2AM: Weakly Supervised Tumor Segmentation in Pathology via CAM and SAM Synergy
Abstract
1. Introduction
- To the best of our knowledge, this is the first work to combine Class Activation Mapping (CAM) and the Segment Anything Model (SAM) for pathological image analysis, and we propose a simple but effective framework, termed as 2AM, for tumor segmentation on pathological images in a weakly supervised manner.
- We present an adaptive point selection (APS) module for providing more reliable initial prompts for the subsequent SAM by designing three priors of basic appearance, space distribution, and feature difference, which achieves effective integration of CAM and SAM.
- Experimental results on two independent datasets show that our proposed method boosts tumor segmentation accuracy by nearly 25% compared with the baseline method, and achieves more than 15% improvement compared with previous state-of-the-art segmentation methods in a weakly supervised manner.
2. Related Work
2.1. Pathological Image Segmentation
2.2. Class Activation Mapping
2.3. Segment Anything Model
3. Methods
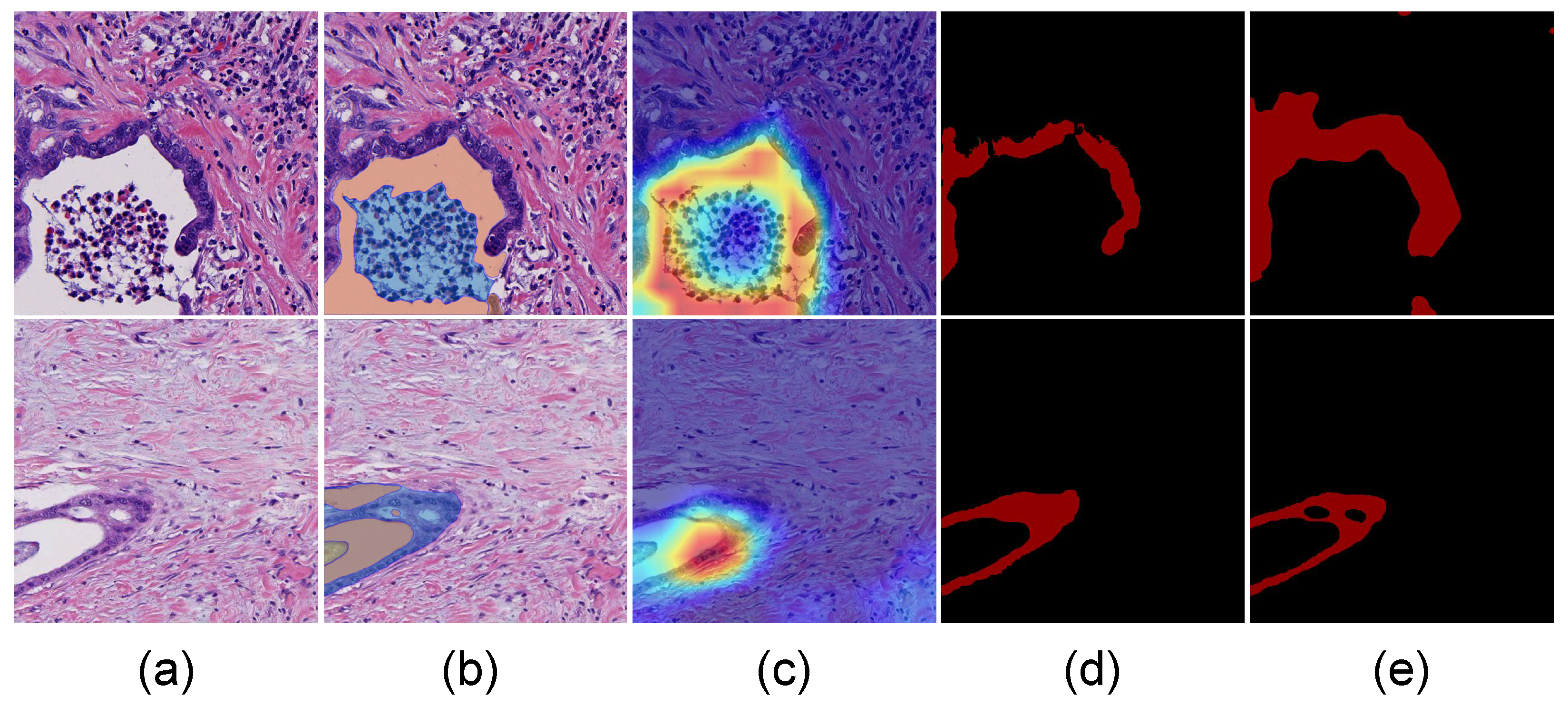
3.1. Pipeline
3.2. CAM Module
3.3. Adaptive Point Selection (APS) Module
| Algorithm 1 Adaptive Point Selection (APS) for Tumor Segmentation |
| 1: Input: CAM heatmap, Image
2: Output: Positive points , Negative point N 3: Compute color prior: Calculate mean tumor color from training images 4: Select positive points: Extract high-response region A (CAM score ) Apply K-means++ clustering on A (, 10 iterations) to select Ensure minimum distance d between points using distance prior 5: Select negative point: Randomly choose N from region with CAM score |
3.4. SAM Module
4. Experiments
4.1. Datasets
4.2. Metrics
4.3. Implementation Details
4.4. Computational Efficiency Analysis
4.5. Ablation Study
4.6. Comparison with Other Weakly Supervised Segmentation Methods
5. Discussion
5.1. Failure Case Analysis and Limitations
5.2. Future Directions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kiemen, A.; Braxton, A.M.; Grahn, M.P.; Han, K.S.; Babu, J.M.; Reichel, R.; Amoa, F.; Hong, S.-M.; Cornish, T.C.; Thompson, E.D.; et al. In situ characterization of the 3d microanatomy of the pancreas and pancreatic cancer at single cell resolution. bioRxiv 2020. [Google Scholar] [CrossRef]
- Jiao, Y.; Li, J.; Qian, C.; Fei, S. Deep learning-based tumor microenvironment analysis in colon adenocarcinoma histopathological whole-slide images. Comput. Methods Programs Biomed. 2021, 204, 106047. [Google Scholar] [CrossRef]
- Li, Y.; Ping, W. Cancer metastasis detection with neural conditional random field. arXiv 2018, arXiv:1806.07064. [Google Scholar] [CrossRef]
- Liu, Y.; Gadepalli, K.; Norouzi, M.; Dahl, G.E.; Kohlberger, T.; Boyko, A.; Venugopalan, S.; Timofeev, A.; Nelson, P.Q.; Corrado, G.S.; et al. Detecting cancer metastases on gigapixel pathology images. arXiv 2017, arXiv:1703.02442. [Google Scholar] [CrossRef]
- Madabhushi, A.; Lee, G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med. Image Anal. 2016, 33, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Cai, Z.; Mao, L.; Nie, Z.; Qiu, Y.; Yang, X. Automated peripancreatic vessel segmentation and labeling based on iterative trunk growth and weakly supervised mechanism. Artif. Intell. Med. 2024, 150, 102825. [Google Scholar] [CrossRef]
- Randell, R.; Ruddle, R.A.; Thomas, R.; Treanor, D. Diagnosis at the microscope: A workplace study of histopathology. Cogn. Technol. Work 2012, 14, 319–335. [Google Scholar] [CrossRef]
- Wang, B.; Zou, L.; Chen, J.; Cao, Y.; Cai, Z.; Qiu, Y.; Mao, L.; Wang, Z.; Chen, J.; Gui, L.; et al. A weakly supervised segmentation network embedding cross-scale attention guidance and noise-sensitive constraint for detecting tertiary lymphoid structures of pancreatic tumors. J. Biomed. Health Inform. (J-BHI) 2024, 28, 12. [Google Scholar] [CrossRef]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2921–2929. [Google Scholar]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.-Y.; et al. Segment anything. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 1–6 October 2023; pp. 4015–4026. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Isensee, F.; Petersen, J.; Klein, A.; Zimmerer, D.; Jaeger, P.F.; Kohl, S.; Wasserthal, J.; Koehler, G.; Norajitra, T.; Wirkert, S.; et al. nnu-net: Self-adapting framework for u-net-based medical image segmentation. arXiv 2018, arXiv:1809.10486. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 5998–6008. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; Springer Nature Switzerland: Cham, Switzerland, 2022; pp. 205–218. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.-Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A convnet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 11976–11986. [Google Scholar]
- Lou, W.; Li, H.; Li, G.; Han, X.; Wan, X. Which pixel to annotate: A label-efficient nuclei segmentation framework. IEEE Trans. Med Imaging 2022, 42, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Agarwal, D.; Aggarwal, K.; Safta, W.; Balan, M.M.; Brown, K. Masked image modeling advances 3d medical image analysis. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–7 January 2023; pp. 1970–1980. [Google Scholar]
- Li, W.; Li, J.; Polson, J.; Wang, Z.; Speier, W.; Arnold, C. High resolution histopathology image generation and segmentation through adversarial training. Med. Image Anal. 2022, 75, 102251. [Google Scholar] [CrossRef]
- Chan, L.; Hosseini, M.S.; Rowsell, C.; Plataniotis, K.N.; Damaskinos, S. Histosegnet: Semantic segmentation of histological tissue type in whole slide images. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 10662–10671. [Google Scholar]
- Wang, Y.; Zhang, J.; Kan, M.; Shan, S.; Chen, X. Self-supervised equivariant attention mechanism for weakly supervised semantic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020; pp. 12275–12284. [Google Scholar]
- Xu, L.; Ouyang, W.; Bennamoun, M.; Boussaid, F.; Xu, D. Multi-class token transformer for weakly supervised semantic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 4310–4319. [Google Scholar]
- Ma, J.; He, Y.; Li, F.; Han, L.; You, C.; Wang, B. Segment anything in medical images. Nat. Commun. 2024, 15, 654. [Google Scholar] [CrossRef]
- Cui, C.; Deng, R.; Liu, Q.; Yao, T.; Bao, S.; Remedios, L.W.; Landman, B.A.; Tang, Y.; Huo, Y. All-in-sam: From weak annotation to pixel-wise nuclei segmentation with prompt-based finetuning. J. Phys. Conf. Ser. 2024, 2722, 012012. [Google Scholar] [CrossRef]
- Ke, L.; Ye, M.; Danelljan, M.; Tai, Y.-W.; Tang, C.-K.; Yu, F. Segment anything in high quality. Adv. Neural Inf. Process. Syst. 2024, 36, 29914–29934. [Google Scholar]
- Zhang, J.; Ma, K.; Kapse, S.; Saltz, J.; Vakalopoulou, M.; Prasanna, P.; Samaras, D. Sam-path: A segment anything model for semantic segmentation in digital pathology. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, BC, Canada, 8–12 October 2023; Springer Nature Switzerland: Cham, Switzerland, 2023; pp. 161–170. [Google Scholar]
- Nateghi, R.; Pourakpour, F. Perineural invasion detection in multiple organ cancer based on deep convolutional neural network. arXiv 2021, arXiv:2110.12283. [Google Scholar] [CrossRef]
- Zou, L.; Cai, Z.; Qiu, Y.; Gui, L.; Mao, L.; Yang, X. Ctg-net: An efficient cascaded framework driven by terminal guidance mechanism for dilated pancreatic duct segmentation. Phys. Med. Biol. 2023, 68, 215006. [Google Scholar] [CrossRef]
- Chen, L.-C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Chattopadhay, A.; Sarkar, A.; Howlader, P.; Balasubramanian, V.N. Grad-cam++: Generalized gradient-based visual explanations for deep convolutional networks. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 839–847. [Google Scholar]
- Wang, C.; He, S.; Wu, M.; Lam, S.-K.; Tiwari, P.; Gao, X. Looking clearer with text: A hierarchical context blending network for occluded person re-identification. IEEE Trans. Inf. Forensics Secur. 2025, 20, 4296–4307. [Google Scholar] [CrossRef]
- Wang, C.; Cao, R.; Wang, R. Learning discriminative topological structure information representation for 2d shape and social network classification via persistent homology. Knowl.-Based Syst. 2025, 311, 113125. [Google Scholar] [CrossRef]
- Hayat, M.; Aramvith, S.; Bhattacharjee, S.; Ahmad, N. Attention ghostunet++: Enhanced segmentation of adipose tissue and liver in ct images. arXiv 2025, arXiv:2504.11491. [Google Scholar]
- Xie, Y.; Wu, H.; Tong, H.; Xiao, L.; Zhou, W.; Li, L.; Wanger, T.C. fabsam: A farmland boundary delineation method based on the segment anything model. arXiv 2025, arXiv:2501.12487. [Google Scholar]






| Method | Parameters (M) | FLOPs (G) |
|---|---|---|
| U-Net | 34 | 180 |
| DeepLabV3+ | 58 | 350 |
| Swin Transformer | 60 | 250 |
| 2AM (Ours) | 648 | 260 |
| Datasets | CP | AP | DP | HD ↓ | FPSR ↓ | FNSR ↓ | Dice ↑ |
|---|---|---|---|---|---|---|---|
| NDTH | 215.29 ± 47.76 | 33.24 ± 11.48 | 12.14 ± 5.77 | 54.62 ± 11.86 | |||
| ✓ | 211.03 ± 45.58 | 17.60 ± 11.81 | 25.75 ± 10.29 | 56.65 ± 14.47 | |||
| ✓ | 221.61 ± 46.16 | 25.06 ± 12.57 | 19.68 ± 13.20 | 55.26 ± 11.04 | |||
| ✓ | 238.61 ± 31.13 | 28.38 ± 11.38 | 12.77 ± 6.78 | 58.85 ± 12.07 | |||
| ✓ | ✓ | 185.48 ± 54.87 | 6.53 ± 2.93 | 23.15 ± 12.57 | 66.32 ± 11.33 | ||
| ✓ | ✓ | 197.54 ± 51.31 | 22.54 ± 12.38 | 14.51 ± 4.81 | 62.96 ± 14.67 | ||
| ✓ | ✓ | 197.91 ± 37.83 | 25.44 ± 13.48 | 11.78 ± 5.78 | 62.78 ± 10.87 | ||
| ✓ | ✓ | ✓ | 176.92 ± 47.77 | 6.59 ± 2.96 | 17.47 ± 5.35 | 70.94 ± 9.64 | |
| JHCM | 170.10 ± 93.53 | 30.23 ± 13.50 | 11.72 ± 5.18 | 58.05 ± 15.72 | |||
| ✓ | 143.90 ± 58.09 | 27.23 ± 11.96 | 6.77 ± 2.75 | 65.99 ± 12.91 | |||
| ✓ | 180.99 ± 93.86 | 22.45 ± 13.91 | 14.70 ± 9.94 | 62.85 ± 16.66 | |||
| ✓ | 183.58 ± 95.42 | 32.66 ± 12.15 | 6.78 ± 4.91 | 63.56 ± 10.57 | |||
| ✓ | ✓ | 139.06 ± 59.96 | 19.44 ± 11.24 | 9.20 ± 4.43 | 65.36 ± 12.33 | ||
| ✓ | ✓ | 121.47 ± 60.65 | 32.19 ± 9.70 | 4.49 ± 2.45 | 66.02 ± 9.86 | ||
| ✓ | ✓ | 190.41 ± 97.00 | 35.22 ± 11.50 | 4.74 ± 2.49 | 63.03 ± 11.04 | ||
| ✓ | ✓ | ✓ | 121.65 ± 65.29 | 29.95 ± 9.66 | 3.64 ± 2.52 | 68.41 ± 9.29 |
| Datasets | Methods | HD ↓ | FPSR ↓ | FNSR ↓ | Dice ↑ |
|---|---|---|---|---|---|
| NDTH | CAM | 257.61 ± 62.01 * | 12.99 ± 6.38 * | 32.52 ± 11.06 * | 46.50 ± 8.58 * |
| Grad-CAM | 219.80 ± 36.44 * | 9.99 ± 2.98 + | 39.50 ± 4.09 * | 47.75 ± 6.15 * | |
| Grad-CAM++ | 226.79 ± 46.08 * | 12.91 ± 4.43 * | 39.34 ± 5.54 * | 50.51 ± 5.06 * | |
| 2AM(Ours) | 176.92 ± 47.77 | 6.59 ± 2.96 | 17.47 ± 5.35 | 70.94 ± 9.64 | |
| JHCM | CAM | 212.97 ± 84.14 * | 15.24 ± 6.51 | 17.10 ± 8.72 * | 42.66 ± 6.93 * |
| Grad-CAM | 222.41 ± 79.88 * | 16.69 ± 6.42 | 29.98 ± 12.42 * | 43.33 ± 8.37 * | |
| Grad-CAM++ | 218.27 ± 75.48 * | 16.40 ± 7.03 | 25.26 ± 12.41 * | 48.34 ± 8.75 * | |
| 2AM(Ours) | 121.65 ± 65.29 | 29.95 ± 9.66 | 3.64 ± 2.52 | 68.41 ± 9.29 |
| Datasets | Methods | HD ↓ | FPSR ↓ | FNSR ↓ | Dice ↑ |
|---|---|---|---|---|---|
| NDTH | DeepLabV3+ | 169.13 ± 59.79 | 1.04 ± 0.48 | 50.34 ± 14.67 * | 48.42 ± 14.41 * |
| Swin Transformer | 167.51 ± 47.72 | 2.41 ± 1.18 | 32.33 ± 11.85 * | 55.26 ± 10.92 * | |
| U-Net | 140.37 ± 40.58 | 1.79 ± 0.69 | 31.13 ± 9.83 * | 57.09 ± 9.65 * | |
| 2AM(Ours) | 176.92 ± 47.77 | 6.59 ± 2.96 | 17.47 ± 5.35 | 70.94 ± 9.64 | |
| JHCM | DeepLabV3+ | 198.67 ± 90.70 * | 17.42 ± 6.26 | 36.39 ± 19.15 * | 46.19 ± 14.52 * |
| Swin Transformer | 249.00 ± 73.80 * | 25.78 ± 6.72 | 24.72 ± 14.83 * | 49.50 ± 11.28 * | |
| U-Net | 258.79 ± 80.68 * | 23.71 ± 7.73 | 31.56 ± 14.11 * | 44.73 ± 11.77 * | |
| 2AM(Ours) | 121.65 ± 65.29 | 29.95 ± 9.66 | 3.64 ± 2.52 | 68.41 ± 9.29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, C.; Zou, L.; Gui, L. 2AM: Weakly Supervised Tumor Segmentation in Pathology via CAM and SAM Synergy. Electronics 2025, 14, 3109. https://doi.org/10.3390/electronics14153109
Ren C, Zou L, Gui L. 2AM: Weakly Supervised Tumor Segmentation in Pathology via CAM and SAM Synergy. Electronics. 2025; 14(15):3109. https://doi.org/10.3390/electronics14153109
Chicago/Turabian StyleRen, Chenyu, Liwen Zou, and Luying Gui. 2025. "2AM: Weakly Supervised Tumor Segmentation in Pathology via CAM and SAM Synergy" Electronics 14, no. 15: 3109. https://doi.org/10.3390/electronics14153109
APA StyleRen, C., Zou, L., & Gui, L. (2025). 2AM: Weakly Supervised Tumor Segmentation in Pathology via CAM and SAM Synergy. Electronics, 14(15), 3109. https://doi.org/10.3390/electronics14153109






