Abstract
Ovarian cancer remains a significant global health concern, and its diagnosis heavily relies on whole-slide images (WSIs). Due to their gigapixel spatial resolution, WSIs must be split into patches and are usually modeled via multi-instance learning (MIL). Although previous studies have achieved remarkable performance comparable to that of humans, in clinical practice WSIs are distributed across multiple hospitals with strict privacy restrictions, necessitating secure, efficient, and effective federated MIL. Moreover, heterogeneous data distributions across hospitals lead to model heterogeneity, requiring a framework flexible to both data and model variations. This paper introduces HFed-MIL, a heterogeneous federated MIL framework that leverages gradient-based attention distillation to tackle these challenges. Specifically, we extend the intuition of Grad-CAM to the patch level and propose Patch-CAM, which computes gradient-based attention scores for each patch embedding, enabling structural knowledge distillation without explicit attention modules while minimizing privacy leakage. Beyond conventional logit distillation, we designed a dual-level objective that enforces both class-level and structural-level consistency, preventing the vanishing effect of naive averaging and enhancing the discriminative power and interpretability of the global model. Importantly, Patch-CAM scores provide a balanced solution between privacy, efficiency, and heterogeneity: they contain sufficient information for effective distillation (with minimal membership inference risk, MIA AUC ≈ 0.6) while significantly reducing communication cost (0.32 MB per round), making HFed-MIL practical for real-world federated pathology. Extensive experiments on multiple cancer subtypes and cross-domain datasets (Camelyon16, BreakHis) demonstrate that HFed-MIL achieves state-of-the-art performance with enhanced robustness under heterogeneity conditions. Moreover, the global attention visualizations yield sharper and clinically meaningful heatmaps, offering pathologists transparent insights into model decisions. By jointly balancing privacy, efficiency, and interpretability, HFed-MIL improves the practicality and trustworthiness of deep learning for ovarian cancer WSI analysis, thereby increasing its clinical significance.
1. Introduction
Ovarian cancer remains a significant global health concern affecting millions of women, and the timely and accurate diagnosis of ovarian cancer plays a crucial role in improving patient outcomes and reducing mortality rates. Ovarian cancer diagnosis heavily relies on whole-slide images (WSIs) [1,2,3,4]. Pathologists scan microscope pathological slides to obtain gigapixel WSIs, which require several gigabytes or more to maintain high-resolution features for accurate diagnosis. Even 500 gigapixel WSIs can be as large as the whole ImageNet. Pathologists meticulously analyze slides in clinical practice, which significantly burdens healthcare resources. However, with the surge in big data and artificial intelligence, especially in vast medical image datasets, deep learning algorithms have emerged as pivotal tools to automate pathological image analysis.
Nevertheless, the massive size and spatial consumption of WSIs pose challenges for all deep networks. Typically, existing approaches divide WSIs into patches and model them as multiple instance learning, where each patch is treated as an instance, and the entire WSI is considered a bag because patches within the same WSI are correlated. Common implementations encode the features of each patch using a pre-trained backbone network, then aggregate these patch features to obtain the WSI’s features, followed by classification using a classifier [5,6,7,8,9,10,11,12].
Although existing studies have achieved remarkable performance comparable to humans, they often focus on centralized learning scenarios and overlook that gigapixel WSIs are distributed across multiple hospitals with privacy restrictions in clinical practice. Federated learning [13,14,15,16] was specifically developed to address this issue. This method learns from decentralized data distributed across various sites while ensuring the protection of privacy. Model training and validation are conducted locally at each participating institution in this paradigm. Only essential information is transmitted, thus eliminating the demand for data owners to communicate their private datasets [17,18,19]. Moreover, data distribution variation in multiple hospitals, namely, data heterogeneity, poses additional challenges to federated learning, prompting the development of federated learning methods that transmit different types of information. The types of information transmitted in federated learning can be categorized into parameter transmission, gradient transmission, and proxy dataset prediction knowledge transmission.
Parameter transmission is the most prevalent category. FedAvg [13] employs an approach that averages the model parameters for global model aggregation. However, numerous studies have demonstrated that FedAvg’s performance deteriorates in non-IID settings [20,21,22]. Subsequent works, such as FedProx [23] and Xie et al. [24], have been introduced to address these challenges, incorporating global model parameters into the local optimization process as a regularization term to align local models with the global model.
Gradient transmission can also optimize performance declines under non-IID conditions. Initially, FedSGD was proposed as a baseline for FedAvg, referring to the implementation of previous methods [25]. This approach closely resembles centralized training, eliminating the heterogeneity caused by distributed training, but it leads to high communication costs. Therefore, subsequent methods have adopted strategies that approximate the global gradient, such as Scaffold [20], Fed-grab [26], and AOCC [27].
Proxy dataset prediction knowledge transmission is also named distillation-based federated learning [28]. Unlike the above methods, it does not solely focus on the heterogeneity of data distributions. Because data heterogeneity can ultimately lead to heterogeneity in the preferred models at different sites, methods of transmitting proxy dataset prediction knowledge have been developed. Proxy datasets can be either public or synthetic [29]. The transmission of public dataset prediction results is commonly modeled in the paradigm of distillation learning. Typically, local models are regarded as teacher models. In contrast, the global model is viewed as the student model, which completes knowledge aggregation by distilling knowledge from predictions on public data [30,31]. Since only information independent of the model structure is transmitted, past research has shown that this can adapt to model and data heterogeneity [9,32,33].
Parameter and gradient transmissions could indirectly expose sensitive personal information if leaked or attacked. Merely anonymizing data does not provide sufficient protection [34] as parts of the training data can be reconstructed through gradients [35,36] or model parameters [37,38]. Pathology images are often considered capable of indirect identity recognition, posing a threat to patient privacy and usually subject to legal restrictions. Generally, communicating less data is more conducive to privacy protection, and transmitting knowledge from public datasets is more secure. Moreover, federated learning must also consider the cost of communication, necessitating that the fewer data shared during communication, the better. Combined with the demand for relative flexibility to model heterogeneity, transmitting only the knowledge of predictions on public datasets is the most appropriate choice compared with model parameters or gradient transmission. On the other hand, for the effectiveness of the training model, we need to share as much information as possible to convey knowledge that is as rich as possible to mitigate the performance decline brought by heterogeneity, especially when models process vasts numbers of slices in WSIs. Merely using non-resultant prediction knowledge, namely, pseudo-labels, is insufficient, and we also demand secure, lightweight, structured prediction knowledge as distillation knowledge.
For additional distillation information selection, directly aggregating intermediate parameters or their gradients for distillation is not possible due to model heterogeneity, and sharing raw input data is also prohibited due to privacy concerns. Therefore, we selected patch feature gradient-based attention in the proxy dataset during training, which commonly exists in any MIL framework. We drew inspiration from Grad-CAM [39], measuring the importance of each patch based on the average gradient and then distilling its structural knowledge. This approach was chosen partly because fully exposing gradient information could increase the risk of model leakage and partly because lesion tissue patterns generally differ with healthy tissues, leading to variations in gradient-based patch attention scores. This represents a form of knowledge that generalizes well, even across heterogeneous ovarian cancer WSI data distributions. We name the proposed mechanism Patch-CAM.
To address these issues, this study presents HFed-MIL, which uses federated learning based on gradient attention measured distillation. To be more precise, we start by inducing and formulating a widely used variant of MIL. Next, we distill the structural knowledge of each patch based on its average gradient and use it to measure its importance. This allows us to be relatively flexible when modeling heterogeneity and achieve a balance between security and effectiveness in HFed-MIL. This method enables sufficient information distillation without compromising privacy issues since patch gradient significance scores contain more information than labels but less than features, and they are also more dimensionally adjustable than features. Our results show that the suggested model outperforms the state-of-the-art. HFed-MIL raises the clinical relevance of deep learning by making it more applicable to ovarian cancer and more trustworthy in the medical field.
The key contributions of this work are summarized as follows:
- Patch-CAM-based structural distillation. We extend the intuition of Grad-CAM to the patch level in WSIs and propose Patch-CAM, which computes gradient-based attention scores for each patch embedding. This design enables knowledge distillation in heterogeneous MIL frameworks without requiring explicit attention modules, while avoiding the privacy risks associated with directly sharing gradients or features.
- Dual-level distillation objective (class-level and structural-level). Beyond conventional logit distillation, we introduce structural distillation that enforces consistency of patch-level attention distributions across clients. This prevents the vanishing effect of naive averaging and strengthens the discriminative power and interpretability of the global model.
- Balanced trade-off between privacy, efficiency, and heterogeneity. Patch-CAM scores lie between logits and raw features: they provide sufficient information for effective distillation while leaking minimal private data (MIA AUC ≈ 0.6, close to random guessing). In addition, HFed-MIL reduces communication cost to 0.32 MB per round, which is orders of magnitude smaller than parameter or gradient sharing, making it practical for real-world federated pathology.
- Enhanced interpretability and generalization in heterogeneous federated WSI analysis. Extensive experiments on multiple cancer subtypes and cross-domain datasets (Camelyon16, BreakHis) demonstrate that HFed-MIL yields more robust performance across heterogeneous models. Moreover, the global attention visualizations show sharper and clinically meaningful heatmaps, offering pathologists transparent insights into model decisions.
The rest of the study is outlined as follows: Section 2 discusses literature work followed by Section 3, which discusses the dataset and the proposed work’s methodology together. In Section 4 experiments are presented. Lastly, Section 5 concludes the current study and discusses limitations with possible future directions.
2. Related Work
In this section, we briefly discuss current advancements in ovarian cancer, followed by a discussion on attention mechanisms, and finally, we present challenges in multi-site data distribution.
2.1. Advancements in Ovarian Cancer
Ovarian cancer remains a significant health concern, with its late-stage diagnosis contributing to high mortality due to complex subtype classification [1,40,41]. Traditional diagnostic methods, including histopathological examination of whole-slide images (WSIs), have been enhanced by deep learning techniques, allowing for improved accuracy and efficiency in cancer detection. Earlier works demonstrate the potential of CNNs in histopathological image classification—subsequent studies, such as those by [2,3,42,43,44,45], have focused on the application of deep learning frameworks to the analysis of ovarian cancer, achieving remarkable predictive performances.
However, despite these advancements, several critical gaps remain unaddressed. Notably, the heterogeneity of the data across different clinical sites poses challenges for the generalizability of machine learning models [46,47]. A few studies have highlighted that variability in image quality and annotation standards across institutions can lead to biased model performance [48]. Furthermore, privacy concerns limit the sharing of sensitive patient data, necessitating innovative approaches that can leverage decentralized data for training while adhering to ethical guidelines [49]. HFed-MIL aims to bridge these gaps by utilizing federal learning to facilitate collaborative model training without compromising patient privacy.
2.2. Attention Mechanisms in Deep Learning
Attention mechanisms have emerged as a powerful tool in deep learning, particularly for enhancing model interpretability and performance in complex tasks such as image segmentation [50], breast cancer diagnosis [51], image captioning [52], generative modeling [52], remote sensing [53], foreign object detection [54], and facial micro-expression recognition [55]. These methods showcase their versatility and effectiveness in handling diverse challenges in computer vision. Further advancements in attention in WSI-based works have yielded significant results, demonstrating applicability across multiple domains, including ovarian cancer. Vaswani et al. [56] initially proposed the Transformer architecture, fundamentally altering how attention is integrated into neural networks. The dynamics of attention allow the model to focus on relevant parts of an image, enhancing its ability to capture intricate patterns.
Notably, Vision Transformer (ViT) [57] and its hierarchical variant, Swin Transformer [58], have demonstrated strong capability in modeling global and multi-scale context. Transformer-based attention techniques have been adapted for WSI analysis, offering superior modeling of long-range dependencies [7,59]. Furthermore, cross-attention networks have been developed to leverage inter-slide relationships, particularly beneficial for rare ovarian cancer subtypes [60,61]. Gupta et al. [62] integrated gradient-based visualization, enabling efficient training of very large images and potentially advancing pathologists’ trust in model predictions. Beyond canonical Transformer designs, hybrid architectures combining CNN and Transformer modules have also shown strong potential. For instance, Huo et al. [53] proposed FCIHMRT, which employs a ViT branch to capture global dependencies and a Res2Net branch for multi-scale local feature representation, merging their outputs to enhance remote sensing scene classification.
Multiple instance learning (MIL) is central to WSI analysis, allowing models to learn from bags of image patches with only slide-level labels. Incorporating attention within MIL enhances focus on diagnostically relevant regions, improving interpretability and performance [5,6]. Recent frameworks integrate probabilistic attention [63], multi-head attention [8], and cascaded attention [64] to refine feature extraction and localization. However, most of these approaches neglect the federated setting, particularly the heterogeneous nature of multi-institutional WSI data, where variations in staining protocols, scanner types, and population distributions pose significant challenges. In such scenarios, striking a balance between effective knowledge sharing and strict privacy preservation remains a significant challenge. Moreover, gradients derived from attention weights can inadvertently expose sensitive patient data [35,36]. To address these issues, HFed-MIL introduces a privacy-preserving patch gradient attention distillation framework tailored for heterogeneous federated MIL, ensuring that the benefits of attention mechanisms are retained without compromising robustness across sites or patient confidentiality.
2.3. Federated Learning with WSIs
Whole-slide images (WSIs) have revolutionized digital pathology by enabling high-resolution visualization of tissue samples. However, due to their inherent complexity, direct centralized learning on WSIs is computationally expensive and raises substantial data privacy concerns, particularly in medical applications, where patient confidentiality is paramount. In response, federated learning (FL) has gained attention in computational pathology as a privacy-preserving approach to train models on distributed data points without data sharing [16,65].
While existing works have shown significant results within the intersection of federated learning with WSIs, this intersection introduces several unique challenges, which are summarized as follows:
- Meaningful Gigapixel Analysis: WSIs require patch-wise modeling and MIL-based aggregation, which complicates communication and synchronization in federated settings.
- Data and Model Heterogeneity: Differences in data distributions, annotation quality, and model architecture across hospitals cause a decrease in performance in conventional FL pipelines, further discussed in the subsequent Section 2.4.
- Privacy vs. Performance trade-off: Existing methods often compromise diagnostic performance for stronger privacy or vice versa.
To address these issues, we propose HFed-MIL, a novel gradient attention-based knowledge distillation framework for federated MIL in ovarian cancer WSI analysis. Instead of relying solely on logits or weight updates, HFed-MIL extracts and transfers gradient-derived patch attention scores, which provide richer semantic context than predictions but are less privacy-sensitive than raw features. This design balances model interpretability, data privacy, and robustness to non-IID distributions—making HFed-MIL particularly suited for real-world multi-institutional pathology workflows.
2.4. Challenges in Multi-Site Data Distribution
The deployment of machine learning models in multi-site healthcare settings presents unique challenges, particularly concerning data distribution. As discussed above, federated learning has emerged as a promising solution, allowing models to be trained across decentralized data sources while maintaining privacy. However, the heterogeneous nature of medical data—characterized by variations in imaging protocols, demographic distributions, and annotation practices—further complicates the training process. Several studies have outlined these difficulties, emphasizing the challenges of achieving significant results and model convergence under such conditions.
Moreover, existing federated learning frameworks often struggle with knowledge distribution, where the goal is to effectively convey insights from diverse data sources. Techniques such as federated averaging have been widely adopted; however, they may not adequately address the complexities of multi-instance learning scenarios inherent in whole-slide image (WSI) analysis. Our method seeks to overcome these limitations by introducing a standardized approach that not only accommodates the variability of multi-site data but also enhances the efficiency of knowledge transfer across federated networks, facilitating broader insights.
In conclusion, while significant advancements have been made in ovarian cancer diagnosis, the integration of patch gradient-based attention mechanisms with federated learning remains an underexplored area. This innovative approach holistically addresses the challenges of privacy, data heterogeneity, and efficient learning. Our method represents a substantial enhancement in this domain, promising to improve the accuracy and applicability of cancer diagnostics across diverse clinical environments.
3. Dataset and Methodology
3.1. Dataset
The study compiled a dataset from the UBC-Ovarian Cancer Challenge, (https://www.kaggle.com/competitions/UBC-OCEAN/overview, (accessed on 14 May 2025)), which included histopathological images categorized into five ovarian cancer subtypes: Clear Cell Carcinoma (CC), Mucinous Carcinoma (MC), Low-Grade Serous Carcinoma (LGSC), High-Grade Serous Carcinoma (HGSC), and Endometrioid Carcinoma (EC). To handle the large WSIs, we performed downscaled patch sampling to reduce the computational load. We chose 50 representative tiles per WSI, each sized pixels. The selection focused on diagnostically significant regions, eliminating extraneous background. The preprocessing stage involved reducing noise and normalizing the data to improve the model’s accuracy without sacrificing efficiency. Figure 1 shows sample WSIs from UBC-Ovarian Cancer, while Figure 2 depicts the distribution of tissue types, and Figure 3 emphasizes the need to effectively preprocess WSI into smaller patches as it shows WSI gigapixel information.
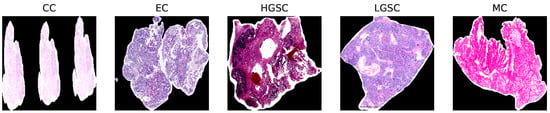
Figure 1.
Representative samples from each class of the UBC Ovarian Cancer Dataset.
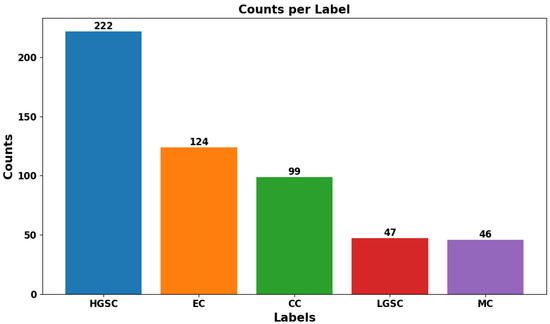
Figure 2.
The ovarian sub-type distribution at actual from source data.
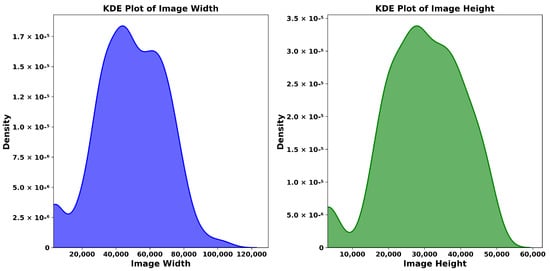
Figure 3.
The size distribution of ovarian WSIs. Most of them are several gigapixels.
Furthermore, this dataset poses unique challenges in the context of federated learning compared to other WSI cancer datasets. The limited availability of ovarian cancer cases results in a smaller dataset, complicating model training. Additionally, class imbalance among different subtypes of ovarian cancer further hinders the performance of FL models. Addressing these specific challenges is crucial for developing practical machine learning approaches in ovarian cancer diagnosis and treatment. The variability in imaging techniques across institutions can lead to inconsistencies in slide quality, while the complexity of histopathological features necessitates the use of advanced feature extraction methods.
3.2. Local WSI MIL Modeling
Let W represent a whole-slide image (WSI), which is decomposed into a collection of patches . Each patch is processed through a backbone neural network f (e.g., ResNet [66], ViT [67]) that serves as an encoder. This network maps each patch to a feature vector in a high-dimensional space:
where denotes the parameters of the backbone network, is the feature vector with any shape or dimension with the total size of d for the i-th patch.
The feature vectors in the W are then aggregated using an aggregation operator g, which could be self-attention, max pooling, or so on, to form a single feature vector that represents the entire WSI [68]:
Here, represents the parameters of the aggregation mechanism, and is the aggregated feature vector for the WSI. And the g can be anonymous to the server.
Finally, a classifier h, parameterized by , takes the aggregated feature vector and produces a prediction y for the WSI:
where z is the predicted logits for the WSI, which have not been normalized with the softmax function.
where is the c-th dimension of z, and predicted probability y is the logits z normalized by the softmax function.
Given the prediction y for the WSI, we define the target label as t, which is a categorical label representing the true class of the WSI. The cross-entropy loss function L is used to quantify the error between the predicted probabilities from the classifier and the true distribution t. The cross-entropy loss for a single instance is given by
where C is the number of classes, is an indicator function that equals 1 if the predicted class y is equal to the true class and 0 otherwise, and is the predicted probability of the class c as output by the softmax layer of the classifier.
3.3. Label Distillation Fed-MIL
In a federated learning setting, consider several local models, each hosted on a client, denoted by where . These models, probability differing in architecture, generate logits for given inputs. The global model, hosted on a server, is denoted by .
The distillation process involves treating the local models as teacher models. The predicted logits from each local model are averaged to form pseudo-labels:
Here, denotes the local dataset on client i, and is the number of WSIs (samples) in that dataset. The term represents the aggregated logits, which serve as pseudo-labels for supervising the global model at the class prediction level.
The global model learns from these pseudo-labels through distillation. The loss function used is the Kullback–Leibler (KL) divergence, which measures the difference between the predictive distribution of the global model and the distilled pseudo-labels:
Here, represents the logits output by the global model, and denotes the temperature-scaled softmax with parameter .
The objective is to minimize this loss, aligning the global model’s predictions with the pseudo-labels derived from the local models, thus leveraging the collective knowledge of all participating models while maintaining data privacy.
3.4. Patch-CAM-Based Distillation
For distillation information selection, directly aggregating intermediate parameters or their gradients is infeasible in federated settings due to model heterogeneity, and sharing raw input data is prohibited due to privacy concerns. Whole-slide images (WSIs) are extremely large and must be divided into hundreds of patches, most of which do not contain lesions. This makes attention mechanisms particularly important as they enable the model to highlight the small fraction of diagnostically relevant patches. However, not all WSI models include explicit attention modules. To ensure generality across heterogeneous clients, we draw inspiration from Grad-CAM [39] and extend it to the patch level, leading to our Patch-CAM design.
Specifically, we compute gradient-based importance scores for each patch feature in the proxy dataset during training. The importance score is derived from the gradients of the maximum predicted logit with respect to each feature component and then averaged across channels to obtain a stable estimate of patch relevance. This adaptation follows the spirit of Grad-CAM but is redefined at the patch level, allowing for universal application to any MIL framework. Moreover, compared with exposing full gradients, sharing only Patch-CAM scores is less privacy-sensitive since they represent coarse structural knowledge rather than fine-grained feature information. Importantly, lesion tissue typically induces different gradient responses compared with normal tissue, which makes these scores both discriminative and generalizable across heterogeneous data distributions. In addition, the Patch-CAM scores provide interpretable heatmaps that help clinicians understand which regions influenced the model’s decision, further enhancing transparency.
Formally, let denote the j-th feature component of the i-th patch , where each patch has a feature dimension d, and the total number of patches in the slide is . The patch importance weight is obtained by averaging the gradient of the maximum predicted logit with respect to all feature components. Following the Grad-CAM logic, we define the Patch-CAM score as
where the multiplication between patch features and their corresponding gradient-derived weights highlights the relative importance of each patch in the global decision.
Every WSI can be regarded as the collection of patch features x. We compute a normalized importance score for each patch and treat it as additional structural distillation knowledge:
The structural distillation process aggregates patch-level attention distributions from clients:
Here, denotes the attention-based importance distribution over patches within a WSI, and is the corresponding distribution computed from client i. Similar to the label distillation, the weighting factor ensures that clients with larger datasets contribute more. Unlike , which aligns class-level predictive distributions, aligns the patch-level attention distributions, thereby transferring structural knowledge across heterogeneous clients.
The final loss is
3.5. Model Architecture Diagram
The architecture diagram as shown in Figure 4 depicts the proposed HFed-MIL, a specialized federated learning framework designed for ovarian cancer diagnosis using whole-slide images (WSIs) from whole-slide images (WSIs) across multiple medical sites. The procedure begins by converting whole-slide images (WSIs) into smaller sections called patches at each location. Data augmentation techniques are then employed to improve the resilience of the model. Each site calculates attention scores based on gradients for its patches in a way that protects privacy. These scores are then used to inform the global model’s process of distilling knowledge. This allows the central model to get valuable information without directly accessing sensitive data, maintaining privacy. The approach achieves a compromise between sharing information and maintaining anonymity. This allows the federated system to effectively manage various data distributions and model preferences while delivering state-of-the-art medical picture analysis performance. The corresponding algorithm is shown in Algorithm 1.
| Algorithm 1: Algorithm for Proposed HFed-MIL |
 |
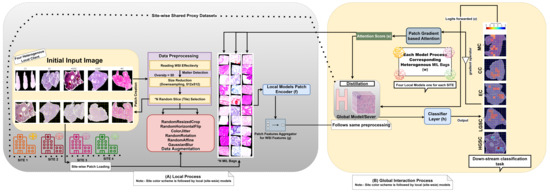
Figure 4.
Overview of the proposed HFed-MIL architecture. Each site (hospital) contains a local MIL-based model with its own WSI patch loader and patch encoder f. Patch-level gradients are used to compute attention scores for each instance, enabling interpretable and privacy-aware representations. These are aggregated via a site-specific aggregator g and passed to the downstream classifier h. *N is the number of slices obtained from a WSI during preprocessing, which later corresponds to the MIL bag size. Gradient-based attention knowledge is distilled into a global server model using a shared proxy dataset processed with the same pipeline. The framework supports heterogeneity across sites, including data distributions and model architectures, while preserving patient privacy and maximizing generalization performance.
4. Experiments and Discussion
In this section, we present the results of our experiments, focusing on the impact of model heterogeneity on the performance of different federated learning methods for ovarian cancer analysis.
4.1. Baseline and Parameter Setting
We compare HFed-MIL with other state-of-the-art FL approaches, including FedAvg [21], FedDF [28], FedMD [30], and FedRAD [32]. FedAvg aggregates local models by averaging their parameters. FedDF distills the ensemble knowledge of all client models into a single global model. FedMD resembles FedDF but maintains no global model; each client independently trains its local model using aggregated soft predictions from the server. FedRAD improves robustness by using the median aggregation of client predictions to filter out low-quality knowledge. Finally, HFed-MIL refines the global model by integrating both class-level and structural knowledge from client models.
The following hyperparameters are used throughout all experiments without special mention:
- Optimizer: Adam with learning rate ;
- Number of communication rounds: 20;
- Distillation temperature: ;
- Patch sampling: 100 patches per WSI, each of size ;
- Number of clients: 4;
- Batch size: 1 WSI bag per iteration (due to memory constraints with large patch sets);
- GPU: 4× Titan RTX.
This setting serves as the default configuration across all compared methods.
4.2. Experiment Type—A: Comparative Analysis
In this subsection, we compare the performance of different federated learning methods in the context of ovarian cancer analysis. The following methods were evaluated: Centralized, FedAvg, FedDF, FedMD, FedRAD, and our proposed method, HFed-MIL, as shown in Table 1 for four clients. For the heterogeneous model setting, we apply ResNet, ViT, EffcientViT [69], and ConVit [70], and for the homogeneous model setting, we apply four ResNet models. Following the non-IID setting of [71], we apply Dirichlet distribution to split the local datasets parameterized by . The homogeneous data setting is 1, and the heterogeneous data setting is 10; further relevant experimental parameters are discussed earlier in Section 4.1.
Starting with the centralized approach, we achieved an accuracy of 76.7% with a loss of 0.80. This serves as an upper bound for comparison with the federated learning methods, which denote performance without any heterogeneity. The experimental results indicate significant variability in performance across different federated learning methods when dealing with heterogeneous data and models. Specifically, FedAvg [21] exhibited a substantial decline in performance with heterogeneous data, recording accuracy of only 37.2%, and it does not support heterogeneous model scenarios, as shown by the absence of results in these cases. This highlights its limitations in environments with diverse data distributions.

Table 1.
Comparison of federated learning methods on different models and data types. The relevant loss and accuracy curves of the heterogeneous model and data are presented in Figure 5 and Figure 6, respectively. Abbreviations: Homo = Homogeneous, Heter = Heterogeneous.
| Method | Model | Data | Accuracy | Loss | Sensitivity | Specificity |
|---|---|---|---|---|---|---|
| Centralized | - | - | 75.7% | 0.70 | 76.2% | 75.0% |
| FedAvg [21] | Homo | Homo | 74.0% | 0.74 | 73.5% | 74.2% |
| Heter | 47.2% | 1.54 | 46.0% | 48.0% | ||
| Heter | Homo | - | - | - | - | |
| Heter | - | - | - | - | ||
| FedDF [28] | Homo | Homo | 56.5% | 1.07 | 57.0% | 56.0% |
| Heter | 42.0% | 1.33 | 41.5% | 42.2% | ||
| Heter | Homo | 61.5% | 1.08 | 62.0% | 61.0% | |
| Heter | 50.1% | 1.32 | 50.5% | 49.8% | ||
| FedMD [30] | Homo | Homo | 54.7% | 1.14 | 55.0% | 54.5% |
| Heter | 47.1% | 1.37 | 47.5% | 46.8% | ||
| Heter | Homo | 55.7% | 1.25 | 56.0% | 55.2% | |
| Heter | 49.3% | 1.31 | 49.0% | 49.5% | ||
| FedRAD [32] | Homo | Homo | 48.3% | 1.31 | 48.5% | 48.0% |
| Heter | 43.5% | 1.45 | 44.0% | 43.0% | ||
| Heter | Homo | 54.0% | 1.23 | 54.5% | 53.8% | |
| Heter | 50.0% | 1.20 | 50.5% | 49.8% | ||
| HFed-MIL | Homo | Homo | 66.1% | 0.91 | 66.5% | 65.8% |
| Heter | 57.3% | 1.03 | 57.0% | 57.5% | ||
| Heter | Homo | 57.2% | 0.88 | 57.5% | 57.0% | |
| Heter | 53.9% | 0.95 | 54.0% | 53.8% |
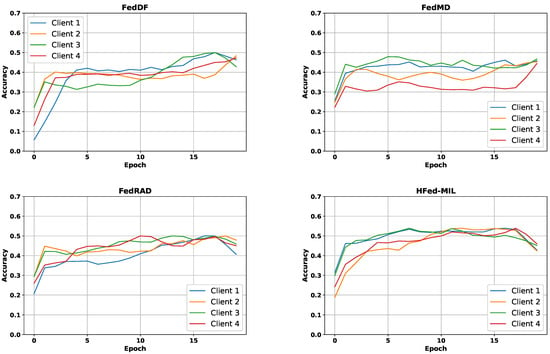
Figure 5.
Comparative plots of accuracy curves for different federated aggregation methods. Each colored curve corresponds to one client.
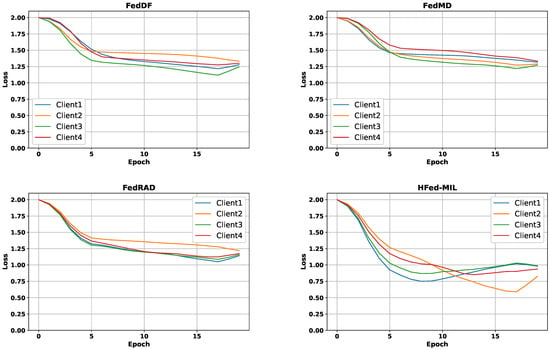
Figure 6.
Comparative plots of training loss curves for different federated aggregation methods. Each colored curve corresponds to one client.
In contrast, methods employing knowledge distillation, such as FedDF [28] and our proposed HFed-MIL, demonstrated more robust performance regarding data and model heterogeneity. Notably, HFed-MIL outperformed other methods in heterogeneous settings, achieving an accuracy of 63.9% on heterogeneous data within heterogeneous models. This superior performance can be attributed to effectively utilizing diverse knowledge inherent in heterogeneous models, which is crucial for distillation-based approaches. The distillation process benefits significantly from these models’ rich and varied information, enhancing learning efficacy and model robustness across diverse federated networks.
Finally, our proposed method, HFed-MIL, outperformed all other federated learning methods regarding accuracy and loss. When a homogeneous model with homogeneous data was used, HFed-MIL achieved an accuracy of 66.1% with a loss of 0.91, surpassing the performance of other methods. However, when dealing with heterogeneous data, HFed-MIL, like other methods, faced challenges and experienced a drop in performance, achieving an accuracy of 63.9% with a loss of 0.95. Nevertheless, HFed-MIL demonstrated superior performance when employing a heterogeneous model with homogeneous data, achieving an accuracy of 67.2% with a loss of 0.88.
In the context of model heterogeneity, we emphasize that model diversity contributes to improved performance in our distillation-based models. In contrast, the performance of FedAvg, which uses “transmit” parameters, suffers when models are heterogeneous. Notably, incorporating diverse knowledge becomes crucial for models employing distillation and transfer learning with proxy datasets, particularly when dealing with heterogeneous data. This diversity enhances the overall performance of our proposed approach.
4.3. Experiment Type—B: Global Model Attention Visualization
To further investigate the interpretability of the proposed method, we visualize the attention maps produced by the global model. The motivation is to examine whether the gradient-based attention design can highlight discriminative regions in whole-slide images (WSIs) and provide consistent structural cues across heterogeneous clients. By comparing the visualization results with those from baseline methods, we aim to demonstrate that HFed-MIL not only achieves competitive accuracy but also yields more coherent and semantically meaningful attention patterns, thereby enhancing the transparency of the federated aggregation process.
As illustrated in Figure 7, the top row corresponds to the proposed HFed-MIL, the middle row shows the results of FedDF, and the bottom row presents FedRAD. HFed-MIL produces sharper and more distinctive attention patterns that emphasize discriminative lesion regions, thereby mitigating the vanishing effect often caused by averaging attention across heterogeneous clients. In contrast, FedDF yields smoother but less informative responses due to the averaging of knowledge distillation. At the same time, FedRAD exhibits an even more uniform distribution of attention, which may further dilute critical diagnostic cues. These findings indicate that HFed-MIL not only improves predictive performance but also enhances interpretability by providing pathologists with more precise and clinically meaningful visual evidence of the model’s decision rationale.
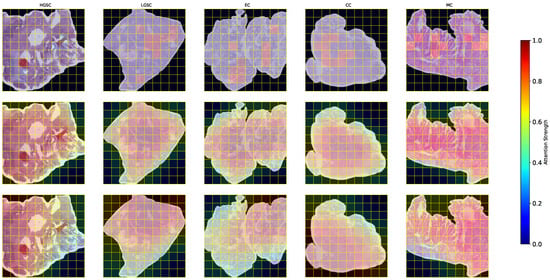
Figure 7.
Comparison of attention maps between HFed-MIL (top row), FedDF (middle row), and FedRAD (bottom row) across different ovarian cancer subtypes. HFed-MIL produces sharper and more distinctive attention patterns, FedDF yields smoother but less informative responses, while FedRAD exhibits even more uniform distributions, potentially diluting critical diagnostic cues.
4.4. Experiment Type—C: Ablation Study of Heterogeneity
As shown in Figure 8, We tested the three methods previously mentioned—FedDF, FedRAD, and HFed-MIL—with the same settings as the heterogeneous models discussed earlier, except for variation, to validate the model performance with various degrees of heterogeneity. The experiments indicate that all methods experience a decline in performance as heterogeneity increases. However, HFed-MIL consistently achieves the best results regardless of the level of heterogeneity.
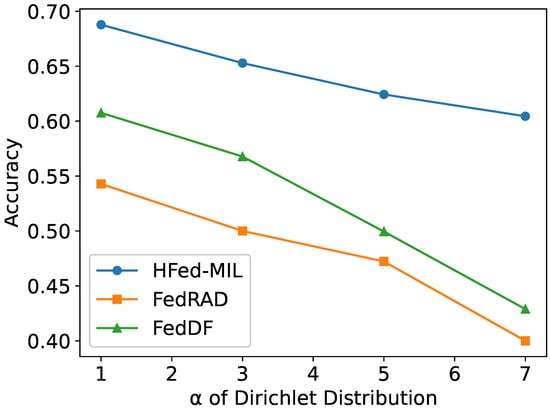
Figure 8.
Ablation study on the effect of under heterogeneous client conditions.
4.5. Experiment Type—D: Ablation Study of Client Number
Figure 9 presents the ablation study investigating how the number of clients affects model performance under heterogeneous conditions. As the number of clients increases with the same settings as the heterogeneous models discussed earlier except the client number, the data becomes more fragmented, and the heterogeneity among clients is exacerbated, leading to a general decline in accuracy for all methods. Despite this trend, HFed-MIL consistently outperforms both FedDF and FedRAD across all settings, demonstrating superior robustness and generalization capability in the presence of increasing client numbers.
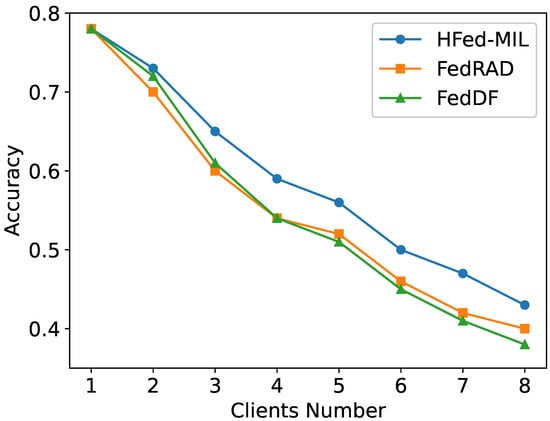
Figure 9.
Ablation study on the effect of the client number under heterogeneous client conditions.
4.6. Experiment Type—E: Generalization Test with Proxy Dataset from Another Domain
To further evaluate cross-domain generalization, we constructed proxy datasets by mixing a small portion of in-domain data with external sources. Specifically, in addition to of the original training data, we consider two external domains: Camelyon16 and BreakHis. Three proxy compositions are tested with the same total size N = 100: (i) Camelyon-only, where the remaining samples are from Camelyon16; (ii) BreakHis-only, where the remaining samples are from BreakHis; and (iii) both, where the remaining samples are equally split between Camelyon16 and BreakHis. In all cases, label spaces are aligned with the target task by mapping external samples to the closest categories. We keep the backbone (ResNet-18), client number (4), and all optimization hyperparameters identical to previous experiments for a fair comparison. Distillation-based methods use the proxy only at the server side, while FedAvg does not rely on a proxy. The experiement result is shown in Table 2.

Table 2.
Cross-domain proxy generalization (10% in-domain + external). Metrics are percentages (%).
This setup simulates realistic scenarios where publicly available proxy data may derive from different sources or acquisition conditions than the clients’ local data. We expect Camelyon-only to benefit tumor-region localization due to structural similarity, while BreakHis-only emphasizes appearance shift at the patch level. The mixed proxy stabilizes performance under distribution shift conditions, and HFed-MIL is expected to retain stronger robustness by leveraging structural attention in addition to class-level logits.
4.7. Experiment Type—F: Privacy and Security Analysis
To substantiate our claim that gradient-based attention scores are less privacy-sensitive than raw features, we conducted a dedicated privacy analysis within the federated setting. The objective is to examine whether an adversary can exploit the information shared in HFed-MIL to infer private data from local clients. We compared four types of shared information: (1) raw gradients, (2) class logits, (3) gradient-based attention scores, and (4) our proposed HFed-MIL, which combines logits with attention.
Among various privacy attacks, we focus on the Membership Inference Attack (MIA) [72], where an adversary attempts to determine whether a given sample belongs to the training set. MIA is the most broadly applicable since it can exploit any type of shared signal. Other attack families such as gradient inversion [35,36] or GAN-based reconstruction are not applicable in our setting: gradient inversion requires direct access to raw gradients, which HFed-MIL never exposes, while GAN-based conditional generation could at best produce vague class-level prototypes rather than patient-specific WSI patches. Therefore, we only evaluated MIA, which remains the most realistic threat with our federated design.
To evaluate MIA, we followed the standard shadow-model setup. For each type of shared information, we trained a binary attack classifier using shadow clients with disjoint datasets, where positive examples correspond to training samples and negative examples to held-out samples. The attack model was then applied to target clients, and MIA performance was reported in terms of AUC. We do not report results for reconstruction-style attacks since such methods require raw gradients or embeddings that are not shared in our design.
As shown in Table 3, exposing raw gradients leads to severe privacy leakage with MIA AUC∼0.85. Sharing logits alone reduces leakage to nearly random guessing (AUC∼0.55), while attention scores exhibit slightly higher vulnerability (AUC∼0.60). HFed-MIL, which combines logits and attention, yields AUC∼0.62. Although this is marginally higher than either signal alone, it remains close to the random baseline of 0.5 and substantially lower than the gradient case. This indicates that HFed-MIL achieves a practical balance between utility and privacy preservation: it improves predictive performance and interpretability without incurring significant privacy risks.

Table 3.
Membership inference performance for different shared information types. Higher AUC indicates greater privacy leakage. An AUC of 0.5 corresponds to random guessing, meaning no privacy leakage. Reconstruction-style attacks are infeasible without direct access to the gradient.
4.8. Experiment Type—G: Computation and Communication Cost Analysis
To ensure applicability across heterogeneous local model architectures, HFed-MIL includes gradient-based attention scores. While this design provides a unified way of distilling structural knowledge across clients, it may introduce additional computational overhead. Therefore, we conducted an experiment to quantify the computation and communication cost, where all methods were evaluated with the same setting using four clients with ResNet-18 as the backbone model. For distillation-based methods, a proxy dataset of size 100 was used, while for FedAvg, each client was assigned 100 local samples.
The results in Table 4 show that HFed-MIL incurs only a modest additional overhead, with about higher training time and around higher GPU memory usage per round compared with other distillation-based methods (FedDF, FedMD, and FedRAD). This gap mainly derives from the computation of gradient-based attention scores, which requires intermediate gradients to be retained. Nevertheless, since the global model can also be equipped with an explicit attention head to directly align with the distilled structural targets, the overhead can be bounded without doubling the training cost.

Table 4.
Comparison of computation and communication cost across different federated aggregation methods. Reported values are averaged over 20 communication rounds.
Regarding communication, all distillation-based methods drastically reduce the cost compared with FedAvg since they exchange only predictions rather than full model parameters. Among them, HFed-MIL requires larger communication than FedDF/FedMD/FedRAD because in addition to class-level logits, it uploads patch-level attention scores. Concretely, for each client with N proxy samples (each containing P patches and C classes), FedDF/FedRAD only transmit logits per round, whereas HFed-MIL transmits values (i.e., one patch attention vector per sample). Aggregated across K clients, the server receives values each round. This explains the ∼0.32 MB per client per round cost of HFed-MIL, which still remains orders of magnitude smaller than FedAvg (∼0.37 GB).
Overall, the results demonstrate that the proposed gradient-based attention design is computationally feasible in practice, while providing additional robustness and interpretability.
5. Conclusions
Ovarian cancer remains a significant global health concern, and its diagnosis heavily relies on whole-slide images (WSIs). Due to WSI gigapixel spatial consumption and medical privacy restrictions, the Fed-MIL framework is highly appropriate. This study summarizes and categorizes the current federated learning into three types based on the information transmitted: parameters, gradients, and proxy dataset prediction knowledge. However, due to model heterogeneity, we can only adopt the transmission of proxy dataset prediction knowledge, namely, the distillation-based method. Furthermore, the performance of distillation-based federated learning highly depends on the effectiveness of the distilled information. For the substantial spatial consumption of WSIs, in addition to general unstructured result information, we also require attention to information that can be measured within heterogeneous models to ensure performance. This paper introduces HFed-MIL, which applies gradient attention measured-distillation-based federated learning to tackle these challenges. Specifically, we first induce and formulate a commonly used form of MIL. Then, to enable relative flexibility to model heterogeneity and reach the balance between security and effectiveness in HFed-MIL, we not only distill the information of prediction results but also measure the importance of each patch based on the average gradient and then distill its structural knowledge. Since the patch gradient importance scores contain more information than labels but less than features and are more dimensionally flexible than features, this method allows for sufficient information distillation without compromising privacy risks. Our experiments demonstrate that the proposed model achieves state-of-the-art performance. HFed-MIL improves the practicality of deep learning in ovarian cancer and further enhances the trustworthiness of medical deep learning, thereby increasing its clinical significance.
Although HFed-MIL achieves notable performance improvements on ovarian cancer WSIs under data and model heterogeneity conditions, this study’s limitation is that the shared proxy dataset must have uniformly sampled patch positions and sizes, which reduces the framework’s flexibility. We plan to improve on this aspect in future work. Furthermore, comprehensive evaluation on an external multi-institutional dataset is necessary to fully validate HFed-MIL’s generalization ability and robustness in practical clinical scenarios.
Author Contributions
Conceptualization, X.Z.; methodology, X.Z.; software, X.Z.; validation, X.Z. and A.A.; formal analysis, X.Z.; investigation, X.Z.; resources, X.Z.; data curation, X.Z.; writing—original draft preparation, X.Z.; writing—review and editing, A.A.; visualization, M.H.T.; supervision, X.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
Correction Statement
This article has been republished with a minor correction to the Data Availability Statement. This change does not affect the scientific content of the article.
References
- Che, M.; Yin, R. Analysis of the global burden of ovarian cancer in adolescents. Int. J. Gynecol. Cancer 2025, 35, 101620. [Google Scholar] [CrossRef]
- Farahani, H.; Boschman, J.; Farnell, D.; Darbandsari, A.; Zhang, A.; Ahmadvand, P.; Jones, S.J.; Huntsman, D.; Köbel, M.; Gilks, C.B.; et al. Deep learning-based histotype diagnosis of ovarian carcinoma whole-slide pathology images. Mod. Pathol. 2022, 35, 1983–1990. [Google Scholar] [CrossRef]
- Breen, J.; Allen, K.; Zucker, K.; Adusumilli, P.; Scarsbrook, A.; Hall, G.; Orsi, N.M.; Ravikumar, N. Artificial intelligence in ovarian cancer histopathology: A systematic review. NPJ Precis. Oncol. 2023, 7, 83. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, C.; Zhou, S. Artificial intelligence-based risk stratification, accurate diagnosis and treatment prediction in gynecologic oncology. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2023; Volume 96, pp. 82–99. [Google Scholar]
- Ilse, M.; Tomczak, J.M.; Welling, M. Attention-based deep multiple instance learning. In Proceedings of the 35th International Conference on Machine Learning, Stockholm, Sweden, 10–15 July 2018; pp. 2127–2136. [Google Scholar]
- Lu, M.Y.; Williamson, D.F.; Chen, T.Y.; Chen, R.J.; Barbieri, M.; Mahmood, F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 2021, 5, 555–570. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, Y.; Eliceiri, K.W. Dual-stream multiple instance learning network for whole slide image classification with self-supervised contrastive learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 14318–14328. [Google Scholar]
- Shao, Z.; Bian, H.; Chen, Y.; Wang, Y.; Zhang, J.; Ji, X. Transmil: Transformer based correlated multiple instance learning for whole slide image classification. Adv. Neural Inf. Process. Syst. 2021, 34, 2136–2147. [Google Scholar]
- Zhang, H.; Meng, Y.; Zhao, Y.; Qiao, Y.; Yang, X.; Coupland, S.E.; Zheng, Y. Dtfd-mil: Double-tier feature distillation multiple instance learning for histopathology whole slide image classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 18802–18812. [Google Scholar]
- Lin, T.; Yu, Z.; Hu, H.; Xu, Y.; Chen, C.W. Interventional bag multi-instance learning on whole-slide pathological images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 19830–19839. [Google Scholar]
- Tang, W.; Huang, S.; Zhang, X.; Zhou, F.; Zhang, Y.; Liu, B. Multiple instance learning framework with masked hard instance mining for whole slide image classification. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 2–3 October 2023; pp. 4078–4087. [Google Scholar]
- Zhong, L.; Wang, G.; Liao, X.; Zhang, S. HAMIL: High-Resolution Activation Maps and Interleaved Learning for Weakly Supervised Segmentation of Histopathological Images. IEEE Trans. Med. Imaging 2023, 42, 2912–2923. [Google Scholar] [CrossRef] [PubMed]
- McMahan, B.; Moore, E.; Ramage, D.; Hampson, S.; y Arcas, B.A. Communication-efficient learning of deep networks from decentralized data. In Proceedings of the Artificial Intelligence and Statistics, PMLR, Fort Lauderdale, FL, USA, 20–22 April 2017; pp. 1273–1282. [Google Scholar]
- Konečnỳ, J.; McMahan, H.B.; Yu, F.X.; Richtárik, P.; Suresh, A.T.; Bacon, D. Federated learning: Strategies for improving communication efficiency. arXiv 2016, arXiv:1610.05492. [Google Scholar]
- Yang, Q.; Liu, Y.; Chen, T.; Tong, Y. Federated machine learning: Concept and applications. ACM Trans. Intell. Syst. Technol. (TIST) 2019, 10, 1–19. [Google Scholar] [CrossRef]
- Rieke, N.; Hancox, J.; Li, W.; Milletari, F.; Roth, H.R.; Albarqouni, S.; Bakas, S.; Galtier, M.N.; Landman, B.A.; Maier-Hein, K.; et al. The future of digital health with federated learning. NPJ Digit. Med. 2020, 3, 1–7. [Google Scholar] [CrossRef]
- Liu, Y.; Kang, Y.; Zou, T.; Pu, Y.; He, Y.; Ye, X.; Ouyang, Y.; Zhang, Y.Q.; Yang, Q. Vertical Federated Learning: Concepts, Advances, and Challenges. IEEE Trans. Knowl. Data Eng. 2024, 36, 3615–3634. [Google Scholar] [CrossRef]
- Guo, W.; Zhuang, F.; Zhang, X.; Tong, Y.; Dong, J. A Comprehensive Survey of Federated Transfer Learning: Challenges, Methods and Applications. arXiv 2024, arXiv:2403.01387. [Google Scholar] [CrossRef]
- Li, Q.; Wen, Z.; Wu, Z.; Hu, S.; Wang, N.; Li, Y.; Liu, X.; He, B. A survey on federated learning systems: Vision, hype and reality for data privacy and protection. IEEE Trans. Knowl. Data Eng. 2021, 35, 3347–3366. [Google Scholar] [CrossRef]
- Karimireddy, S.P.; Kale, S.; Mohri, M.; Reddi, S.; Stich, S.; Suresh, A.T. Scaffold: Stochastic controlled averaging for federated learning. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual Event, 13–18 July 2020; pp. 5132–5143. [Google Scholar]
- Li, X.; Huang, K.; Yang, W.; Wang, S.; Zhang, Z. On the convergence of fedavg on non-iid data. arXiv 2019, arXiv:1907.02189. [Google Scholar]
- Zhao, Y.; Li, M.; Lai, L.; Suda, N.; Civin, D.; Chandra, V. Federated learning with non-iid data. arXiv 2018, arXiv:1806.00582. [Google Scholar] [CrossRef]
- Li, T.; Sahu, A.K.; Zaheer, M.; Sanjabi, M.; Talwalkar, A.; Smith, V. Federated optimization in heterogeneous networks. In Proceedings of the Machine Learning and Systems, Seoul, Republic of Korea, 25–28 October 2020; Volume 2, pp. 429–450. [Google Scholar]
- Xie, H.; Xia, M.; Wu, P.; Wang, S.; Huang, K. Decentralized Federated Learning with Asynchronous Parameter Sharing for Large-scale IoT Networks. IEEE Internet Things J. 2024, 11, 34123–34139. [Google Scholar] [CrossRef]
- Chen, J.; Pan, X.; Monga, R.; Bengio, S.; Jozefowicz, R. Revisiting distributed synchronous SGD. arXiv 2016, arXiv:1604.00981. [Google Scholar]
- Xiao, Z.; Chen, Z.; Liu, S.; Wang, H.; Feng, Y.; Hao, J.; Zhou, J.T.; Wu, J.; Yang, H.; Liu, Z. Fed-grab: Federated long-tailed learning with self-adjusting gradient balancer. Adv. Neural Inf. Process. Syst. 2024, 36, 77745–77757. [Google Scholar]
- Wang, H.; Xu, W.; Fan, Y.; Li, R.; Zhou, P. AOCC-FL: Federated Learning with Aligned Overlapping via Calibrated Compensation. In Proceedings of the IEEE INFOCOM 2023-IEEE Conference on Computer Communications, New York, NY, USA, 17–20 May 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 1–10. [Google Scholar]
- Lin, T.; Kong, L.; Stich, S.U.; Jaggi, M. Ensemble distillation for robust model fusion in federated learning. Adv. Neural Inf. Process. Syst. 2020, 33, 2351–2363. [Google Scholar]
- Zhu, Z.; Hong, J.; Zhou, J. Data-free knowledge distillation for heterogeneous federated learning. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual Event, 18–24 July 2021; pp. 12878–12889. [Google Scholar]
- Li, D.; Wang, J. Fedmd: Heterogenous federated learning via model distillation. arXiv 2019, arXiv:1910.03581. [Google Scholar] [CrossRef]
- Wang, H.; Yurochkin, M.; Sun, Y.; Papailiopoulos, D.; Khazaeni, Y. Federated learning with matched averaging. arXiv 2020, arXiv:2002.06440. [Google Scholar] [CrossRef]
- Sturluson, S.P.; Trew, S.; Muñoz-González, L.; Grama, M.; Passerat-Palmbach, J.; Rueckert, D.; Alansary, A. Fedrad: Federated robust adaptive distillation. arXiv 2021, arXiv:2112.01405. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, Y.; Zheng, Y.; Tian, X.; Peng, H.; Liu, T.; Han, B. FedFed: Feature distillation against data heterogeneity in federated learning. Adv. Neural Inf. Process. Syst. 2024, 36, 60397–60428. [Google Scholar]
- Rocher, L.; Hendrickx, J.M.; De Montjoye, Y.A. Estimating the success of re-identifications in incomplete datasets using generative models. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Geiping, J.; Bauermeister, H.; Drozdzal, M.; Moeller, M. Inverting gradients–How easy is it to break privacy in federated learning? Adv. Neural Inf. Process. Syst. 2020, 33, 16937–16947. [Google Scholar]
- Zhu, L.; Liu, Z.; Han, S. Deep leakage from gradients. Adv. Neural Inf. Process. Syst. 2019, 32, 14774–14784. [Google Scholar]
- Zhang, Y.; Jia, R.; Pei, H.; Wang, W.; Li, B.; Song, D. The secret revealer: Generative model-inversion attacks against deep neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 253–261. [Google Scholar]
- Carlini, N.; Liu, C.; Erlingsson, Ú.; Kos, J.; Song, D. The secret sharer: Evaluating and testing unintended memorization in neural networks. In Proceedings of the 28th USENIX Security Symposium (USENIX Security 19), Santa Clara, CA, USA, 14–16 August 2019; pp. 267–284. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Beg, A.; Shah, S.N.A.; Parveen, R. Deep learning approaches for interpreting Non-coding regions in Ovarian cancer. In Deep Learning in Genetics and Genomics; Elsevier: Amsterdam, The Netherlands, 2025; pp. 71–86. [Google Scholar]
- Hong, M.K.; Ding, D.C. Early diagnosis of ovarian cancer: A comprehensive review of the advances, challenges, and future directions. Diagnostics 2025, 15, 406. [Google Scholar] [CrossRef]
- Xu, Y.; Jia, Z.; Wang, L.B.; Ai, Y.; Zhang, F.; Lai, M.; Chang, E.I.C. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinform. 2017, 18, 281. [Google Scholar] [CrossRef]
- Nahid, A.A.; Kong, Y. Histopathological breast-image classification using local and frequency domains by convolutional neural network. Information 2018, 9, 19. [Google Scholar] [CrossRef]
- Ahmed, A.; Xiaoyang, Z.; Tunio, M.H.; Butt, M.H.; Shah, S.A.; Chengxiao, Y.; Pirzado, F.A.; Aziz, A. OCCNET: Improving Imbalanced Multi-Centred Ovarian Cancer Subtype Classification in Whole Slide Images. In Proceedings of the 2023 20th International Computer Conference on Wavelet Active Media Technology and Information Processing (ICCWAMTIP), Chengdu, China, 15–17 December 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 1–8. [Google Scholar]
- Wang, Y.; Lin, W.; Zhuang, X.; Wang, X.; He, Y.; Li, L.; Lyu, G. Advances in artificial intelligence for the diagnosis and treatment of ovarian cancer. Oncol. Rep. 2024, 51, 1–17. [Google Scholar] [CrossRef]
- Jahanifar, M.; Raza, M.; Xu, K.; Vuong, T.T.L.; Jewsbury, R.; Shephard, A.; Zamanitajeddin, N.; Kwak, J.T.; Raza, S.E.A.; Minhas, F.; et al. Domain generalization in computational pathology: Survey and guidelines. ACM Comput. Surv. 2025, 57, 1–37. [Google Scholar] [CrossRef]
- Yoon, J.S.; Oh, K.; Shin, Y.; Mazurowski, M.A.; Suk, H.I. Domain generalization for medical image analysis: A review. Proc. IEEE 2024, 112, 1583–1609. [Google Scholar] [CrossRef]
- Herath, H.; Herath, H.; Madusanka, N.; Lee, B.I. A Systematic Review of Medical Image Quality Assessment. J. Imaging 2025, 11, 100. [Google Scholar] [CrossRef] [PubMed]
- Koçak, B.; Ponsiglione, A.; Stanzione, A.; Bluethgen, C.; Santinha, J.; Ugga, L.; Huisman, M.; Klontzas, M.E.; Cannella, R.; Cuocolo, R. Bias in artificial intelligence for medical imaging: Fundamentals, detection, avoidance, mitigation, challenges, ethics, and prospects. Diagn. Interv. Radiol. 2025, 31, 75. [Google Scholar] [CrossRef] [PubMed]
- Xie, E.; Wang, W.; Yu, Z.; Anandkumar, A.; Alvarez, J.M.; Luo, P. SegFormer: Simple and efficient design for semantic segmentation with transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 12077–12090. [Google Scholar]
- Xi, R.; Ahmed, A.; Zeng, X.; Hou, M. A novel transformers-based external attention framework for breast cancer diagnosis. Biomed. Signal Process. Control 2025, 110, 108065. [Google Scholar] [CrossRef]
- Cornia, M.; Stefanini, M.; Baraldi, L.; Cucchiara, R. Meshed-memory transformer for image captioning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 10578–10587. [Google Scholar]
- Huo, Y.; Gang, S.; Guan, C. FCIHMRT: Feature cross-layer interaction hybrid method based on Res2Net and transformer for remote sensing scene classification. Electronics 2023, 12, 4362. [Google Scholar] [CrossRef]
- Tan, F.; Zhai, M.; Zhai, C. Foreign object detection in urban rail transit based on deep differentiation segmentation neural network. Heliyon 2024, 10, e37072. [Google Scholar] [CrossRef]
- Tang, Y.; Yi, J.; Tan, F. Facial micro-expression recognition method based on CNN and transformer mixed model. Int. J. Biom. 2024, 16, 463–477. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 6000–6010. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. In Proceedings of the International Conference on Learning Representations (ICLR), Vienna, Austria, 3–7 May 2021. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Paris, France, 2–3 October 2021; pp. 10012–10022. [Google Scholar]
- Alahmadi, A. Towards ovarian cancer diagnostics: A vision transformer-based computer-aided diagnosis framework with enhanced interpretability. Results Eng. 2024, 23, 102651. [Google Scholar] [CrossRef]
- Adak, D.; Sonawane, S.; Verma, G. OvarianNet-Ca: A Hybrid Cross-Attention Ensemble Model Approach Using MixTransformer and EfficientNet. In Proceedings of the 2025 4th OPJU International Technology Conference (OTCON) on Smart Computing for Innovation and Advancement in Industry 5.0, Raigarh, India, 9–11 April 2025; IEEE: Piscataway, NJ, USA, 2025; pp. 1–6. [Google Scholar]
- Liu, P.; Fu, B.; Ye, F.; Yang, R.; Ji, L. DSCA: A dual-stream network with cross-attention on whole-slide image pyramids for cancer prognosis. Expert Syst. Appl. 2023, 227, 120280. [Google Scholar] [CrossRef]
- Gupta, D.K.; Mago, G.; Chavan, A.; Prasad, D.K. Patch gradient descent: Training neural networks on very large images. arXiv 2023, arXiv:2301.13817. [Google Scholar] [CrossRef]
- Yufei, C.; Liu, Z.; Liu, X.; Liu, X.; Wang, C.; Kuo, T.W.; Xue, C.J.; Chan, A.B. Bayes-MIL: A New Probabilistic Perspective on Attention-based Multiple Instance Learning for Whole Slide Images. In Proceedings of the the Eleventh International Conference on Learning Representations, Kigali, Rwanda, 25–29 April 2022. [Google Scholar]
- Chen, R.J.; Chen, C.; Li, Y.; Chen, T.Y.; Trister, A.D.; Krishnan, R.G.; Mahmood, F. Scaling vision transformers to gigapixel images via hierarchical self-supervised learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 16144–16155. [Google Scholar]
- Sheller, M.J.; Edwards, B.; Reina, G.A.; Martin, J.; Pati, S.; Kotrotsou, A.; Milchenko, M.; Xu, W.; Marcus, D.; Colen, R.R.; et al. Federated learning in medicine: Facilitating multi-institutional collaborations without sharing patient datao. Sci. Rep. 2020, 10, 12598. [Google Scholar] [CrossRef]
- Xu, W.; Fu, Y.L.; Zhu, D. ResNet and its application to medical image processing: Research progress and challenges. Comput. Methods Programs Biomed. 2023, 240, 107660. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Wang, Y.; Chen, H.; Chen, X.; Guo, J.; Liu, Z.; Tang, Y.; Xiao, A.; Xu, C.; Xu, Y.; et al. A survey on vision transformer. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 45, 87–110. [Google Scholar] [CrossRef] [PubMed]
- Hassanin, M.; Anwar, S.; Radwan, I.; Khan, F.S.; Mian, A. Visual attention methods in deep learning: An in-depth survey. Inf. Fusion 2024, 108, 102417. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, PMLR, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- d’Ascoli, S.; Touvron, H.; Leavitt, M.L.; Morcos, A.S.; Biroli, G.; Sagun, L. Convit: Improving vision transformers with soft convolutional inductive biases. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual Event, 18–24 July 2021; pp. 2286–2296. [Google Scholar]
- Hsu, T.M.H.; Qi, H.; Brown, M. Measuring the effects of non-identical data distribution for federated visual classification. arXiv 2019, arXiv:1909.06335. [Google Scholar] [CrossRef]
- Shokri, R.; Stronati, M.; Song, C.; Shmatikov, V. Membership inference attacks against machine learning models. In Proceedings of the 2017 IEEE Symposium on Security and Privacy (SP), San Jose, CA, USA, 22–26 May 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 3–18. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).