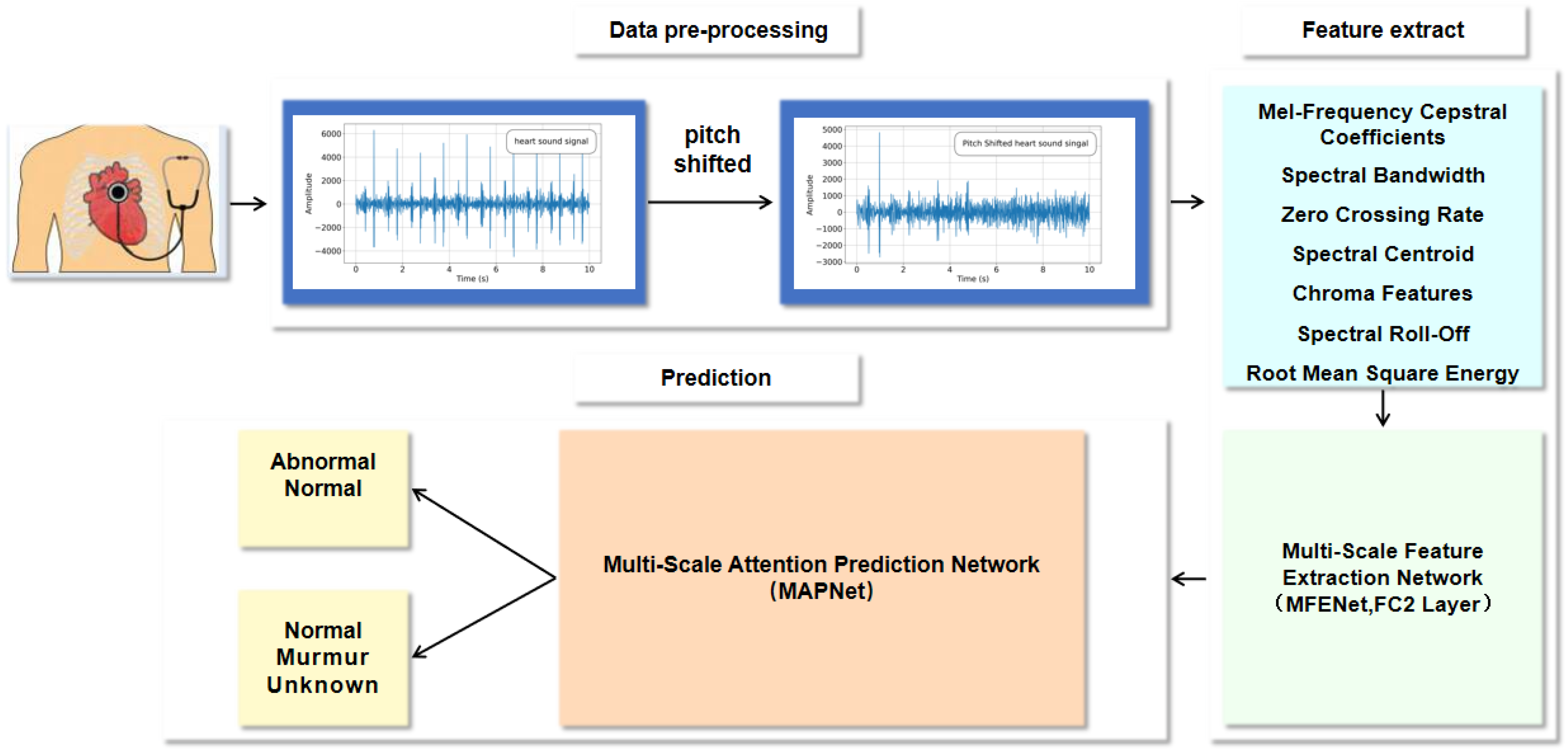
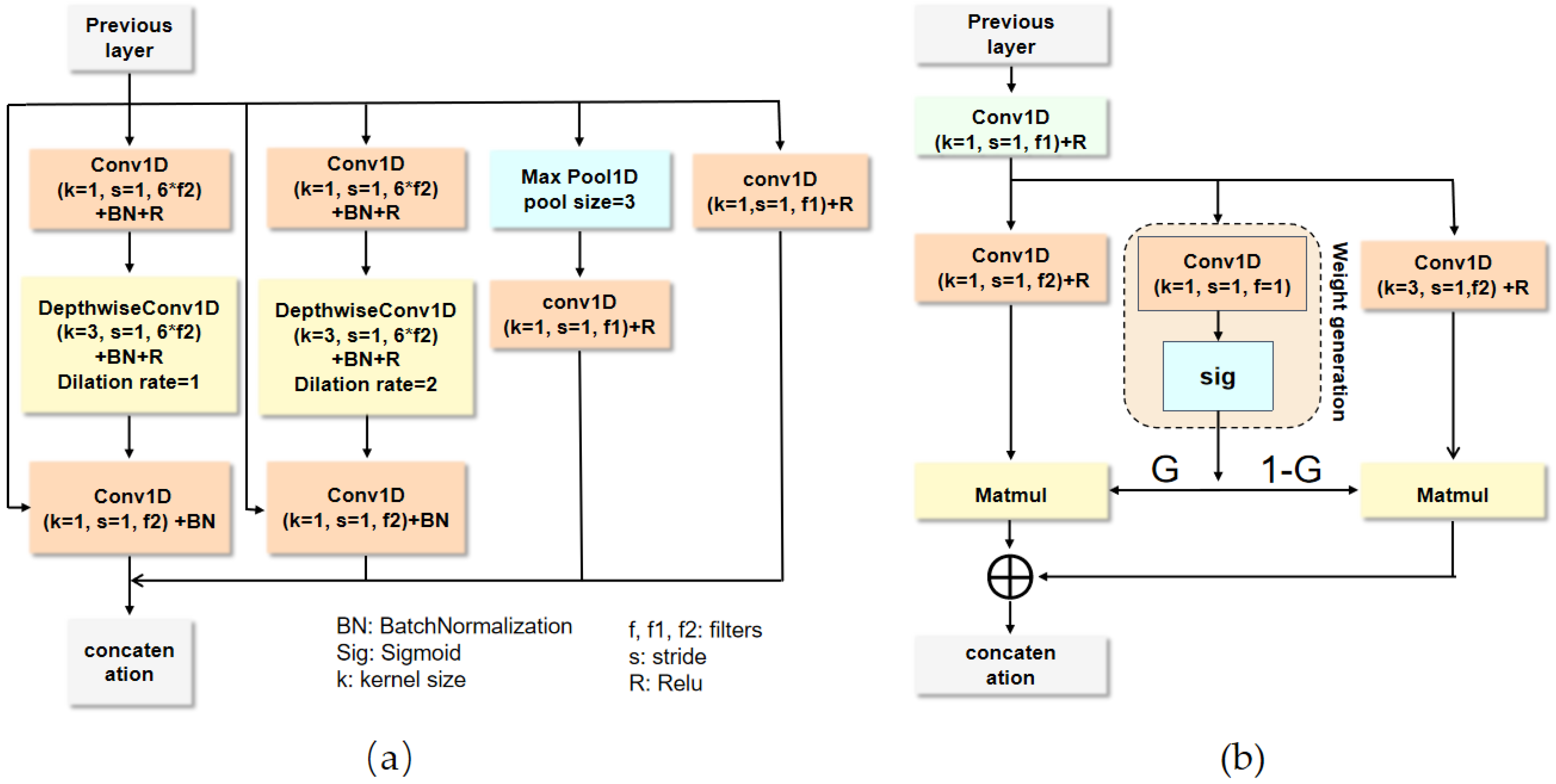
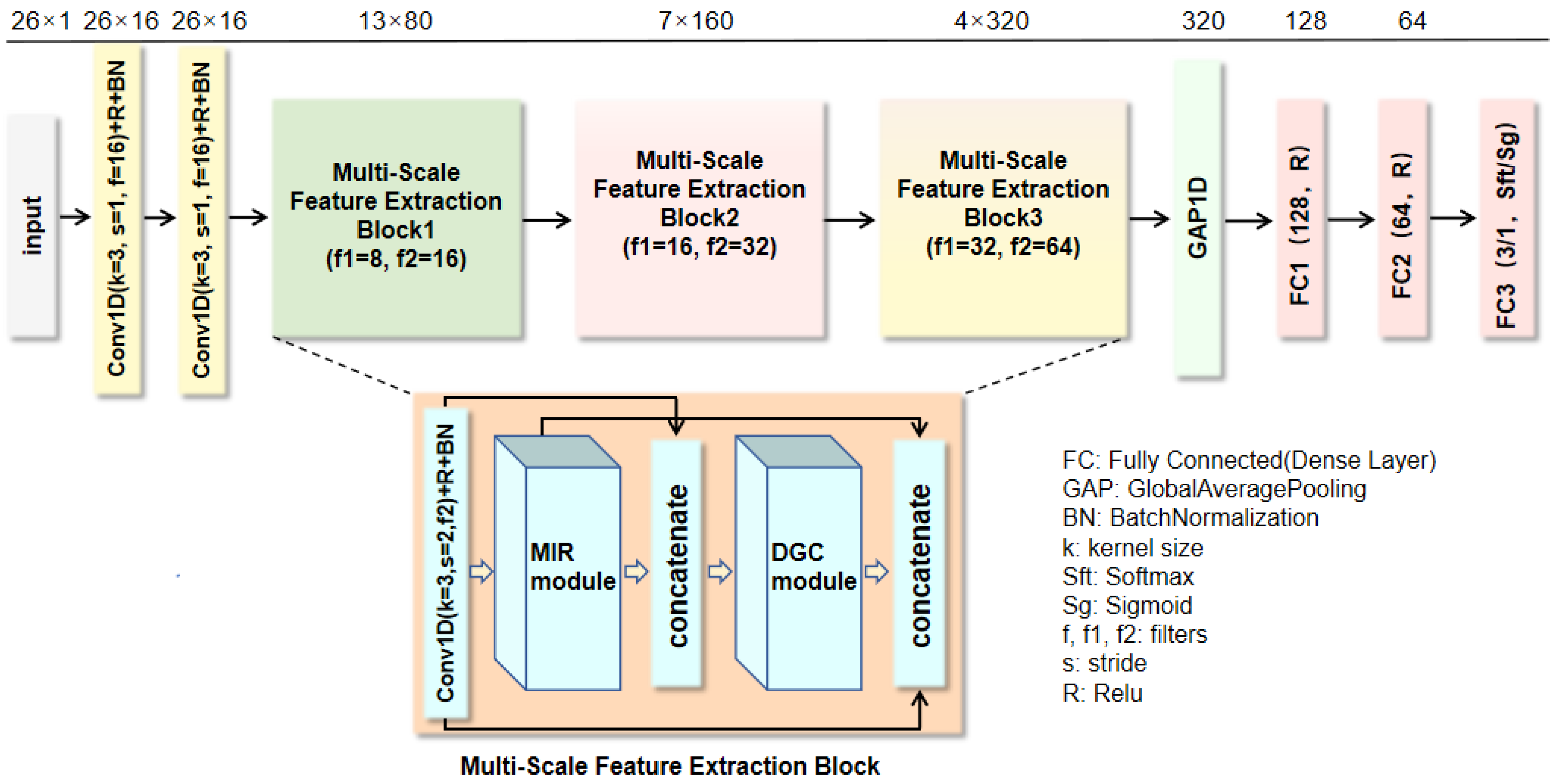
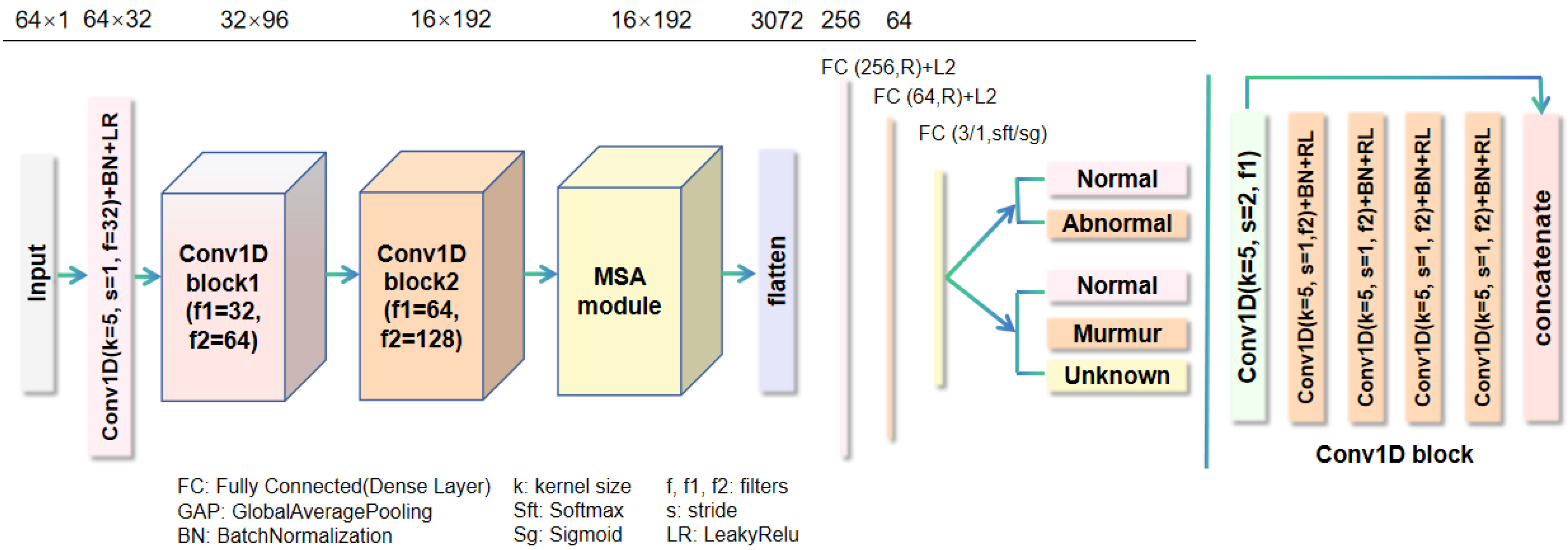
In this section, we conduct a comprehensive ablation experiment on two public heart sound datasets to analyze our proposed model. Subsequently, the proposed algorithm is evaluated by comparing it with the state-of-the-art methods.
4.2. Comparative Experiment
To comprehensively evaluate our algorithm’s performance, we conducted comparative experiments with other advanced methods using both the 2016 and 2022 PhysioNet/CinC Challenge datasets. For fair comparison, we directly adopted the experimental results reported in the original publications. When the F1-score or sensitivity was not provided in certain studies, we rigorously calculated these metrics based on the available data from the respective papers. However, due to some literature not providing complete information, especially AUC values, the AUC comparison for the 2016 PhysioNet/CinC Challenge dataset remains incomplete. To address this, we presents a more complete AUC comparison for the 2022 PhysioNet/CinC Challenge dataset, allowing our method to be directly compared with other methods on this metric, thereby ensuring that the overall evaluation is more comprehensive and fair.
Table 6 presents the comparison results of our model with other advanced methods on the 2016 PhysioNet/CinC Challenge dataset, in which our model outperforms other state-of-the-art methods. Specifically, the accuracy, F1-score, sensitivity, specificity, and AUC of our model after five-fold cross-validation were 98.89%, 97.20%, 97.34%, 99.27%, 97.11% and 98.30%, respectively. The accuracy, F1-score, specificity, and AUC achieved the highest rankings among all comparison algorithms. Among them, SAR-CardioNet [
46] achieved the third highest accuracy and F1-score by leveraging its split-self attention mechanism to enhance the capture of critical temporal features and its multi-path residual design to address gradient vanishing issues. MK-RCNN [
26] employed a multi-kernel residual convolutional structure to fuse phonocardiogram features at different scales, combined with the gradient stabilization properties of residual learning, achieving the second-highest accuracy on the PhysioNet Challenge 2016 dataset. In contrast, our MFENet leverages the MIR module to learn features from diverse regions and scales at a low computational cost, while the DGC module captures the interrelationships between features and dynamically adjusts information flow to deeply extract heart sound features, overcoming the insufficient feature extraction and representation capabilities of existing networks. Subsequently, the MAPNet is employed for heart sound classification, where the MSA module acquires multi-scale features and enables cross-feature learning, allowing it to focus on subtle variations between heart sound features while integrating both global and local information. This effectively addresses the insufficient integration of global and local feature information in existing heart sound attention mechanisms. Compared with SAR-CardioBNet, our model’s accuracy improved from 96.37% to 98.89% (a 2.52% improvement), and the F1-score enhanced from 96.33% to 97.20% (a 0.87% improvement). Through comparative evaluation with MK-RCNN [
26], our method achieves higher accuracy and specificity. The five-fold cross-validation results are detailed in
Table 7. Specifically, the proposed model reached the highest accuracy of 99.38%, F1-score of 98.63%, MCC of 98.23%, and G-Mean of 99.12% in the three-fold cross-validation. These results strongly demonstrate the effectiveness and robustness of our model across different evaluation metrics.
To further evaluate our method’s performance, we performed an external comparison using the Wilcoxon signed-rank test and Cohen’s d effect size on the PhysioNet/CinC Challenge 2016 dataset, as summarized in
Table 8. In this comparison, the proposed method achieved an AUC of 98.30%, outperforming existing methods such as MDFNet [
2] and SAR-CardioBNet [
46], which both achieved an AUC of 98.00%. The Wilcoxon signed-rank test revealed no significant difference between our method and these two models (
p = 0.2188), with Cohen’s d of 0.331 for both, indicating a small effect size. However, compared to KNN [
47], which achieved an AUC of 95.00%, our method showed a statistically significant improvement (
p = 0.0313) with a large effect size (Cohen’s
d = 3.637). This demonstrates that our method provides a significant performance advantage over KNN and is on par with other advanced methods, further reinforcing the robustness and reliability of our model in heart sound abnormality recognition.
Table 6.
Performance comparison with other state-of-the-art methods on the PhysioNet/CinC Challenge 2016 dataset.
Table 6.
Performance comparison with other state-of-the-art methods on the PhysioNet/CinC Challenge 2016 dataset.
| Methods | Accuracy (%) | F1-Score (%) | Sensitivity (%) | Specificity (%) | AUC (%) |
|---|
| LEPCNet [24] | | | | | - |
| MDFNet [2] | 94.44 | 86.90 | 89.77 | 95.65 | 98.00 |
| Le-LWTNet [4] | 94.50 | 86.40 | 86.40 | 96.60 | - |
| PU learning technique using CNN [48] | 94.83 | - | 94.90 | 94.42 | - |
| DDA [16] | 95.15 | 94.05 | 95.24 | 97.59 | - |
| Semi-supervised NMF + ABS-GP + SVM [49] | 95.26 | - | - | 95.21 | - |
| MACNN [30] | 95.49 | 97.17 | 97.32 | 88.42 | - |
| HS-Vectors [3] | 95.60 | 89.20 | 82.06 | 87.60 | - |
| KNN [47] | 95.78 | 96.00 | - | - | 95.00 |
| 1D + 2D-CNN [20] | 96.30 | 91.10 | 90.60 | 97.90 | - |
| SAR-CardioBNet [46] | 96.37 | 96.33 | 96.37 | 90.25 | 98.00 |
| MK-RCNN [26] | 98.53 | - | 99.08 | 98.04 | - |
| Proposed method | 98.89 | 97.20 | 97.34 | 99.27 | 98.30 |
Table 7.
Our method achieved results using 5-fold cross-validation on the PhysioNet/CinC Challenge 2016 dataset.
Table 7.
Our method achieved results using 5-fold cross-validation on the PhysioNet/CinC Challenge 2016 dataset.
| Fold | Accuracy | F1-Score | Sensitivity | Specificity | AUC (%) | MCC (%) | G-Mean (%) |
|---|
| | (%) | (%) | (%) | (%) | | | |
|---|
| 1 | 99.07 | 97.81 | 97.81 | 99.41 | 98.61 | 97.22 | 98.60 |
| 2 | 98.61 | 96.07 | 94.83 | 99.44 | 97.13 | 95.24 | 97.10 |
| 3 | 99.38 | 98.63 | 98.63 | 99.60 | 99.12 | 98.23 | 99.12 |
| 4 | 99.38 | 98.56 | 98.56 | 99.61 | 99.08 | 98.17 | 99.08 |
| 5 | 97.99 | 94.94 | 96.85 | 98.27 | 97.56 | 93.75 | 97.56 |
| Average | 98.89 | 97.20 | 97.34 | 99.27 | 98.30 | 96.52 | 98.29 |
Table 8.
External comparison with existing methods based on the AUC metric using the Wilcoxon signed-rank test and Cohen’s d effect size on the PhysioNet/CinC Challenge 2016 dataset.
Table 8.
External comparison with existing methods based on the AUC metric using the Wilcoxon signed-rank test and Cohen’s d effect size on the PhysioNet/CinC Challenge 2016 dataset.
| Method | AUC (%) | Wilcoxon p-Value | Cohen’s d | Statistical Significance |
|---|
| Proposed (Ours) | 98.30 | - | - | - |
| MDFNet [2] | 98.00 | 0.2188 | 0.331 | Not Significant |
| SAR-CardioBNet [46] | 98.00 | 0.2188 | 0.331 | Not Significant |
| KNN [47] | 95.00 | 0.0313 | 3.637 | Significant |
To further evaluate and compare our method and verify its generalization ability, we conducted experiments on the 2022 PhysioNet/CINC challenge dataset.
Table 9 shows the outcomes of our model compared with other advanced methods. Our model remains superior to other advanced methods in the field of cardiac murmur recognition. Specifically, the accuracy, F1-score, sensitivity, specificity, and AUC of our model after five-fold cross-validation were 98.86%, 98.13%, 97.92%, 98.82% and 99.83%, respectively. The accuracy, F1-score, sensitivity, specificity, and AUC all ranked first among the comparison algorithms. This comprehensive superiority in all key metrics underscores the robustness and reliability of our method in detecting cardiac murmurs, an essential task in clinical diagnostics. This comprehensive superiority in all key metrics underscores the robustness and reliability of our method in detecting cardiac murmurs, an essential task in clinical diagnostics. Among these methods, MK-RCNN achieved the second-highest overall accuracy and F1-score. Compared with MK-RCNN, the accuracy of our model improved from 98.33% to 98.86% (a 0.53% improvement), and the F1-score enhanced from 97.05% to 98.13%(a 1.08% improvement). Comparative evaluation with MK-RCNN [
26] demonstrates our method’s superior performance in handling class imbalance. Notably, our model achieves a more balanced clinical performance with 97.92% sensitivity and 98.82% specificity, effectively reducing both false negatives and false positives compared to MK-RCNN [
26]. The specifics of the five-fold cross-validation are detailed in
Table 10. Specifically, the proposed model reached the peak accuracy of 99.21%, sensitivity of 98.80%, and specificity of 99.21% during the three-fold cross-validation, where it also achieved the highest MCC of 97.91% and G-Mean of 98.79%. In addition, the model attained the highest F1-score of 98.79% and AUC of 99.92% in the four-fold cross-validation. These outcomes strongly demonstrate the effectiveness and generalization of our model, indicating that it can maintain high accuracy and reliability across different datasets.
To evaluate the performance of the proposed method in real-world clinical settings, we conducted experiments on the ZCHSound noisy dataset. As shown in
Table 11, the detailed performance metrics of the proposed method on the ZCHSound noisy dataset using five-fold cross-validation are presented. The results indicate that the model maintains strong performance on noisy data, with all core metrics averaging above 97% (average accuracy: 97.17%, F1-score: 97.14%, AUC: 97.21%, and G-Mean: 97.20%). This consistent high performance across multiple evaluation metrics highlights the robustness of the model, particularly in handling noisy inputs commonly encountered in real-world clinical environments. Importantly, the average MCC reaches 94.33%, providing a more informative measure of classification performance under class imbalance and noisy conditions. This demonstrates that the model not only achieves high predictive accuracy but also maintains strong consistency between predicted and true labels across different classes. The fluctuation range of the metrics across the five cross-validations is very small (for example, accuracy ranges from 96.83% to 98.44%). This high degree of consistency indicates that the model’s performance is not significantly affected by the randomness of data partitioning and exhibits significant stability when facing internal data diversity. Sensitivity, which is crucial for disease screening applications, averages 96.89% (with three out of five experiments exceeding 97% and one reaching 100%), indicating that the model can effectively identify positive cases in noisy environments and minimize false negatives. Meanwhile, the specificity averages as high as 97.52%, demonstrating a very low rate of false positives. This high performance on noisy data highlights the model’s capability to accurately detect positive cases and maintain robust predictive accuracy even under challenging real-world conditions, which is essential for clinical applications where noise is ubiquitous.
In order to better visualize the classification performance, we provide the confusion matrices for the three-fold test datasets in the five-fold cross-validation of our model on the 2016 and 2022 PhysioNet/CinC Challenge datasets, as shown in
Table 12. In
Table 12a, for the 2016 dataset, 500 out of 502 healthy subjects were correctly predicted (accuracy: 99.60%), and 144 out of 146 abnormal subjects were correctly identified (accuracy: 98.63%). The misclassified cases mainly involved two normal subjects incorrectly predicted as abnormal and two abnormal subjects misclassified as normal. These errors may be attributed to borderline pathological signals, where murmur intensity is weak or masked by noise, making them acoustically similar to normal patterns.
Table 12b presents the confusion matrix of the 2022 dataset. Among 484 normal samples, 483 were correctly predicted (accuracy: 99.79%). Of the 118 murmur samples, 114 were classified correctly (accuracy: 96.64%), while 3 were misclassified as normal and 1 as an unknown murmur. The misclassified murmur cases are likely due to the subtlety of pathological acoustic features, particularly when murmurs overlap with respiratory noise or occur intermittently, reducing their spectral distinctiveness. Importantly, all 31 samples of unknown murmur were correctly predicted (100%), suggesting that the model effectively distinguishes this class despite its limited sample size. Overall, the confusion matrices not only highlight the excellent diagnostic capability of our model but also reveal that most misclassifications occur between normal and murmur/abnormal categories. This suggests that enhancing sensitivity to subtle pathological cues, such as low-intensity or transient murmurs, could further reduce error rates and improve clinical reliability.
To further demonstrate the diagnostic ability of our network for heart abnormalities, we provide the receiver operating characteristic (ROC) curves for the 2016 and 2022 PhysioNet/CinC Challenge datasets under five-fold cross-validation, as shown in
Figure 6. For the 2016 dataset (
Figure 6a), the ROC curves of all five folds are presented with their respective AUC values. The AUC values range from 97.13% (Fold 2) to 99.12% (Fold 3), with a mean AUC of 98.30%. This small fluctuation range illustrates the stability and robustness of our model across different data partitions. Misclassifications in this dataset primarily occurred between normal and abnormal classes, likely due to weak murmurs that blur the acoustic distinction between these categories. This challenge emphasizes the need for fine-grained feature extraction and the importance of robust models that can capture subtle variations in heart sounds, which are often critical for accurate classification. The model’s high stability across folds indicates that it is not sensitive to particular data splits, further reinforcing its reliability. For the 2022 dataset (
Figure 6b–f), the average AUCs reached 99.78% for normal, 99.77% for murmur, and 99.95% for unknown, yielding an overall mean of 99.83%. The few misclassifications were primarily murmur samples mislabeled as normal or unknown, likely caused by subtle murmurs or overlapping respiratory noise. Overall, the ROC curves not only highlight the strong diagnostic capability of our model but also demonstrate its stability across folds and robustness in handling different categories. The in-depth analysis of misclassified cases further suggests that enhancing sensitivity to weak or transient murmurs, along with refining the detection of subtle heart sound variations, could reduce errors and improve clinical applicability, enabling more reliable and accurate diagnosis in real-world settings.
To provide a more comprehensive evaluation of computational efficiency and practical applicability in clinical scenarios, we compared the model parameters, FLOPs, and average inference time of our proposed method with baseline approaches, as presented in
Table 13. The proposed method achieves the highest classification accuracy (98.89%), surpassing LEPCNet (93.1%) and MK-RCNN (98.53%). In terms of computational complexity, our model has 1.57M parameters and 22.53M FLOPs, which are higher than those of LEPCNet (52.7K parameters, 1.6M FLOPs) but comparable to MK-RCNN (217K parameters). Importantly, the proposed method requires an average inference time of only 98.1 ms per recording, which is sufficiently low for real-time or near real-time deployment in clinical and wearable auscultation devices. These results demonstrate that, despite a moderate increase in model size, the MIR-enhanced architecture achieves a favorable trade-off between accuracy and efficiency, ensuring both high diagnostic reliability and practical feasibility in resource-constrained environments.