Abstract
Teledermatology relies on digital transfer of dermatological images, but compression and resolution differences compromise diagnostic quality. Image enhancement techniques are crucial to compensate for these differences and improve quality for both clinical assessment and AI-based analysis. We developed a customized image degradation pipeline simulating common artifacts in dermatological images, including blur, noise, downsampling, and compression. This synthetic degradation approach enabled effective training of DermaSR-GAN, a super-resolution generative adversarial network tailored for dermoscopic images. The model was trained on 30,000 high-quality ISIC images and evaluated on three independent datasets (ISIC Test, Novara Dermoscopic, PH2) using structural similarity and no-reference quality metrics. DermaSR-GAN achieved statistically significant improvements in quality scores across all datasets, with up to 23% enhancement in perceptual quality metrics (MANIQA). The model preserved diagnostic details while doubling resolution and surpassed existing approaches, including traditional interpolation methods and state-of-the-art deep learning techniques. Integration with downstream classification systems demonstrated up to 14.6% improvement in class-specific accuracy for keratosis-like lesions compared to original images. Synthetic degradation represents a promising approach for training effective super-resolution models in medical imaging, with significant potential for enhancing teledermatology applications and computer-aided diagnosis systems.
1. Introduction
Skin cancer incidence has increased significantly over the past decade, underscoring the vital role of early diagnosis through routine screening of skin lesions [1]. Regular skin checks are recommended for high-risk individuals, including those with fair skin, a history of sunburns, or a family history of skin cancer [2].
Teledermatology has established a pivotal role in diagnosing skin cancers, particularly through teledermoscopy [3]. This approach has revolutionized dermatological care delivery, especially in rural or underserved areas where specialized access is limited. Teledermoscopy allows remote examination of skin lesions using dermoscopes, providing improved convenience and accessibility [4]. This digitization has proven especially significant in pandemic contexts where in-person consultations are limited. Studies have explored smartphone cameras for skin lesion acquisition, envisioning a future where general practitioners or patients can consult expert dermatologists in ambiguous cases [5].
In the last decade, artificial intelligence (AI) methods have been extensively applied in medical imaging, enabling advancements in tasks such as segmentation [6], classification [7], anomaly detection [8], and synthetic data generation [9]. AI-based tools are increasingly valuable in dermatoscopic image analysis, demonstrating promising results in improving diagnostic speed, accuracy, and early detection efficacy [10,11,12]. These tools can analyze large datasets and identify patterns difficult for human detection. However, AI development requires high-quality data [13], and not all images in open databases present suitable clarity and resolution. In teledermatology contexts, patient-acquired images often suffer from artifacts related to environment, camera quality, and acquisition techniques, resulting in poor-quality, blurry, and out-of-focus images. Such inconsistencies hamper effective diagnosis for both medical professionals and AI-based diagnostic tools, emphasizing the need for methodologies that improve image quality and enable accurate diagnosis.
Generative adversarial networks (GANs) have shown promise in image enhancement applications [14], including super-resolution [15,16,17]. However, current GAN approaches often fail to address the unique challenges of dermoscopic images. Dermoscopic images exhibit distinct features such as color variations, irregular patterns, and specific texture details, which require specialized modeling to preserve diagnostic information effectively. Existing GAN models designed for general enhancement may struggle to capture these essential dermatological details, leading to suboptimal results [18,19].
To bridge this gap, we present DermaSR-GAN, a super-resolution GAN tailored specifically for dermoscopic images. Our approach overcomes current limitations by incorporating domain-specific knowledge and a controllable dermatological image degradation pipeline. The main contributions of this study can be summarized as follows:
- We introduce DermaSR-GAN, a novel super-resolution generative adversarial network tailored through specialized training and architecture optimizations to address the unique challenges and image characteristics encountered in dermatology.
- We develop a customized image degradation pipeline to simulate common quality issues in real-world dermatological images, allowing DermaSR-GAN to learn effective mappings from low to high-quality counterparts during training.
- We comprehensively evaluate DermaSR-GAN on three external dermatology image datasets, demonstrating consistent and statistically significant improvements in objective image quality metrics compared to original images and state-of-the-art super-resolution techniques.
- We showcase DermaSR-GAN’s capabilities in boosting downstream classification performance, achieving up to 14.6% improvement in a dermatological image classifier when using DermaSR-GAN-enhanced images, highlighting its utility for computer-aided diagnosis.
The primary innovation of our work lies not in the network architecture itself, but in the synergistic combination of realistic degradation modeling with state-of-the-art super-resolution techniques, demonstrating that domain-specific data synthesis is crucial for effective medical image enhancement.
2. Related Works
2.1. SR Techniques in Medical Imaging
Super-resolution (SR) techniques have evolved significantly from classical enhancement methods to sophisticated deep learning approaches. Early approaches in medical imaging relied on traditional enhancement techniques, such as histogram equalization, contrast stretching, and unsharp masking to improve image visibility. Classical super-resolution methods employed bicubic interpolation and dictionary-learning techniques, but these approaches often produced blurry outputs, lacking high-frequency details essential for medical imaging applications [20].
Deep learning revolutionized super-resolution through convolutional neural networks (CNNs). SRCNN [21] introduced the first CNN-based approach, while subsequent works like VDSR and EDSR achieved improved performance through deeper architectures and residual connections. However, CNN-based methods optimizing pixel-wise fidelity metrics often produce perceptually unsatisfying results, as the synthetic images often lack sharpness and realistic textures.
Generative adversarial networks have addressed these limitations by incorporating perceptual loss functions. SRGAN [22] was the first to pioneer GAN-based super-resolution by combining adversarial loss with perceptual loss to generate visually appealing results. ESRGAN [23] further improved on SRGAN through enhanced generator architecture and refined training strategies, achieving state-of-the-art performance in natural image super-resolution. In 2021, Real-ESRGAN [17] introduced realistic degradation modeling during training: instead of using simple bicubic downsampling, it employed complex degradation pipelines including blur, noise, and compression artifacts to better simulate real-world image degradation. This approach significantly improved generalization to real-world scenarios.
In medical imaging, super-resolution techniques have shown promise across various domains. Several studies have explored super-resolution for dermatological images specifically. Alwakid et al. [24] proposed a GAN-based approach for skin lesion image enhancement, while Mukadam et al. [25] investigated deep learning techniques for improving dermoscopic image quality. These works suggest that better input image quality through SR can positively impact automated diagnosis, presumably by making features like border irregularities or texture patterns more discernible to both algorithms and clinicians. Recently, Veeramani et al. [26] developed a multi-scale approach for skin lesion super-resolution, incorporating multiple resolution levels to capture both fine-grained texture details and global lesion structure. Their method employed a cascaded architecture with progressive refinement stages. However, their approach relies on simplified degradation models using basic downsampling operations, which may not accurately represent real-world image quality degradation scenarios encountered in clinical practice. Additionally, their evaluation was limited to a single dataset, without assessment of generalization across diverse dermatological image collections or evaluating the impact on downstream diagnostic performance.
2.2. Motivation of Our Work
Despite recent advances, existing super-resolution approaches face key limitations in dermatological applications. General-purpose models are typically not optimized for the specific visual characteristics of dermoscopic images, such as fine-grained textures, color patterns, and diagnostically relevant structures. Moreover, many lack systematic evaluation on diverse dermatological datasets or assessment of their impact on downstream clinical tasks.
To overcome these issues, we propose a domain-specific approach that leverages a GAN-based architecture tailored for dermatological imaging. Our pipeline integrates a customized degradation pipeline that goes beyond simplistic assumptions like Gaussian noise or bicubic downsampling. Instead, it models realistic artifacts commonly found in dermatological workflows (such as motion blur, Poisson noise, compression distortions, and heterogeneous downsampling), resulting in more effective training and improved diagnostic relevance across multiple datasets.
The choice of GAN-based methods in this study is further motivated by their efficiency and suitability for real-world applications. GANs generally have shorter training and inference times compared to other generative models, such as diffusion models, making them ideal for scenarios requiring rapid image processing. Additionally, the increasing availability of frameworks like Executorch [27] enables GAN models to run directly on mobile devices. This capability opens up possibilities for integrating super-resolution models into portable dermoscopes, providing real-time high-resolution imaging capabilities without relying on external hardware. Such advancements could significantly enhance the accessibility and efficiency of dermatological imaging workflows, making high-quality imaging available even in resource-limited settings.
To facilitate reproducibility and further research in dermatological super-resolution, we have made publicly available the training/validation splits, training codes, and all experimental results on Mendeley Data: https://doi.org/10.17632/27xk9v8z73.1.
3. Materials and Methods
3.1. Dataset Description
The primary dataset was sourced from the International Skin Imaging Collaboration (ISIC) Archive [28,29]. An expert operator evaluated the ISIC images, excluding those of unsuitable quality or with excessive artifacts, as our model requires high-quality images for effective training. The image selection process, including manual evaluation and filtering of unsuitable images, required approximately 3 weeks. We retained images with a resolution of 1024 × 1024 or higher, ensuring diverse and robust data for training. The images that did not meet this criterion and did not present invasive artifacts formed our test set. Hence, the initial selection of 33,000 images was refined to include 18,884 high-quality samples (15,857 for training and 3027 for validation), ensuring diverse and robust data for training without requiring additional data augmentation. The evaluation of the DermaSR-GAN model extended beyond the ISIC Archive. In addition to using the ISIC Test set, we also employed a private dataset of dermatoscopic images sourced from the Hospital Maggiore della Carità di Novara [3,14] and the PH2 dataset [30] to assess our model’s ability to handle varying image conditions and quality. Table 1 provides a detailed breakdown of the number of images included in each dataset used in this study. The complete framework is shown in Figure 1.

Table 1.
Distribution and quantification of images from each dataset used in this study. The ISIC Archive dataset is divided and employed during both the construction phase and the testing phase. Additionally, two external datasets are used exclusively for testing purposes.
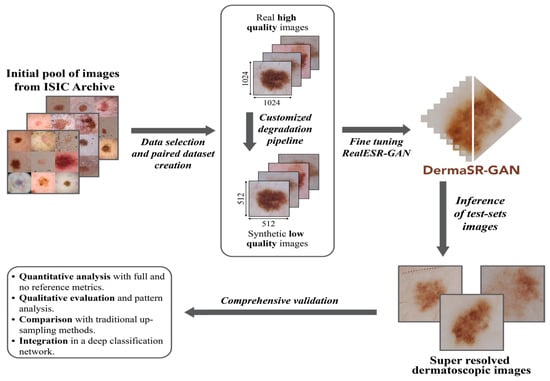
Figure 1.
Overview of the proposed super-resolution framework for dermatological imaging. The framework consists of three main stages: (1) a customized degradation pipeline that simulates realistic artifacts commonly observed in dermoscopic images; (2) a GAN-based generator that reconstructs high-resolution images from low-resolution inputs; and (3) integration with downstream diagnostic tasks to evaluate the impact on classification accuracy.
3.2. Custom Degradation Pipeline for Dermatological Images
The customized degradation pipeline is a fundamental step that ensures that the DermaSR-GAN model is trained on images that closely resemble real-world scenarios, thereby improving its overall performance. Unlike previous works that rely on simplistic degradation models (e.g., bicubic downsampling or basic Gaussian noise), our pipeline simulates the complex artifacts commonly encountered in teledermatology. Our degradation pipeline implements a sequential multi-stage process that more accurately reflects the image quality issues observed in clinical practice:
- Image blur is simulated by randomly applying one of three types: Gaussian, motion, or defocus blur, each with equal probability. Kernel sizes are sampled from odd values between 7 × 7 and 21 × 21. For Gaussian blur, the standard deviation σ ranges between 0.1 and 3.0. This variability aims to capture the diversity of acquisition conditions (e.g., handheld devices, shaky images).
- To replicate sensor-related degradation or transmission noise, we added either Gaussian noise (p = 0.20), Poisson noise (p = 0.20), or salt-and-pepper noise (p = 0.05). Gaussian noise uses a σ randomly sampled between 1 and 30. Poisson noise mimics low-light image acquisition and sensor variability. Salt-and-pepper noise is applied with a lower probability to simulate digital artifacts or faulty pixel sensors.
- Image resolution is reduced using one of four interpolation methods: bilinear, bicubic, area, or nearest-neighbor, each selected with equal likelihood. This step ensures that the model becomes robust to various image resizing operations, as commonly seen when images are shared or viewed on different devices.
- Finally, lossy compression is applied using JPEG compression [31] with a quality factor randomly chosen between 50 and 100, with high probability (p = 0.80). This emulates compression artifacts resulting from storage limitations or transfer via mobile or web platforms, which is frequent in real-world teledermatology workflows.
The stochastic nature of this pipeline ensures diverse training samples that better prepare the model for real-world scenarios. These degradation stages provided a paired dataset with high-resolution (HR) images at 1024 × 1024 pixels matched with their degraded low-resolution (LR) counterparts with a resolution of 512 × 512 pixels. Figure 2 provides a visual comparison between HR dermoscopic images and their LR counterparts obtained with our degradation pipeline (Figure 2).
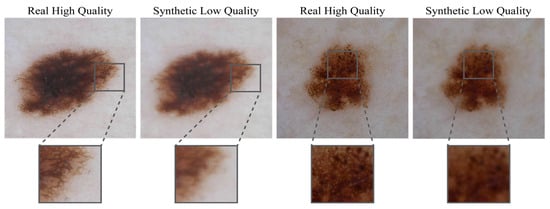
Figure 2.
Comparative showcase of high-quality dermoscopic images and their low-quality synthetic counterparts. High-quality images, essential for accurate dermatological diagnosis, are shown alongside their degraded versions generated using the proposed degradation pipeline. The synthetic low-quality images exhibit artifacts such as motion blur, Poisson noise, and compression distortions, simulating real-world challenges encountered in teledermatology.
3.3. DermaSR-GAN Architecture and Training
Building upon our custom degradation pipeline, we employ a modified Real-ESRGAN architecture optimized for dermatological images. The choice of Real-ESRGAN as our base architecture is motivated by its demonstrated superiority over previous super-resolution methods, achieving state-of-the-art performance in both quantitative metrics and visual quality assessments [17]. While the base architecture follows established super-resolution principles, our key contribution lies in the synergistic combination with domain-specific degradation modeling tailored for dermatological images.
The generator network employs residual-in-residual dense blocks (RRDBs) [23] (Figure 3a) for hierarchical feature extraction, with the following specifications:
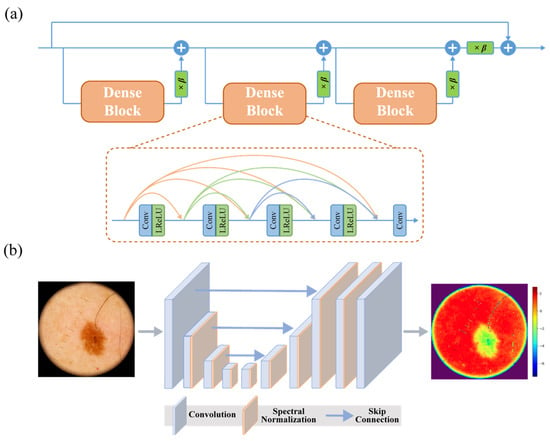
Figure 3.
Architectural highlights of DermaSR-GAN: (a) scheme of a residual-in-residual dense block (RRDB), which forms the core building block of the generator network; (b) U-Net with skip connections and spectral normalization, which stabilizes training and improves adversarial loss optimization.
- Twenty-three RRDB blocks with growth channel of 32;
- Initial convolution layer with 64 feature channels;
- Upsampling via pixel shuffle with scale factor of 2;
- Final convolution layer producing 3-channel RGB output.
The RRDB design removes all batch normalization layers to improve generalization and reduce computational complexity, and integrates dense skip connections with residual scaling (β = 0.2), which stabilizes the training of deeper networks. LeakyReLU activations are used throughout the generator.
The discriminator utilizes a U-Net architecture [32,33] enhanced with spectral normalization [34], which has proven more effective than VGG-style discriminators for providing detailed per-pixel feedback (Figure 3b). It is composed of 9 convolutional layers with progressively increasing feature channels (from 64 up to 512), using strided convolutions (stride 2) for downsampling, followed by 3 additional convolutional layers and a final 1-channel output map. All other architectural settings were left unchanged from those provided in the original Real-ESRGAN repository [17], as they have demonstrated robust performance across multiple studies. This configuration results in a model with approximately 26.2 M parameters, capable of processing a 512 × 512 input tile in ~48 ms on an NVIDIA (NVIDIA Corporation, Santa Clara, CA, USA) RTX 3090 GPU (fp16 inference).
The training employs a two-stage approach. First, we train a PSNR-oriented model (Real-ESRNet) using L1 loss. This pre-trained model then serves as initialization for the full GAN training. The complete loss function for DermaSR-GAN combines multiple components:
where
- is the pixel-wise L1 loss ensuring content consistency,
- uses pre-trained VGG19 features,
- is the adversarial loss for generation of high-frequency details.
For perceptual loss, we extract features from conv1 to conv5 layers of the discriminator, with layer weights wi = {0.1, 0.1, 1, 1, 1}, respectively [22]. The total loss weights are set as = 1.0, = 1.0, and = 0.1, following the optimal configuration identified in [17].
Training employed the Adam optimizer [25] with β1 = 0.9, β2 = 0.999, and an initial learning rate of 2 × 10−4 for the PSNR-oriented model and 1 × 10−4 for GAN training. A learning rate scheduler reduces the rate by a factor of 0.5 every 10 epochs. Training used a batch size of 16 images and employed data augmentation techniques, including random rotations, horizontal flips, and brightness adjustments, to improve model robustness. The model was trained for 100 epochs on an NVIDIA RTX 3090 GPU, requiring approximately 48 h of training time. To ensure training stability, no gradient penalty or label smoothing was applied, but checkpoints were saved regularly to retain the best-performing models based on validation loss. The curated dataset of over 18,000 paired high- and low-resolution dermoscopic images, substantially larger than those used in previous super-resolution works in dermatology, which typically rely on fewer than 2000 samples [35]. This extensive dataset from the ISIC archive includes a wide variety of skin lesions, anatomical locations, illumination conditions, and imaging artifacts. This preset ensures that DermaSR-GAN can generalize effectively across real-world clinical scenarios and lesion types, enhancing robustness and diagnostic applicability.
3.4. Evaluation Criteria
The quality of the DermaSR-GAN model was assessed using a combination of full-reference and no-reference metrics. Full-Reference metrics include the feature similarity (FSIM) [36], the structural similarity (SSIM) [35], and the peak signal-to-noise ratio (PSNR) [37]. These metrics compare the super-resolution (SR) generated image to a reference image, such as the high-resolution counterpart or the original image from the test sets. They evaluate the preservation of original image features and the fidelity of the SR image.
To independently assess image quality, we used the recent no-reference multi-dimension attention network for no-reference image quality assessment (MANIQA) metric [38], which employs a multi-dimension attention network to predict quality scores in line with human subjective perception. Higher scores indicate an enhanced perceived quality of the image.
In addition to comparing the DermaSR-GAN results against two state-of-the-art heuristic upsampling algorithms, bicubic and bilinear [39] interpolation, we further benchmarked the MANIQA values with images generated by five different generative models, to provide a more extensive comparative analysis. These methods include the following:
- Pix2Pix: A Pixel-To-Pixel HD [40] architecture trained using the same high-resolution images, but with low-resolution images obtained solely by downsampling the high-resolution ones.
- Pix2PixDP: Another Pixel-To-Pixel-HD network trained using the same dataset utilized for the DermaSR-GAN, where low-resolution images were obtained through the customized degradation pipeline (DP), ensuring a fair comparison under identical conditions.
- VDSR: A single-image SR method [41] based on a very deep convolutional network (20 layers) inspired by VGG-Net.
- ResShift: A diffusion-based SR model [42] that overcomes the slow inference of traditional diffusion methods (which require hundreds of steps) by reducing sampling to just 15 steps.
- An ESR-GAN [23] pretained model tailored for real-world applications, providing a benchmark against a renowned model in super-resolution tasks beyond the specific domain of dermatological imagery.
Finally, we integrated and evaluated the impact of the images generated by DermaSR-GAN and the other generative models in a deep learning-based classification framework.
4. Results
4.1. Quantitative Evaluation and Benchmark
Significant improvements were observed in the generated SR images for all three test datasets. Table 2 presents the full-reference metrics (FSIM, SSIM, PSNR) and no-reference MANIQA scores. These metrics demonstrate the model’s superior performance in enhancing the perceptual quality and maintaining strong structural fidelity.

Table 2.
Full-reference metrics and MANIQA scores for DermaSR-GAN across different datasets. MANIQA: multi-dimension attention network for no-reference image quality assessment (OR and SR are referred to original images and DermaSR-GAN processed images, respectively). The asterisk (*) indicates a statistically significant difference from the MANIQA values of the original images, as determined by a paired t-test (p < 0.001).
Analysis of the ISIC Test dataset revealed an FSIM score of 0.952 ± 0.019, an SSIM score of 0.940 ± 0.002, and a PSNR value of 33.828 ± 2.340, all indicative of strong structural and perceptual similarities, as well as high fidelity between the SR and the original HR images. The FSIM values above 0.95 indicate excellent preservation of structural features, while SSIM values above 0.94 demonstrate strong perceptual similarity to reference images, confirming that diagnostic features are maintained during enhancement. These metrics, coupled with a rise in the MANIQA score from 0.504 ± 0.053 to 0.591 ± 0.048, point towards a substantial 17.3% enhancement in perceptual quality. Importantly, the small standard deviation values illustrate the model’s performance consistency across diverse images.
The Novara Dermoscopic dataset also showed significant improvements in image quality, as demonstrated by an FSIM score of 0.987 ± 0.004, an SSIM score of 0.919 ± 0.001, and a PSNR value of 32.360 ± 1.243. The MANIQA score for SR images increased from 0.463 ± 0.049 to 0.570 ± 0.043. Similarly, for the PH2 dataset, the FSIM score was 0.920 ± 0.016, the SSIM score of 0.868 ± 0.018, and a PSNR value of 30.380 ± 1.654, further reinforcing the efficacy of DermaSR-GAN in significantly enhancing dermoscopic image quality. The MANIQA score for SR images increased from 0.543 ± 0.041 to 0.614 ± 0.047, indicating improved perceptual quality. Paired t-test comparisons between the MANIQA scores of the original and SR images yielded p-values less than 0.001, indicating that the improvements are statistically significant.
To visualize the distribution of MANIQA scores for the original and enhanced images across the different datasets, we employed kernel density estimation (KDE). This is a non-parametric technique that estimates the probability density function of a random variable based on a set of observed data. Figure 4 presents the KDE curves of the MANIQA scores for the original and enhanced images across the different datasets. The figure also provides a visual representation of the density of scores, with higher peaks indicating a higher concentration of scores in a particular range. In the ISIC Test dataset, we observe that the peak corresponding to the DermaSR-enhanced images not only surpasses the original in height but also exhibits a rightward shift, signifying an improvement in perceptual quality with a denser concentration of higher MANIQA scores.
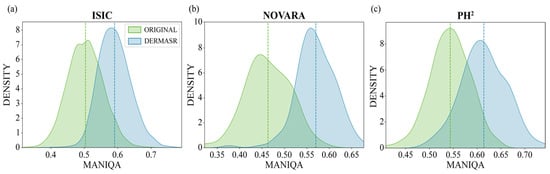
Figure 4.
Kernel density estimation (KDE) of MANIQA scores for original and enhanced images across different datasets. The KDE plots illustrate the distribution of perceptual quality scores (MANIQA) for both original low-quality images and their super-resolved counterparts generated by DermaSR-GAN. A clear shift towards higher MANIQA scores is observed for the enhanced images, indicating consistent improvements in perceptual quality across the ISIC (a), Novara (b), and PH2 (c) datasets.
The results on the Novara Dermoscopic dataset show a more pronounced difference, where the peak mean opinion score for the DermaSR-enhanced images is significantly higher and shifted further to the right compared to the original images. This indicates that DermaSR-GAN offers a considerable enhancement in the perceived quality of images in this dataset. The notable separation between the peaks suggests a marked improvement achieved by DermaSR-GAN in enhancing the perceptual quality of images from the Novara dataset.
In contrast, for the PH2 dataset, the peak mean opinion score for the original images is higher, while the peak for the DermaSR-enhanced images is right-shifted. This indicates that while the highest concentration of scores is larger for the original PH2 images, the DermaSR-enhanced PH2 images tend to receive consistently higher scores across the range in the MANIQA assessment. This reflects an enhanced quality that is more diffuse across the score distribution for the PH2 dataset.
4.2. Comparison with Heuristic Methods
To further demonstrate the effectiveness of our model, we conducted a performance comparison with two heuristic upsampling algorithms: bilinear and bicubic interpolation. Using these traditional methods, we doubled the resolution of the images and calculated their corresponding MANIQA scores, as shown in Figure 5. The MANIQA scores for the upsampling methods are reported in the third and fourth columns of Table 3.
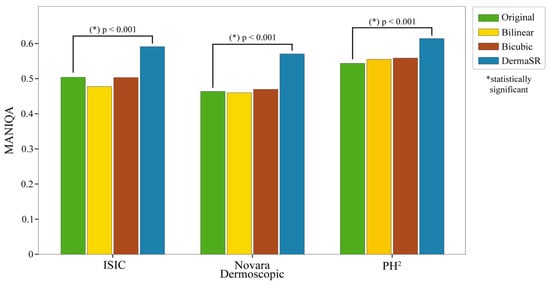
Figure 5.
Comparative MANIQA scores of original images vs. bilinear and bicubic interpolation methods vs. DermaSR-GAN across different test subsets. Paired t-tests confirm statistically significant enhancements (p-values < 0.001) in MANIQA scores for DermaSR-GAN.

Table 3.
MANIQA scores for different upscaling methods across test datasets. Best results are highlightedin bold. The asterisk (*) indicates a statistically significant difference from the MANIQA values of the original images, as determined by a paired t-test (p < 0.001).
The poor performance of traditional interpolation methods stems from their inability to reconstruct high-frequency details, resulting in blurry outputs that fail to enhance perceptual quality. Bicubic and bilinear interpolation merely increase pixel count through mathematical interpolation without adding visual information or recovering lost details. In contrast, DermaSR-GAN consistently outperformed both methods, exhibiting statistically significantly higher MANIQA scores in all cases. This highlights the capability of our specialized DermaSR-GAN not only to upscale images to a higher pixel count but also to enhance their overall quality, particularly in terms of image sharpness and detail recovery. For a comprehensive quantitative analysis comparing DermaSR-GAN with the upsampling algorithms, please refer to Figure 5.
Figure 6 presents a side-by-side comparison of an image processed by bilinear interpolation, bicubic interpolation, and DermaSR-GAN. While the interpolation methods increase the resolution in terms of pixel count, they do not enhance image details and quality. In contrast, DermaSR-GAN noticeably sharpens image features, as evidenced by the higher MANIQA score. This example highlights DermaSR-GAN’s capabilities beyond simply increasing resolution but also improving perceptual quality.
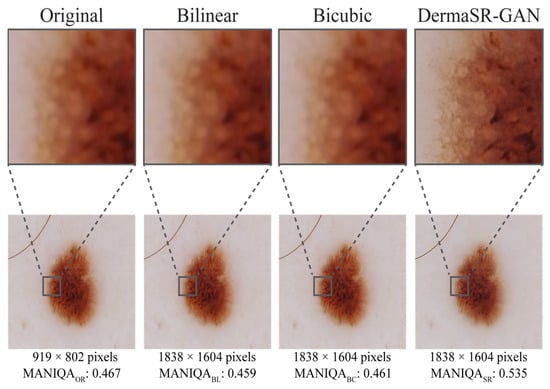
Figure 6.
Comparison of original and upsampled versions of the same image with bilinear interpolation, bicubic interpolation, and DermaSR-GAN in the test set. This figure visually highlights the differences between the methods, with DermaSR-GAN showing superior capability in recovering fine textures, lesion boundaries, and color patterns compared to traditional interpolation techniques.
4.3. Comparison with Deep Learning Methods
Comparison with deep learning-based super-resolution methods reveals that DermaSR-GAN achieves significantly higher image quality enhancement compared to alternative models (Table 4).

Table 4.
MANIQA scores (higher is better) for different deep-learning-based super-resolution methods across test datasets. Best results are highlightedin bold. The asterisk (*) indicates a statistically significant difference from the MANIQA values of the original images, as determined by a paired t-test (p < 0.001).
In the ISIC Test dataset, Pix2PixDP achieved the lowest mean MANIQA score (0.394), while the other methods averaged around 0.454. VDSR performed comparatively well (0.501), approaching the original image score (0.504). In the Novara Dermoscopic dataset, VDSR obtained the highest score among the alternative models (0.494), surpassing Pix2PixDP (0.409), ESRGAN (0.380), and ResShift (0.376). Similarly, in the PH2 dataset, VDSR again ranked first among the alternatives (0.533), followed by ResShift (0.492) and Pix2Pix (0.451), whereas Pix2PixDP recorded the lowest score (0.396).
DermaSR-GAN significantly outperformed all comparison methods (ISIC: 0.591 ± 0.048; Novara: 0.570 ± 0.043; PH2: 0.614 ± 0.047; all p < 0.001), confirming its consistent advantage over both conventional (VDSR, Pix2Pix) and diffusion-based (ResShift) approaches in dermatological applications.
Visual inspection highlights clear differences in the level of detail achieved by the various models. Pix2Pix produces images very similar to the originals, with minimal enhancement in diagnostic details (Figure 7). VDSR tends to over-enhance contrast, occasionally generating non-existent patterns, while ResShift produces generative artifacts, especially at higher magnifications.
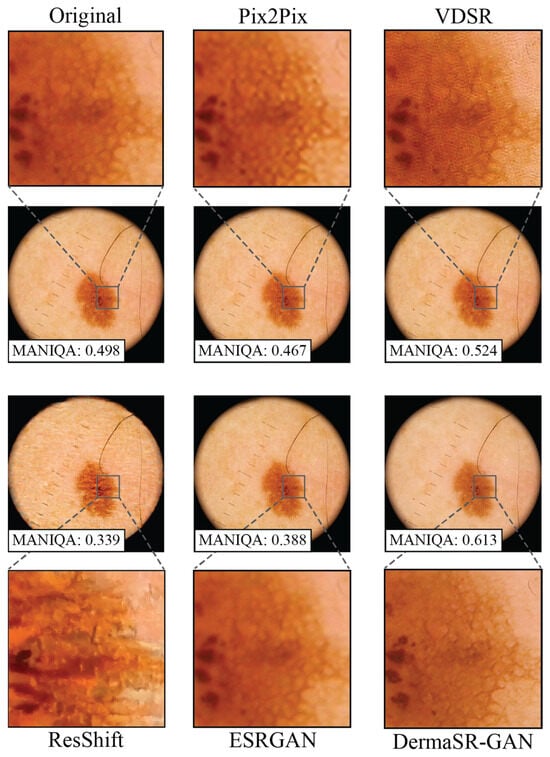
Figure 7.
Comparison of original and upsampled versions of the same image with Pix2Pix, VDSR, ResShift, ESRGAN, and DermaSR-GAN in the test set. DermaSR-GAN is able to recover lesion texture from blurry images and highlight lesion boundaries.
In contrast, DermaSR-GAN not only increases the level of detail but also enhances key diagnostic features essential for dermatological assessment. This qualitative advantage is consistent with the significantly higher MANIQA scores reported in Table 4.
4.4. Qualitative Evaluation and Impact on Downstream Tasks
Qualitative assessment visually demonstrates the effectiveness of DermaSR-GAN in enhancing image resolution and quality. Side-by-side comparisons in Figure 8 highlight the increased level of detail achieved, without distorting original dermatoscopic patterns. When zoomed in, the enhanced complexity is evident. Beneath each panel, intensity profiles generated from bisecting horizontal lines further substantiate the model’s precision. These profiles closely align with trends of the original images, with peaks indicating enhanced contrast and detail from DermaSR-GAN. This confirms the model’s ability to intensify image quality while preserving dermatological pattern integrity.
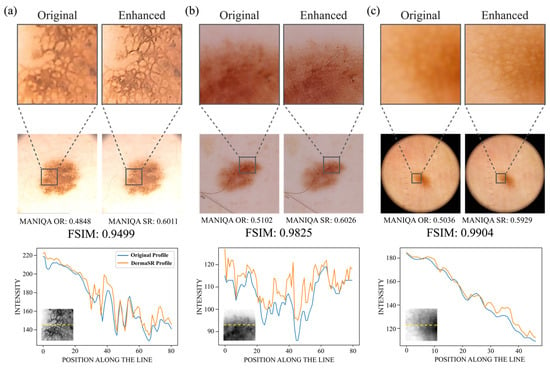
Figure 8.
Visual comparison of super-resolution images generated by DermaSR-GAN for each test set. (a) An example image from the PH2 demonstrating light yellow skin tone, (b) an example image from the ISIC test set showcasing a darker rosy skin tone, and (c) an example image from the Novara test set highlighting a dark orange skin tone. Beneath each image, grayscale intensity profiles calculated across a zoomed region of interest (ROI) are shown. These profiles illustrate the precision of DermaSR-GAN in enhancing image details while preserving original dermatoscopic patterns, such as lesion boundaries and internal structures.
Importantly, FSIM values verify that lesion structures are preserved after DermaSR-GAN enhancement, which is crucial to avoid introducing distortions or anomalies that could mislead clinical evaluation. Moreover, effectiveness across diverse skin tones is demonstrated through the comparison. Panel (a) presents very light yellow skin, panel (b) shows darker, rosier skin, and panel (c) displays darker orange skin. This demonstrates versatility and potential applicability across clinical settings by ensuring accurate enhancements regardless of skin tone variation.
Finally, we extended our investigation to include the integration of super-resolution images into a deep learning-based classification framework. Specifically, we trained seven distinct vision transformer (ViT) models: one using the original images and six using images enhanced by the compared methods listed in Table 4. The ViT architecture divides the image into patches, applies multi-headed attentions, incorporates a learnable class token for final prediction, and includes a learnable position embedding to capture relative positional information between patches. We employed a transfer learning strategy to train the network, utilizing categorical cross-entropy as the loss function and a learning rate of 10−4.
The experimental outcomes revealed notable performance differences. The DermaSR-GAN-enhanced images facilitated the most significant increase in classification accuracy, marking an 8.1% improvement compared to the original images when considering the average accuracy (i.e., 0.677 vs. 0.758). When considering individual classes, all classes noted an increase in accuracy compared with the original images, with the benign keratosis-like lesion (BKL) class seeing the most marked 14.6% increase (i.e., 0.628 vs. 0.774). This substantial improvement in BKL classification is particularly significant, as keratosis-like lesions often present subtle textural features and irregular patterns that benefit considerably from enhanced resolution and detail recovery. The enhanced visibility of fine-grained texture variations and border characteristics likely contributes to more accurate automated classification of these challenging lesions. For an exhaustive comparison of the classification accuracies facilitated by each super-resolution method, please refer to Table 5.

Table 5.
Comparison of class-specific accuracy in the test set (932 images) of a ViT for a five-class classification task trained on original and Pix2Pix, Pix2PixDP, VDSR, ResShift, ESRGAN, and DermaSR-GAN enhanced images. Best results are highlightedin bold.
5. Discussion
High-resolution dermatological imaging is critical for accurate diagnosis and treatment planning in telemedicine dermoscopy applications. Clear and detailed images enable dermatologists to accurately visualize skin conditions, lesions, and other dermatological features remotely, facilitating more precise diagnoses and treatment recommendations. High-resolution images also improve the ability to detect subtle changes in skin texture, color, and morphology, enhancing the overall quality of telemedicine consultations and ensuring better patient care outcomes.
The relevance of SR techniques in healthcare has been increasingly recognized in recent years [43,44]. Studies such as [24,25,44] have demonstrated the effectiveness of SR methods for improving dermoscopic image quality, reporting substantial gains in image clarity and diagnostic metrics. Similarly, Veeramani et al. [26] and Ding et al. [45] showed that the synthesis of high-resolution dermoscopy images improves the visibility of fine-grained skin lesion features, leading to better classification accuracy in automated systems. By providing clearer and more detailed images, super-resolution not only aids dermatologists but also enhances AI-driven diagnostic tools, underscoring its potential to revolutionize teledermatology workflows.
In this work, we introduce DermaSR-GAN, a super-resolution GAN specifically adapted to manage the characteristics and patterns of dermoscopic images. The key innovation of our approach lies in the combination of two fundamental components: a realistic degradation pipeline that accurately simulates artifacts commonly encountered in teledermatology applications, and a state-of-the-art super-resolution architecture optimized for dermatological image enhancement [17]. This synergistic approach addresses the gap between controlled laboratory conditions and real-world clinical scenarios where image quality is often compromised by acquisition conditions, transmission limitations, and compression artifacts.
Our custom degradation pipeline represents a significant advancement over simplified downsampling approaches used in previous works. By incorporating blur, noise, compression, and downsampling operations that closely resemble artifacts occurring in teledermatology practice, we enable DermaSR-GAN to learn robust mappings from degraded to high-quality images. This realistic modeling is crucial for teledermatology applications where patient-captured images often suffer from suboptimal lighting conditions, camera shake, focus issues, and transmission-related compression. The effectiveness of this approach is demonstrated by our superior performance compared to models trained with basic downsampling degradation (P2PDS vs. P2PDP). This highlights the importance of realistic training data synthesis.
The quantitative and qualitative results demonstrate DermaSR-GAN’s capabilities in enhancing resolution while preserving critical diagnostic details across diverse dermatological images. Our comprehensive evaluation across three independent datasets provides robust evidence of real-world applicability. The statistically significant improvements of 17.3%, 23.1%, and 13.0% in MANIQA scores for ISIC, Novara, and PH2 datasets, respectively, demonstrate consistent performance across different imaging conditions and patient populations. The variation in improvement levels across datasets suggests that DermaSR-GAN may be particularly effective on images with specific acquisition characteristics, highlighting the importance of diverse evaluation protocols in medical imaging applications.
The superior performance of DermaSR-GAN compared to existing approaches validates our core hypothesis that realistic degradation modeling is crucial for effective dermatological super-resolution. Our results demonstrate that the combination of domain-specific degradation synthesis and architectural optimization yields substantial improvements over both general-purpose methods and previous dermatological approaches, with consistent enhancements across diverse datasets and imaging conditions.
The significant improvement in downstream classification performance, particularly the 14.6% increase for keratosis-like lesions, demonstrates the clinical utility of our approach beyond simple image enhancement. By enhancing low-quality images to match in-clinic standards, DermaSR-GAN could improve both automated diagnostic systems and physician confidence in teledermatology contexts.
Despite the promising results, our method is not free from limitations. First, our evaluation was restricted to images with initial resolutions ≥512 × 512 pixels, and performance on severely degraded images (<256 × 256 pixels) remains unexplored. Second, while our degradation pipeline models common artifacts encountered in teledermatology, it may not capture all real-world scenarios such as severe motion blur, extreme lighting variations, or device-specific distortions.
Future research directions should focus on several key areas. Given the promising results of diffusion-based models in image generation and restoration tasks, their adaptation to dermatological imaging could provide further improvements. Diffusion models’ ability to generate high-quality, diverse outputs while maintaining fine-grained control over the generation process makes them particularly attractive for medical imaging applications where preservation of diagnostic information is paramount. Additionally, exploring transformer-based architectures and vision-language models could enable more sophisticated understanding of dermatological features and context-aware enhancement.
6. Conclusions
This work presents DermaSR-GAN, a specialized super-resolution framework for dermatological imaging that addresses critical quality limitations in teledermatology applications. Our approach demonstrates significant improvements in both quantitative metrics and clinical utility, with substantial gains in downstream classification performance. The integration of dermatology-specific design choices and comprehensive evaluation across multiple datasets validates the clinical relevance of our method. DermaSR-GAN represents a practical solution for enhancing low-quality dermatological images while preserving diagnostic information, potentially improving screening accuracy and expanding access to quality dermatological care through telemedicine platforms.
Author Contributions
Conceptualization, M.S.; methodology, F.B.; software, F.B.; validation, F.B. and K.M.M.; formal analysis, F.B., E.Z., P.S. and M.S.; investigation, F.B. and K.M.M.; data curation, F.B., E.Z. and P.S.; writing—original draft preparation, F.B., and M.S.; writing—review and editing, E.Z., P.S. and K.M.M.; visualization, F.B.; supervision, M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The training codes, the training/validation splits, and all experimental results on the three test sets are publicly available on Mendeley Data: https://doi.org/10.17632/27xk9v8z73.1.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AI | Artificial Intelligence |
| BKL | Benign Keratosis-like Lesions |
| CNN | Convolutional Neural Network |
| DP | Degradation Pipeline |
| DS | Downsampling |
| EDSR | Enhanced Deep Super-Resolution |
| ESRGAN | Enhanced Super-Resolution Generative Adversarial Network |
| FSIM | Feature Similarity Index |
| GAN | Generative Adversarial Network |
| HR | High-Resolution |
| ISIC | International Skin Imaging Collaboration |
| LR | Low-Resolution |
| MANIQA | Multi-Dimension Attention Network for No-Reference Image Quality |
| P2P-HD | Pixel-To-Pixel High Definition |
| PSNR | Peak Signal-to-Noise Ratio |
| SR | Super-Resolution |
| SRGAN | Super-Resolution Generative Adversarial Network |
| SSIM | Structural Similarity Index |
| VDSR | Very Deep Super-Resolution |
| ViT | Vision Transformer |
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA A Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Helfand, M.; Mahon, S.M.; Eden, K.B.; Frame, P.S.; Orleans, C.T. Screening for Skin Cancer. Am. J. Prev. Med. 2001, 20, 47–58. [Google Scholar] [CrossRef]
- Veronese, F.; Tarantino, V.; Zavattaro, E.; Biacchi, F.; Airoldi, C.; Salvi, M.; Seoni, S.; Branciforti, F.; Meiburger, K.M.; Savoia, P. Teledermoscopy in the Diagnosis of Melanocytic and Non-Melanocytic Skin Lesions: NurugoTM Derma Smartphone Microscope as a Possible New Tool in Daily Clinical Practice. Diagnostics 2022, 12, 1371. [Google Scholar] [CrossRef]
- Pala, P.; Bergler-Czop, B.S.; Gwiżdż, J. Teledermatology: Idea, Benefits and Risks of Modern Age—A Systematic Review Based on Melanoma. Pdia 2020, 37, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Coates, S.J.; Kvedar, J.; Granstein, R.D. Teledermatology: From Historical Perspective to Emerging Techniques of the Modern Era. J. Am. Acad. Dermatol. 2015, 72, 563–574. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, H.; Zeng, T.; Yang, G.; Shi, Z.; Gao, Z. Bridging Multi-Level Gaps: Bidirectional Reciprocal Cycle Framework for Text-Guided Label-Efficient Segmentation in Echocardiography. Med. Image Anal. 2025, 102, 103536. [Google Scholar] [CrossRef]
- Wang, W.; Liang, D.; Chen, Q.; Iwamoto, Y.; Han, X.-H.; Zhang, Q.; Hu, H.; Lin, L.; Chen, Y.-W. Medical Image Classification Using Deep Learning. In Deep Learning in Healthcare; Chen, Y.-W., Jain, L.C., Eds.; Intelligent Systems Reference Library; Springer International Publishing: Cham, Switzerland, 2020; Volume 171, pp. 33–51. ISBN 978-3-030-32605-0. [Google Scholar]
- Fernando, T.; Gammulle, H.; Denman, S.; Sridharan, S.; Fookes, C. Deep Learning for Medical Anomaly Detection—A Survey. ACM Comput. Surv. 2022, 54, 1–37. [Google Scholar] [CrossRef]
- Konz, N.; Chen, Y.; Dong, H.; Mazurowski, M.A. Anatomically-Controllable Medical Image Generation with Segmentation-Guided Diffusion Models. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2024; Linguraru, M.G., Dou, Q., Feragen, A., Giannarou, S., Glocker, B., Lekadir, K., Schnabel, J.A., Eds.; Lecture Notes in Computer Science; Springer Nature: Cham, Switzerland, 2024; Volume 15007, pp. 88–98. ISBN 978-3-031-72103-8. [Google Scholar]
- Datta, S.K.; Shaikh, M.A.; Srihari, S.N.; Gao, M. Soft Attention Improves Skin Cancer Classification Performance. In Interpretability of Machine Intelligence in Medical Image Computing, and Topological Data Analysis and Its Applications for Medical Data; Reyes, M., Henriques Abreu, P., Cardoso, J., Hajij, M., Zamzmi, G., Rahul, P., Thakur, L., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2021; Volume 12929, pp. 13–23. ISBN 978-3-030-87443-8. [Google Scholar]
- Goceri, E. Automated Skin Cancer Detection: Where We Are and The Way to The Future. In Proceedings of the 2021 44th International Conference on Telecommunications and Signal Processing (TSP), Brno, Czech Republic, 26–28 July 2021; pp. 48–51. [Google Scholar]
- Branciforti, F.; Meiburger, K.M.; Zavattaro, E.; Veronese, F.; Tarantino, V.; Mazzoletti, V.; Cristo, N.D.; Savoia, P.; Salvi, M. Impact of Artificial Intelligence-based Color Constancy on Dermoscopical Assessment of Skin Lesions: A Comparative Study. Ski. Res. Technol. 2023, 29, e13508. [Google Scholar] [CrossRef]
- Li, Z.; Koban, K.C.; Schenck, T.L.; Giunta, R.E.; Li, Q.; Sun, Y. Artificial Intelligence in Dermatology Image Analysis: Current Developments and Future Trends. J. Clin. Med. 2022, 11, 6826. [Google Scholar] [CrossRef] [PubMed]
- Salvi, M.; Branciforti, F.; Molinari, F.; Meiburger, K.M. Generative Models for Color Normalization in Digital Pathology and Dermatology: Advancing the Learning Paradigm. Expert Syst. Appl. 2024, 245, 123105. [Google Scholar] [CrossRef]
- Zhang, M.; Ling, Q. Supervised Pixel-Wise GAN for Face Super-Resolution. IEEE Trans. Multimed. 2021, 23, 1938–1950. [Google Scholar] [CrossRef]
- Liu, L.; Jiang, Q.; Jin, X.; Feng, J.; Wang, R.; Liao, H.; Lee, S.-J.; Yao, S. CASR-Net: A Color-Aware Super-Resolution Network for Panchromatic Image. Eng. Appl. Artif. Intell. 2022, 114, 105084. [Google Scholar] [CrossRef]
- Wang, X.; Xie, L.; Dong, C.; Shan, Y. Real-ESRGAN: Training Real-World Blind Super-Resolution with Pure Synthetic Data. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision Workshops (ICCVW), Montreal, BC, Canada, 11–17 October 2021; pp. 1905–1914. [Google Scholar]
- Abd El-Fattah, I.; Ali, A.M.; El-Shafai, W.; Taha, T.E.; Abd El-Samie, F.E. Deep-Learning-Based Super-Resolution and Classification Framework for Skin Disease Detection Applications. Opt. Quant. Electron. 2023, 55, 427. [Google Scholar] [CrossRef]
- Gohil, Z.M.; Desai, M.B. Revolutionizing Dermatology: A Comprehensive Survey of AI-Enhanced Early Skin Cancer Diagnosis. Arch. Comput. Methods Eng. 2024, 31, 4521–4531. [Google Scholar] [CrossRef]
- Freeman, W.T.; Jones, T.R.; Pasztor, E.C. Example-Based Super-Resolution. IEEE Comput. Grap. Appl. 2002, 22, 56–65. [Google Scholar] [CrossRef]
- Dong, C.; Loy, C.C.; Tang, X. Accelerating the Super-Resolution Convolutional Neural Network. In Computer Vision—ECCV 2016; Leibe, B., Matas, J., Sebe, N., Welling, M., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2016; Volume 9906, pp. 391–407. ISBN 978-3-319-46474-9. [Google Scholar]
- Ledig, C.; Theis, L.; Huszár, F.; Caballero, J.; Cunningham, A.; Acosta, A.; Aitken, A.; Tejani, A.; Totz, J.; Wang, Z.; et al. Photo-Realistic Single Image Super-Resolution Using a Generative Adversarial Network. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 4681–4690. [Google Scholar]
- Wang, X.; Yu, K.; Wu, S.; Gu, J.; Liu, Y.; Dong, C.; Qiao, Y.; Change Loy, C. Esrgan: Enhanced Super-Resolution Generative Adversarial Networks. In Computer Vision—ECCV 2018 Workshops; Springer: Berlin/Heidelberg, Germany, 2018; pp. 63–79. [Google Scholar]
- Alwakid, G.; Gouda, W.; Humayun, M.; Sama, N.U. Melanoma Detection Using Deep Learning-Based Classifications. Healthcare 2022, 10, 2481. [Google Scholar] [CrossRef]
- Mukadam, S.B.; Patil, H.Y. Skin Cancer Classification Framework Using Enhanced Super Resolution Generative Adversarial Network and Custom Convolutional Neural Network. Appl. Sci. 2023, 13, 1210. [Google Scholar] [CrossRef]
- Veeramani, N.; Jayaraman, P. A Promising AI Based Super Resolution Image Reconstruction Technique for Early Diagnosis of Skin Cancer. Sci. Rep. 2025, 15, 5084. [Google Scholar] [CrossRef]
- Meta AI. Executorch. Available online: https://docs.pytorch.org/executorch-overview (accessed on 1 August 2025).
- Rotemberg, V.; Kurtansky, N.; Betz-Stablein, B.; Caffery, L.; Chousakos, E.; Codella, N.; Combalia, M.; Dusza, S.; Guitera, P.; Gutman, D.; et al. A Patient-Centric Dataset of Images and Metadata for Identifying Melanomas Using Clinical Context. Sci. Data 2021, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, B.; Kendrick, C.; Brodzicki, A.; Jaworek-Korjakowska, J.; Yap, M.H. Analysis of the ISIC Image Datasets: Usage, Benchmarks and Recommendations. Med. Image Anal. 2022, 75, 102305. [Google Scholar] [CrossRef]
- Mendonca, T.; Ferreira, P.M.; Marques, J.S.; Marcal, A.R.S.; Rozeira, J. PH2—A Dermoscopic Image Database for Research and Benchmarking. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 5437–5440. [Google Scholar]
- Al-Ani, M.S.; Awad, F.H. The JPEG Image Compression Algorithm. Int. J. Adv. Eng. Technol 2013, 6, 1055–1062. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar] [CrossRef]
- Schonfeld, E.; Schiele, B.; Khoreva, A. A U-Net Based Discriminator for Generative Adversarial Networks. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 8207–8216. [Google Scholar]
- Miyato, T.; Kataoka, T.; Koyama, M.; Yoshida, Y. Spectral Normalization for Generative Adversarial Networks. arXiv 2018, arXiv:1802.05957. [Google Scholar] [CrossRef]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image Quality Assessment: From Error Visibility to Structural Similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Mou, X.; Zhang, D. FSIM: A Feature Similarity Index for Image Quality Assessment. IEEE Trans. Image Process. 2011, 20, 2378–2386. [Google Scholar] [CrossRef]
- Huynh-Thu, Q.; Ghanbari, M. Scope of Validity of PSNR in Image/Video Quality Assessment. Electron. Lett. 2008, 44, 800–801. [Google Scholar] [CrossRef]
- Yang, S.; Wu, T.; Shi, S.; Lao, S.; Gong, Y.; Cao, M.; Wang, J.; Yang, Y. Maniqa: Multi-Dimension Attention Network for No-Reference Image Quality Assessment. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), New Orleans, LA, USA, 19–20 June 2022; pp. 1191–1200. [Google Scholar]
- Gonzalez, R.C. Digital Image Processing; Pearson Education India: Chennai, India, 2009; ISBN 81-317-2695-9. [Google Scholar]
- Wang, T.-C.; Liu, M.-Y.; Zhu, J.-Y.; Tao, A.; Kautz, J.; Catanzaro, B. High-Resolution Image Synthesis and Semantic Manipulation with Conditional GANs. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8798–8807. [Google Scholar]
- Kim, J.; Lee, J.K.; Lee, K.M. Accurate Image Super-Resolution Using Very Deep Convolutional Networks. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 1646–1654. [Google Scholar]
- Yue, Z.; Wang, J.; Loy, C.C. Resshift: Efficient Diffusion Model for Image Super-Resolution by Residual Shifting. In Advances in Neural Information Processing Systems; The MIT Press: Cambridge, MA, USA, 2023; Volume 36, pp. 13294–13307. [Google Scholar]
- Shao, D.; Qin, L.; Xiang, Y.; Ma, L.; Xu, H. Medical Image Blind Super-resolution Based on Improved Degradation Process. IET Image Process. 2023, 17, 1615–1625. [Google Scholar] [CrossRef]
- Lee, D.Y.; Kim, J.Y.; Cho, S.Y. Improving Medical Image Quality Using a Super-Resolution Technique with Attention Mechanism. Appl. Sci. 2025, 15, 867. [Google Scholar] [CrossRef]
- Ding, S.; Zheng, J.; Liu, Z.; Zheng, Y.; Chen, Y.; Xu, X.; Lu, J.; Xie, J. High-Resolution Dermoscopy Image Synthesis with Conditional Generative Adversarial Networks. Biomed. Signal Process. Control 2021, 64, 102224. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).