Abstract
Retinal vessel segmentation plays a critical role in the diagnosis and treatment of various ophthalmic diseases. However, due to poor image contrast, intricate vascular structures, and limited datasets, retinal vessel segmentation remains a long-term challenge. In this paper, based on an encoder–decoder framework, a novel retinal vessel segmentation model called CMP-UNet is proposed. Firstly, the Coarse and Fine Feature Aggregation module decouples and aggregates coarse and fine vessel features using two parallel branches, thus enhancing the model’s ability to extract features for vessels of various sizes. Then, the Multi-Scale Channel Adaptive Fusion module is embedded in the decoder to realize the efficient fusion of cascade features by mining the multi-scale context information from these features. Finally, to obtain more discriminative vascular features and enhance the connectivity of vascular structures, the Pyramid Feature Fusion module is proposed to effectively utilize the complementary information of multi-level features. To validate the effectiveness of the proposed model, it is evaluated on three publicly available retinal vessel segmentation datasets: CHASE_DB1, DRIVE, and STARE. The proposed model, CMP-UNet, reaches F1-scores of 82.84%, 82.55%, and 84.14% on these three datasets, with improvements of 0.76%, 0.31%, and 1.49%, respectively, compared with the baseline. The results show that the proposed model achieves higher segmentation accuracy and more robust generalization capability than state-of-the-art methods.
1. Introduction
The retina performs many critical functions as an essential part of the eye. Morphological and density changes of retinal vessels can represent an important basis for the diagnosis of ophthalmic diseases [1] caused by atherosclerosis, diabetic retinopathy [2], and glaucoma. To analyze the structural properties of retinal vessels, such as branching patterns, angles, and curvature, accurate segmentation of the retinal vessels is required. Traditional retinal vessel extraction requires manual segmentation by experienced ophthalmologists [3], which is time-consuming and laborious [4]. Therefore, automatic segmentation of retinal vessels is of great research value. However, automatic retinal vessel segmentation remains a long-term challenge [5] due to the following reasons: (1) Due to the equipment used to capture the images and the acquisition environment, the quality of fundus images is poor. In retinal images, the large amount of noise, low contrast between the vessels and the background [6], and uneven illumination hinder the accurate segmentation of vessels. (2) Retinal images contain abundant capillaries with complex structures, such as vessel crossing, branching, and centerline reflex. In addition, the width and feature intensity of blood vessels vary greatly. These specific characteristics of retinal vessels lead to undesirable segmentation of fine vessels and poor connectivity of vascular structures. (3) Data with good annotations are limited because of the difficulties of acquiring and annotating fundus images. Moreover, the pixel ratio between background and vessels in retinal images is unbalanced. Limited datasets and category imbalance result in poor generalization performance of models.
Deep learning technology is gradually being used in more and more fields. Currently, the medical field is closely related to deep learning techniques [7] and has achieved many impressive results. Rehman et al. proposed a residual spatial pyramid pooling module based on the U-Net architecture to reduce the loss of location information in different modules. In addition, an attention gate module is utilized to efficiently emphasize and restore the segmented output [8]. Inspired by game theory, Wang et al. proposed an unsupervised model based on the Swin-Unet framework [9]. And an image colorization proxy task was introduced to assist the learning of pixel-level feature representations. Zhao et al. designed the residual ghost block with switchable normalization and the bottleneck transformer to extract fine features [10]. To achieve better tumor identification performance, the feature enhancer block was designed by Rehman et al. Meanwhile, a new loss function was proposed to solve the class imbalance issue [11]. Lyu et al. proposed a novel multiple-tasking Wasserstein generative adversarial network U-shape network and utilized the attention mechanism to enhance the segmentation accuracy of the generator [12]. Wang et al. first proposed the Heart–Lung-Sound classification method SNMF-DCNN and applied U-Net for cardiopulmonary sound separation [13]. Rehman et al. proposed a novel tumor segmentation model, BU-Net [14]. Residual extended skip, wide context, and a customized loss function are used in the U-Net architecture to enhance the model performance.
With the success of U-Net in the medical field, many researchers are investigating automatic retinal vessel segmentation models based on the U-Net architecture. To enhance the feature extraction ability of the model for blood vessels, Wang et al. designed a two-channel encoder, where the context channel uses multi-scale convolution to capture more receptive field and the spatial channel uses a large kernel to retain spatial information [15]. Yang et al. introduced deformable convolution [16] to establish a feature extraction module, which enhances the modeling ability of the model for vessel deformation, and used a residual channel attention [17] module to improve the efficiency of information transfer between U-Net models [18]. Liu et al. proposed ResDO-conv, based on depth-wise over-parameterized convolution [19], as a backbone network for acquiring strong contextual features to enhance feature extraction capabilities [20]. While enhanced feature extraction capability is desirable, the improvement in fine vessel segmentation performance is limited. To further improve a model’s ability to segment fine blood vessels, one approach is to extract coarse and fine vessel features separately. For instance, Xu et al. constructed a thick and thin vessel extraction module based on the morphological differences in retinal blood vessels to separately extract thick and thin vessel features [21]. An alternative approach is to retain maximum vessel information to enable the reuse of vascular features. For instance, Yuan et al. used a dropout dense block to replace the original convolutional blocks in U-Net to preserve maximum vessel information between convolution layers through dense connection [22]. Yue et al. proposed an improved GAN [23] based on R2U-Net. The attention mechanism [24] was added to the generator to reduce information loss, and the dense connection modules were used in the discriminator to mitigate gradient vanishing [25] and achieve feature reuse [26]. In addition, many other researchers have contributed significantly to automatic retinal vessel segmentation. Li et al. proposed a novel multimodule concatenation method using a U-shaped network that combines atrous convolution [27] with multikernel pooling blocks to obtain more contextual information [28]. Deng et al. [29] proposed a segmentation model based on multi-scale attention with residual mechanism [30], D-Mnet, combined with the improved PCNN [31] model to unite the advantages of supervised and unsupervised learning [32]. Su et al. revealed several best practices for achieving state-of-the-art retinal vessel segmentation performance by analyzing the impact of data processing, architecture design, attention mechanism, and regularization strategy on retinal vessel segmentation performance [33].
Although there are many automatic retinal vessel segmentation models [34], most of the methods still have shortcomings in segmenting fine blood vessels and ensuring vascular connectivity. The shortcomings are mainly due to the following reasons: (1) Fundus images contain rich capillaries with large variations in width. Coarse blood vessels with large areas dominate the optimization direction of model parameters, resulting in insufficient feature extraction ability of models for fine blood vessels. (2) For accurate vessel segmentation, the U-shaped structure supplements vessel detail information for decoder features through the encoder features. However, there are information discrepancies between these two types of features, and traditional feature fusion approaches fail to fully utilize the effective information between them [35]. (3) Most existing retinal vessel segmentation models usually use single-level features to achieve prediction. However, single-level features contain limited information and may include more noise due to the introduction of encoder features, leading to mis-segmentation and bad connectivity of vascular structures.
To address the above issues, a new retinal vessel segmentation model, CMP-UNet, is proposed. The main contributions are as follows: (1) The Coarse and Fine Feature Aggregation (CFFA) module is designed based on the morphological discrepancy between thick and thin vessels. This module decouples and aggregates thick and thin vessel features using two branches, which balances the model’s feature extraction ability for vessels of various sizes. (2) The Multi-Scale Channel Adaptive Fusion (MSCAF) module is designed. It leverages parallel atrous convolution to mine multi-scale contextual features in cascade features and refines these features with the adaptive channel attention module to achieve efficient fusion of cascade features. (3) The Pyramid Feature Fusion (PFF) module is proposed to combine the decoder features into multi-level features following the pyramid form. In this way, the complementary information between the multi-level features can be rationally applied to learn more discriminative representations. Experimental results on three publicly available datasets show that CMP-UNet achieves better segmentation accuracy and generalization ability compared with other methods. It could play an important role in the diagnosis of ophthalmic diseases and reduce the consumption of labor and time.
The remaining sections of this paper are organized as follows: Section 2 gives a detailed description of the proposed CMP-UNet. In Section 3, three publicly available datasets and experimental details are presented. In Section 4, we demonstrate the experimental results and analyze the performance of CMP-UNet in vessel segmentation. Finally, the conclusions and outlooks of this paper are given in Section 5.
2. Methods
To achieve automatic segmentation of retinal blood vessels, a novel model, CMP-UNet, is proposed to realize the precise segmentation of retinal vessels. This section provides detailed descriptions of the proposed CMP-UNet and each designed module. For convenience, the symbols used in this work are listed in Table 1.

Table 1.
Symbol descriptions.
2.1. Overall Network Architecture
U-Net has been widely used in various medical image segmentation tasks [36] due to its unique U-shaped structure and suitability for small datasets [37]. In this paper, based on the U-shaped structure, a novel model, CMP-UNet, is proposed for end-to-end retinal vessel segmentation, and its overall structure is shown in Figure 1. The model consists of five parts: encoder, decoder, CFFA module, MSCAF module, and PFF module.
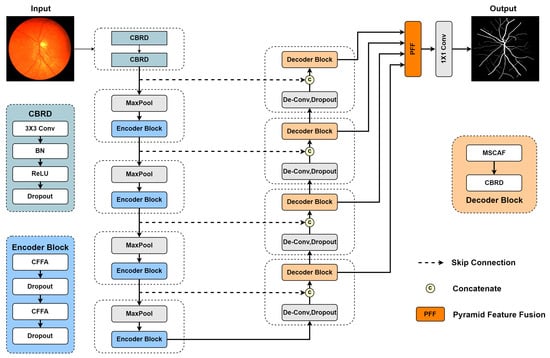
Figure 1.
CMP-UNet.
The input image is first processed by two convolutional layers to increase the number of channels and obtain the initial feature map, . Each convolutional layer is followed by a BN [38] layer for batch normalization to accelerate model training, a ReLU [39] layer to implement nonlinear mapping, and a Dropout [40] layer to mitigate overfitting. For convenience, the combination of convolutional, BN, ReLU, and Dropout layers is named CBRD block. Due to the segmentation task [41] requiring pixel-level prediction results, the initial feature map maintains the original resolution of the input image to avoid information loss. Subsequently, the feature map is fed to the encoder. Several cascade layers, including max pooling with stride 2 and encoder block, are used to map the features into high-dimensional space to extract abstract semantic information. Each encoder block contains two CFFA modules to achieve thick and thin vessel feature extraction and doubles the number of feature channels. In addition, to prevent overfitting, each CFFA module is followed by a Dropout layer. Let denote the original input of the network; then, the output of the ith encoder block can be obtained as follows:
During the decoding stage, deconvolution is used for upsampling and reducing the number of channels by half. Next, feature maps are concatenated with their corresponding ones from the encoder using skip connection. After that, the cascaded features are fed into the decoder block, which consists of an MSCAF module for efficient feature fusion and convolution for feature decoding. Finally, the output features of each decoder block are forwarded to the PFF module for integration and generate probability map P with convolution. Assuming that denotes the output of the ith decoder block, we can acquire the probability map with the following formulas:
2.2. Coarse and Fine Feature Aggregation
In fundus retinal images, thin blood vessels exhibit weak localized features, making them susceptible to background noise interference [42]. In contrast, thick blood vessels possess stronger characteristics that dominate the optimization direction of the model parameters [43]. This leads to the network’s feature extraction ability imbalance for vessels of different sizes, ultimately hindering the segmentation of fine vessels. Most existing retinal vessel segmentation models use a single branch to extract features, which fails to achieve uniform characterization of coarse and fine vessels. Taking inspiration from the method in [21], we propose the CFFA module, as illustrated in Figure 2.
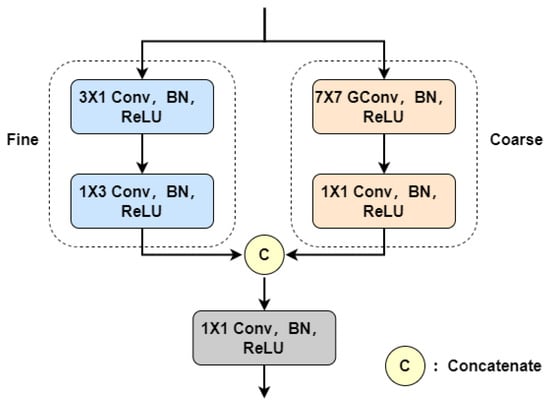
Figure 2.
Coarse and Fine Feature Aggregation (CFFA) module.
For coarse blood vessels, the distance between target pixels is remote due to their large size. To acquire long-range contextual information with lower computational overhead, group convolution (GConv) with a large kernel [44] is used in the coarse branch. However, for fine vessels, using a large convolution kernel may introduce unnecessary background information and noise, resulting in the loss of capillary details. Therefore, we employ heterogeneous convolution with a small kernel for extracting fine vessel features. This strategy helps to reduce the interference of background information and noise, enabling the network to focus more on modeling fine vessel features. The following provides a detailed description of these two branches.
The coarse branch consists of a group convolution operation and a convolution operation. The fine branch includes a convolution operation and a convolution operation. Each convolutional layer is followed by a BN layer and a ReLU layer. Let and be the input and output of the CFFA module, respectively. For the coarse branch, a group convolution operation is first used to capture the long-range context information () of the thick vessels; then, features from each group are fused through convolution to obtain the output () of this branch. In the fine branch, vertical feature is constructed using convolution; then, horizontal feature is further extracted using convolution. Finally, the outputs of the two branches are merged using convolution to aggregate coarse and fine vessel features. The overall procedure of the CFFA module can be formulated as follows:
Ref. [21] employs two parallel U-shaped networks to separately segment coarse and fine blood vessels. During the feature extraction process, there is no exchange of information between these networks, just a simple fusion of their outputs at the end. Instead, we design an independent dual-branch module and use it as the basic component of the encoder. This approach suppresses redundant information among aggregated features by highlighting the differences between thick and thin vessel features, and it also maintains a lower computational burden.
2.3. Multi-Scale Channel Adaptive Fusion
To construct long-range contextual information about retinal vessels, features are gradually downsampled in the encoder to expand the receptive field. However, the slender and variable vessel structure can lead to the loss of fine vessel details and vessel edge information, hindering the accurate segmentation of the vessels. To address this issue, the encoder delivers vessel detail information to the decoder using skip connection. For example, in the U-Net architecture, features from both the encoder and decoder are concatenated, followed by fusion and decoding using two convolution operations. Nevertheless, this approach fails to achieve adequate fusion due to the semantic gap between these types of features. To achieve more efficient feature fusion, the MSCAF module (see Figure 3) is proposed in this paper and embedded into the decoder of the network.
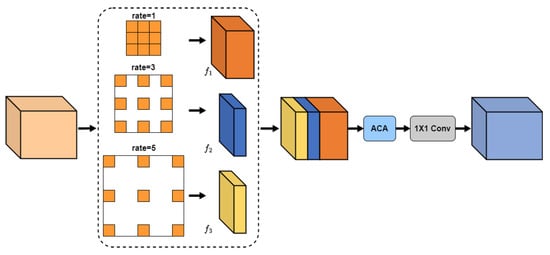
Figure 3.
Multi-Scale Channel Adaptive Fusion (MSCAF) module.
The informational difference between encoder and decoder features is manifested by the fact that they contain contextual features at different scales. Traditional feature fusion methods only consider the commonalities and differences between features at a single scale, causing the fused features to contain a lot of redundant information. Therefore, the proposed MSCAF module needs to acquire the capability of capturing multi-scale contextual features. In addition, considering that contextual features at varying scales have different impacts on vascular segmentation results, it is essential to enable the model to automatically assess the importance of these features. Therefore, the MSCAF module should also be capable of dynamically assigning weights to them. More details on the MSCAF module are given below.
Firstly, multi-scale contextual features are extracted from the cascade features using several atrous convolution operations with rates set to 1, 3, and 5, respectively. To obtain precise segmentation results, more local contextual features should be captured. Let N denote the channel number of F. Then, the channel numbers of , , and are N/2, N/4, and N/4, respectively.
Next, the multi-scale contextual features are forwarded to the adaptive channel attention module (see Figure 4) for adaptive activation, enhancing valuable information while suppressing redundant information. The activation process is as follows: (1) Spatial information is extracted from input utilizing adaptive pooling to generate two different spatial context descriptors, and . They are then reshaped and concatenated along the spatial dimension to form the initial key feature, . (2) The initial key feature is sequentially encoded with the feature encoder and the channel encoder. Subsequently, the encoded result is mapped between 0 and 1 using the sigmoid function, with dimension reshaping, to obtain attention map . (3) The attention map is used to re-weigh the input feature with element-wise multiplication. In addition, to mitigate gradient vanishing, the re-weighed feature is combined with the input feature using element-wise addition to form the final output, O. The process of the ACA module can be described as follows:
where denotes adaptive maximum pooling, denotes adaptive average pooling, and superscript r denotes reshape. Obviously, the increase in channel number leads to more complex inter-channel dependencies. To make the initial key feature adaptively fit in the inter-channel dependencies with different dimensions, we assume that the resolution (K) of the initial key feature is proportional to channel dimension C. Then, K can be calculated with the following equation:
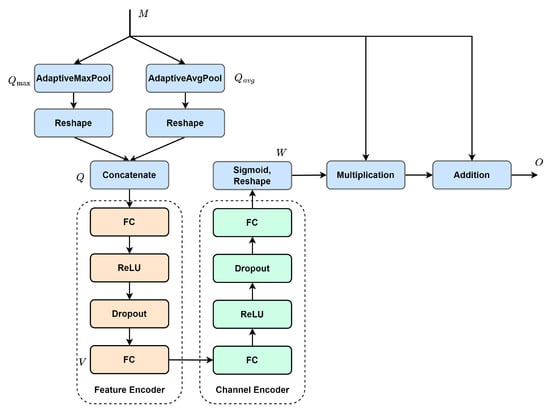
Figure 4.
Adaptive Channel Attention (ACA) module.
Finally, the efficient fusion of activated features is achieved with a convolution operation. The ACA module differs from traditional channel attention. Traditional methods obtain the global feature descriptor, which characterizes overall information using global pooling. The descriptor is then leveraged to generate the attention map. This may not effectively capture global information and is susceptible to noise. In contrast, the ACA module does not extract the global feature descriptor directly. It first extracts several local feature descriptors from the feature map using adaptive pooling. Then, a multi-layer perceptron is used to nonlinearly map these descriptors to obtain the global feature descriptor for generating the attention map. This approach is less susceptible to noise interference and can obtain a more accurate attention map.
2.4. Pyramid Feature Fusion
In the decoder, high-level features contain rich semantic information that helps to localize the vessel trunk and improve the continuity of vessel segmentation results. However, these low-resolution high-level features contain less detailed information, especially fine vessels and vessel edges. Conversely, low-level features possess higher resolution, which includes finer details but also introduces more noise. To attain more discriminative vessel features and enhance the overall continuity of the vessel structure, it is imperative to fully exploit the valuable information within each decoder layer. To address this requirement, we introduce the PFF module, with its specific structure being depicted in Figure 5.
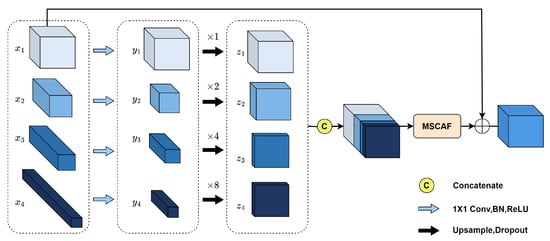
Figure 5.
Pyramid Feature Fusion (PFF) module.
To efficiently capitalize the information from multi-level features, the proposed module needs to satisfy the following demands: Firstly, it should adopt a simple approach to integrating the features of each decoder layer, because the complex module structure can hinder gradient backpropagation, thereby increasing the difficulty of training the network. Secondly, the module should be equipped with efficient feature fusion capability to handle the discrepancy in features in each layer. Lastly, it should be lightweight to avoid a substantial increase in computational overhead and prevent overfitting.
Let denote the generated features at different layers of the decoder, where , , , . Initially, these features are compressed using convolution to avoid a large increase in model computation. To ensure there is enough detailed information to produce precise vessel segmentation results, we expect more low-level features to be included in the multi-level features. Therefore, the numbers of feature channels are set to 16, 8, 4, and 4 for , , , and , respectively. Subsequently, these features are enlarged to the same size as using bilinear interpolation and concatenated along the channel dimensions to generate multi-level features . Then, the multi-level features are fed into the MSCAF module for adaptive fusion to bridge the semantic gap between the features of each layer. Finally, is used to supplement the detailed information of the fused features using element-wise addition.
3. Experiments
3.1. Datasets
The proposed model was evaluated on three datasets (see Table 2): DRIVE, CHASE_DB1, and STARE. The DRIVE dataset [45] contains 40 color retinal images with a resolution of 565 × 584. Seven of these images have mild signs of diabetic retinopathy, and the remaining images were collected from healthy subjects. We adopted the officially assigned dataset split, where the training set contains 20 images, each equipped with only one expert’s labeling results. The testing set also includes 20 images, where each image is equipped with the labeled results of two experts. We chose the first expert’s labeling result as the ground truth. The CHASE_DB1 dataset [46] contains 28 retinal images with a resolution of 999 × 960, and each image is accompanied by two segmentation maps labeled by experts. The first one was taken as the ground truth. The first 20 images were used as training data, and the last 8 images were used as test data [47]. The STARE dataset [48] contains 20 retinal images of 700 × 605 pixels, where 10 images have a retinal disease and the remaining images represent healthy retinas. Each image was annotated by two experts, and we chose the first expert’s annotation as the ground truth. In this paper, the first 10 images were selected as training data, and the remaining 10 images were assigned as test data.

Table 2.
Description of the dataset.
3.2. Data Augmentation
Acquiring and annotating fundus images is challenging, resulting in the limited availability of well-annotated data. To avoid overfitting [49] due to the limited number of data, data augmentation techniques were employed to enrich the dataset. The data augmentation process is described as follows: First, the dataset was doubled by horizontally flipping each image. Next, the expanded dataset from the previous step was rotated by 360 degrees at 2-degree intervals. As a result of these two steps, the final dataset’s size was 360 times the original dataset’s size. The numbers of images before and after data augmentation for each dataset are shown in Table 2.
To decrease memory usage and accelerate model training, fundus images were randomly cropped and scaled to obtain fixed-resolution patches [50]. The CHASE_DB1 and STARE datasets extracted patches with a fixed resolution of 256 × 256 pixels. In comparison, the DRIVE dataset extracted patches with a fixed resolution of 128 × 128 pixels due to the lower image resolution. The specific procedure for random cropping and scaling was as follows (Figure 6): First, patches with arbitrary aspect ratio were randomly cropped from the image (which limited the range of the variation in the aspect ratio); then, they were scaled to the specified resolution (256 × 256 or 128 × 128). Random cropping and scaling can introduce variations in the blood vessel structure, resulting in new samples that are not present in the original dataset. This increases the diversity of data samples, which ultimately enhances the generalization ability of the model.
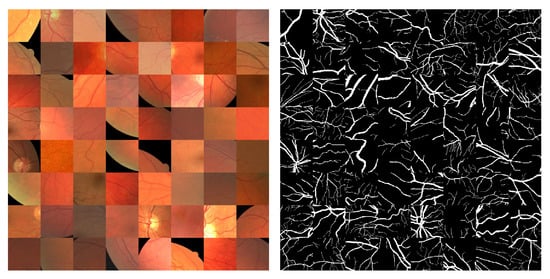
Figure 6.
Data patches.
Furthermore, fundus images are often characterized by low contrast between the vessels and the background, alongside non-uniform brightness and hue. For this reason, data augmentation techniques [51] such as color jittering (random adjustment of image saturation, brightness, contrast, and hue), random sharpening, and random blurring were employed during the training process.
3.3. Training Details
Our model was implemented with the PyTorch framework on the Ubuntu platform, equipped with NVIDIA RTX 8000. In addition, python libraries such as OpenCV, Numpy, and Pillow were used in the development process. We employed the Adam optimizer [52] for model training. Due to the higher image resolution of the CHASE_DB1 dataset, the vessel structure is more obvious, so the epoch number was set to 50. On the other hand, of the three datasets, the DRIVE dataset has the smallest image resolution, and the STARE dataset has the minimum number of training images. To fit the model adequately, the epoch number was set to 100 in these two datasets. The batch size was configured as 16 for the CHASE_DB1 and STARE datasets, and it was set to 32 for the DRIVE dataset due to the lower patch resolution. Initially, we set the learning rate to 0.001 and gradually reduced it during the training process. For the DRIVE and STARE datasets, we implemented a plateau strategy for learning rate adjustment, scaling it down by a factor of 10 at epochs 40 and 80, respectively. For the CHASE_DB1 dataset, the learning rate was decayed at epochs 20 and 40. To prevent gradient vanishing and gradient explosion [53], we applied gradient clipping [54] to make the training process more stable. The binary cross-entropy loss function [55], which is calculated as shown below, was chosen to supervise the model training process.
where N denotes the number of pixel points; denotes the true category of the pixel point (with for the vessel and for the background); denotes the predicted probability that the pixel point belongs to the blood vessel. Finally, we set the category threshold for the model’s prediction to 0.5. If , it was predicted to be the vessel. Otherwise, it was predicted to be the background.
3.4. Evaluation Metrics
To quantify the segmentation performance of the proposed model, several metrics were used to evaluate the segmentation ability of the model, including F1-score (F1), Sensitivity (SE), Specificity (SP), and Accuracy (ACC), as shown in the formulas below:
where , , , and represent true positives, true negatives, false positives, and false negatives, respectively. The values of the above metrics range from 0 to 1, with larger values indicating better model performance.
4. Results
In this section, we systematically analyze the performance of CMP-UNet. First, we show the performance of the model as a whole and give the segmentation results for the test dataset. Second, we compare CMP-UNet with some other retinal vessel segmentation methods proposed in recent years to verify the excellent performance of the model. Then, we conduct ablation experiments on the CHASE_DB1 dataset to demonstrate the effectiveness of the designed modules. Finally, the generalization ability of the proposed model is analyzed with cross-experiments.
4.1. Vessel Segmentation Results
To evaluate the performance of CMP-UNet, we conducted comprehensive qualitative and quantitative analyses on three datasets: CHASE_DB1, DRIVE, and STARE. The retinal vessel segmentation results for the three datasets are shown in Figure 7. We can clearly observe that the proposed model is able to segment both fine and complex vessels more accurately and better maintain the connectivity of the vascular structure. This is crucial for the diagnosis and tracking of diseases such as early diabetic retinopathy. In particular, from the results of the DRIVE dataset, when dealing with U-Net over-segmented samples, the proposed model still achieves accurate segmentation.
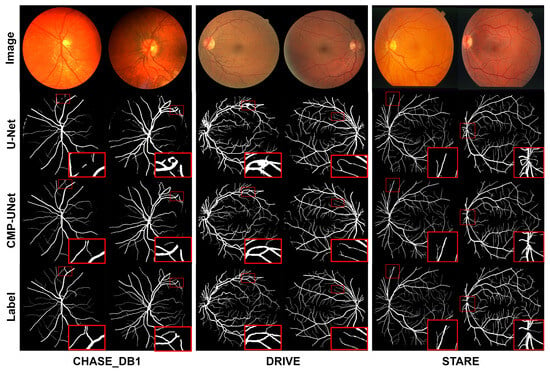
Figure 7.
Test results for the three datasets. The red box enlarges the local detail of the segmentation result.
Table 3 shows the segmentation performance of CMP-UNet on the three datasets. On the CHASE_DB1 and DRIVE datasets, the proposed model outperforms U-Net and the second expert in all metrics. Notably, although our model is trained using the annotations of the first expert, it exceeds the second expert. This indicates that our model is able to provide more consistent and accurate segmentation results while simulating the expert annotations. This difference in results is less than the discrepancy between experts’ annotations, further demonstrating the excellent segmentation performance and generalization ability of our model.

Table 3.
Average metrics for the three datasets.
However, on the STARE dataset, the proposed model has some limitations in vessel discrimination due to the small number of training samples. This limitation is reflected in a reduction in the SE metric. U-Net faces the same problem, resulting in a significant decrease in SE. Therefore, U-Net was unable to accurately detect blood vessels against the background, leading to an increase in background pixels, which is reflected in a larger SP metric.
4.2. Comparison with the Existing Methods
We compared the proposed model with other state-of-the-art retinal vessel segmentation models proposed in recent years on the CHASE_DB1, DRIVE, and STARE datasets, and the experimental results are shown in Table 4, Table 5 and Table 6. Table 4 presents the comparison results of the proposed model, CMP-UNet, with other models on the CHASE_DB1 dataset. It can be seen that our model achieves 97.80%, 84.31%, 98.70%, and 82.84% in the four metrics of ACC, SE, SP, and F1, respectively, which are clearly better results than those of the other models. Table 5 demonstrates the comparison results for the DRIVE dataset. CMP-UNet achieves the best results in the two metrics of ACC and SE, reaching 96.96% and 82.61%, respectively. The SP and F1 results are also superior, with differences of 0.43% and 0.6% from the best results. As shown in Table 6, the ACC, SE, and F1 results of the proposed model are the best on the STARE dataset, reaching 97.62%, 85.36%, and 84.14%, respectively. Compared with the best results of other models, SP has a numerical gap of 0.8%, which is not a significant decrease.

Table 4.
Comparison of CMP-UNet with other models on the CHASE_DB1 dataset.

Table 5.
Comparison of CMP-UNet with other models on the DRIVE dataset.

Table 6.
Comparison of CMP-UNet with other models on the STARE dataset.
Combined with the above, CMP-UNet can achieve the highest values of ACC and SE scores on all three datasets, and the F1 metrics are significantly improved on the CHASE_DB1 and STARE datasets. ACC is a comprehensive metric for all prediction categories, SE measures the proportion of blood vessels correctly recognized, and F1 indicates the similarity between prediction and ground truth. From this, it could be inferred that the proposed model has better vessel discrimination ability and generalization ability.
Nevertheless, on the STARE and DRIVE datasets, the enhancement effect of SP is insignificant. This is because of the lower resolution of the STARE and DRIVE datasets as well as the denser distribution of fine vessels. Although the proposed model can detect more fine blood vessels, it simultaneously incorrectly mis-segments some background near fine blood vessels as vessels, resulting in a smaller SP improvement.
4.3. Ablation Analysis
The proposed model, CMP-UNet, improves performance in retinal vessel segmentation by introducing three independent modules: the CFFA module, the MSCAF module, and the PFF module. In order to evaluate the impact of the three designed modules on the proposed model, ablation experiments were conducted on the CHASE_DB1 dataset. To ensure the fairness of the experiments, all comparison methods used the same training strategy and hyperparameter settings. There were five sets of experiments: (1) baseline (U-Net), (2) C-UNet (adding the CFFA module to the encoder), (3) M-UNet (embedding the MSCAF module in the decoder), (4) CM-UNet (using both CFFA module and MSCAF module), (5) CMP-UNet. Table 7 demonstrates the results of the ablation experiments.

Table 7.
Results of ablation experiments.
With the introduction of the CFFA module in the encoder, the results show improvements in ACC, SE, and F1, especially SE and F1, which improved by 0.86% and 0.45%, respectively, compared with the baseline. This indicates that the CFFA module improves the model’s ability to extract vascular features and helps to detect more fine vessels.
The SE and F1 of our model are separately improved by 1.16% and 0.21% after embedding the MSCAF module into the decoder. This means that the MSCAF module is able to fuse complementary information from multi-scale features in a more efficient way, which reduces the loss of vascular information to segment more vessels.
When both the CFFA module and the MSCAF module are used, the ACC, SE, and F1 of the proposed model are improved. Among them, SE and F1 have greater improvements, 1.58% and 0.66%, respectively. Due to the combined effect of the two modules, the ability of the model to extract vessel features is significantly improved. Nevertheless, this may also lead to mis-classifying some background as vessels, causing a slight decrease in the SP value.
Finally, CMP-UNet outperforms U-Net in ACC, SE, SP, and F1, reaching 97.80%, 84.31%, 98.70%, and 82.84%, respectively. Among them, ACC, SP, and F1 reached the maximum values of several sets of experiments, proving the effectiveness of the model.
Comprehensive analysis results of these ablation experiments strongly confirm that the introduced modules effectively improve the performance of CMP-UNet. The segmentation results for each model are visualized in Figure 8, including slices of the fundus image, the ground truth, and the segmentation results for each model. It can be seen that the performance of the model can be progressively improved by sequentially introducing the designed modules in the baseline model.
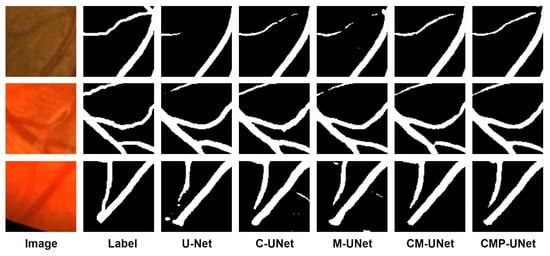
Figure 8.
Visualization of segmentation results of ablation experiments.
4.4. Generalization Analysis
Usually, the scale of the data with good labels is limited, resulting in insufficient generalization ability of a model. To further validate the generalization performance of the proposed model, we conducted cross-experiments on the DRIVE and STARE datasets. Specifically, we trained the model on one dataset and then applied it to the other dataset for vessel segmentation testing.
The experimental results of when the model was trained using the STARE dataset and tested on the DRIVE dataset are shown in Table 8. CMP-UNet achieves 96.74%, 79.77%, and 98.41% in ACC, SE, and SP, respectively, where ACC and SE show to be the best results. Compared with the best results of other models, ACC and SE are improved by 0.61% and 6.64%, respectively. It should be noted that due to the strong vessel detection capability of the proposed model, the background around the vessels may be mis-segmented, resulting in a slight decrease in SP.

Table 8.
Results of cross-experiments.
When trained on the DRIVE dataset and tested on the STARE dataset, CMP-UNet achieves the best results in all three metrics, ACC, SE, and SP, reaching 97.35%, 80.87%, and 98.65%, respectively. Compared with the best results of the other models, ACC and SP are improved by 1.11% and 0.53%, respectively, and SE is slightly improved.
In addition, we present the results of the cross-experiments visually, as shown in Figure 9. From the figure, it can be seen that the model can still segment most of the vessels from the background effectively and maintains good vessel connectivity. Overall, the proposed model, CMP-UNet, has better generalization ability.
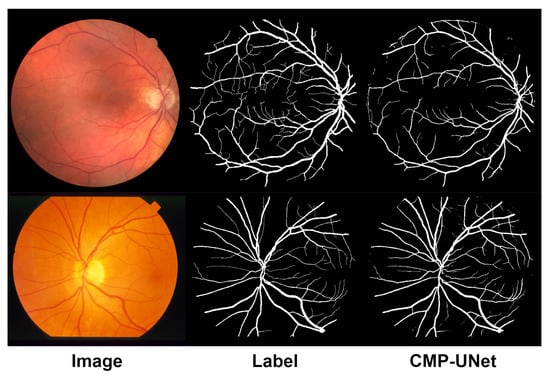
Figure 9.
Visualization results of the cross-experiments. Top: DRIVE (training on STARE). Bottom: STARE (training on DRIVE).
5. Conclusions
In this paper, CMP-UNet is proposed for retinal vessel segmentation. The Coarse and Fine Feature Aggregation module is used to replace the original convolutional block to balance the model’s feature extraction capability for vessels with different sizes. In order to exploit the multi-scale information of cascaded features more efficiently, the Multi-Scale Channel Adaptive Fusion module is embedded in the decoder. In addition, the Pyramid Feature Fusion module is introduced to realize the interaction of multi-level feature information, thus enhancing the discriminability of blood vessels. Experimental results on the CHASE_DB1, DRIVE, and STARE datasets indicate that CMP-UNet outperforms existing models in terms of segmentation performance and generalization ability. The ablation experiments on the CHASE_DB1 dataset demonstrate the effectiveness of the proposed CFFA, MSCAF, and PFF modules. Finally, the cross-experiments further evidence the excellent generalization performance of the proposed model. Although the proposed model in this paper further improves the segmentation accuracy of retinal vessels, there are still shortcomings in vascular structure connectivity, which are mainly due to the following two reasons: Firstly, the proposed model is trained using the cross-entropy loss function. This loss function calculates the prediction error point to point, which ignores the structural information of the blood vessels. Secondly, the proposed model, like the current mainstream models, classifies the vascular probability map in a binary manner by setting a threshold to realize the distinction between blood vessels and background. This method cannot avoid breakage of the vascular structure. In future work, based on the above two points, we will aim to propose a new model to further enhance the connectivity of vascular structures.
Author Contributions
Conceptualization, Y.G.; methodology, R.C.; software, R.C.; validation, Y.G., R.C. and B.L.; formal analysis, D.W.; investigation, Y.G., R.C., D.W. and B.L.; resources, B.L.; data curation, R.C.; writing—original draft preparation, R.C.; writing—review and editing, Y.G., D.W. and B.L.; visualization, R.C.; supervision, Y.G. and D.W.; project administration, Y.G.; funding acquisition, Y.G. and B.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Key Scientific Research Projects of Colleges and Universities in Henan Province (grant number 23B520002); National Natural Science Foundation of China (grant number 42272178); Key Science and Technology Program of Henan Province (grant number 222102210131).
Data Availability Statement
Our code will be available at https://github.com/hpucry/CMP-UNet (accessed on 19 November 2023).
Conflicts of Interest
The authors declare no potential conflicts of interest with respect to the research, authorship, and publication of this article.
References
- Sethuraman, S.; Gopi, V.P. Staircase-Net: A deep learning based architecture for retinal blood vessel segmentation. Sadhana-Acad. Proc. Eng. Sci. 2022, 47, 191. [Google Scholar] [CrossRef]
- Cheng, Y.; Ma, M.; Zhang, L.; Jin, C.; Ma, L.; Zhou, Y. Retinal blood vessel segmentation based on Densely Connected U-Net. Math. Biosci. Eng. 2020, 17, 3088–3108. [Google Scholar] [CrossRef]
- Arsalan, M.; Haider, A.; Koo, J.H.; Park, K.R. Segmenting Retinal Vessels Using a Shallow Segmentation Network to Aid Ophthalmic Analysis. Mathematics 2022, 10, 1536. [Google Scholar] [CrossRef]
- Chen, C.; Chuah, J.H.; Ali, R.; Wang, Y. Retinal Vessel Segmentation Using Deep Learning: A Review. IEEE Access 2021, 9, 111985–112004. [Google Scholar] [CrossRef]
- Tan, Y.; Zhao, S.X.; Yang, K.F.; Li, Y.J. A lightweight network guided with differential matched filtering for retinal vessel segmentation. Comput. Biol. Med. 2023, 160, 106924. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhu, H. M3U-CDVAE: Lightweight retinal vessel segmentation and refinement network. Biomed. Signal Process. Control 2023, 79, 104113. [Google Scholar] [CrossRef]
- Alom, M.Z.; Hasan, M.; Yakopcic, C.; Taha, T.M.; Asari, V.K. Recurrent Residual Convolutional Neural Network based on U-Net (R2U-Net) for Medical Image Segmentation. arXiv 2018, arXiv:1802.06955. [Google Scholar]
- Rehman, M.U.; Ryu, J.; Nizami, I.F.; Chong, K.T. RAAGR2-Net: A brain tumor segmentation network using parallel processing of multiple spatial frames. Comput. Biol. Med. 2023, 152, 106426. [Google Scholar] [CrossRef]
- Wang, T.; Dai, Q. SURVS: A Swin-Unet and game theory-based unsupervised segmentation method for retinal vessel. Comput. Biol. Med. 2023, 166, 107542. [Google Scholar] [CrossRef]
- Zhao, T.; Fu, C.; Tie, M.; Sham, C.W.; Ma, H. RGSB-UNet: Hybrid Deep Learning Framework for Tumour Segmentation in Digital Pathology Images. Bioengineering 2023, 10, 957. [Google Scholar] [CrossRef]
- Rehman, M.U.; Cho, S.; Kim, J.; Chong, K.T. BrainSeg-Net: Brain Tumor MR Image Segmentation via Enhanced Encoder-Decoder Network. Diagnostics 2021, 11, 169. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.; Tian, X. MWG-UNet: Hybrid Deep Learning Framework for Lung Fields and Heart Segmentation in Chest X-ray Images. Bioengineering 2023, 10, 1091. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Qin, D.; Wang, S.; Fang, Y.; Zheng, Y. A multi-channel UNet framework based on SNMF-DCNN for robust heart-lung-sound separation. Comput. Biol. Med. 2023, 164, 107282. [Google Scholar] [CrossRef] [PubMed]
- Rehman, M.U.; Cho, S.; Kim, J.H.; Chong, K.T. BU-Net: Brain Tumor Segmentation Using Modified U-Net Architecture. Electronics 2020, 9, 2203. [Google Scholar] [CrossRef]
- Wang, B.; Wang, S.; Qiu, S.; Wei, W.; Wang, H.; He, H. CSU-Net: A Context Spatial U-Net for Accurate Blood Vessel Segmentation in Fundus Images. IEEE J. Biomed. Health Inform. 2021, 25, 1128–1138. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Qi, H.; Xiong, Y.; Li, Y.; Zhang, G.; Hu, H.; Wei, Y. Deformable Convolutional Networks. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 764–773. [Google Scholar] [CrossRef]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional Block Attention Module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018. [Google Scholar]
- Yang, X.; Li, Z.; Guo, Y.; Zhou, D. DCU-net: A deformable convolutional neural network based on cascade U-net for retinal vessel segmentation. Multimed. Tools Appl. 2022, 81, 15593–15607. [Google Scholar] [CrossRef]
- Cao, J.; Li, Y.; Sun, M.; Chen, Y.; Lischinski, D.; Cohen-Or, D.; Chen, B.; Tu, C. DO-Conv: Depthwise Over-Parameterized Convolutional Layer. IEEE Trans. Image Process. 2022, 31, 3726–3736. [Google Scholar] [CrossRef]
- Liu, Y.; Shen, J.; Yang, L.; Bian, G.; Yu, H. ResDO-UNet: A deep residual network for accurate retinal vessel segmentation from fundus images. Biomed. Signal Process. Control 2023, 79, 104087. [Google Scholar] [CrossRef]
- Xu, Y.; Fan, Y. Dual-channel asymmetric convolutional neural network for an efficient retinal blood vessel segmentation in eye fundus images. Biocybern. Biomed. Eng. 2022, 42, 695–706. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhang, L.; Wang, L.; Huang, H. Multi-Level Attention Network for Retinal Vessel Segmentation. IEEE J. Biomed. Health Inform. 2022, 26, 312–323. [Google Scholar] [CrossRef]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Guo, M.H.; Xu, T.X.; Liu, J.J.; Liu, Z.N.; Jiang, P.T.; Mu, T.J.; Zhang, S.H.; Martin, R.R.; Cheng, M.M.; Hu, S.M. Attention mechanisms in computer vision: A survey. Comput. Vis. Media 2022, 8, 331–368. [Google Scholar] [CrossRef]
- Dai, Z.; Heckel, R. Channel Normalization in Convolutional Neural Network avoids Vanishing Gradients. arXiv 2019, arXiv:1907.09539 2019. [Google Scholar]
- Yue, C.; Ye, M.; Wang, P.; Huang, D.; Lu, X. SRV-GAN: A generative adversarial network for segmenting retinal vessels. Math. Biosci. Eng. 2022, 19, 9948–9965. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, T.; Zhao, Y.; Chen, N.; Zhou, H.; Xu, H.; Guan, Z.; Xue, L.; Yang, C.; Chen, R.; et al. MC-UNet: Multimodule Concatenation Based on U-Shape Network for Retinal Blood Vessels Segmentation. Comput. Intell. Neurosci. 2022, 2022, 9917691. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Ye, J. A retinal blood vessel segmentation based on improved D-MNet and pulse-coupled neural network. Biomed. Signal Process. Control 2022, 73, 103467. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Lian, J.; Yang, Z.; Liu, J.; Sun, W.; Zheng, L.; Du, X.; Yi, Z.; Shi, B.; Ma, Y. An Overview of Image Segmentation Based on Pulse-Coupled Neural Network. Arch. Comput. Methods Eng. 2021, 28, 387–403. [Google Scholar] [CrossRef]
- Raza, K.; Singh, N.K. A Tour of Unsupervised Deep Learning for Medical Image Analysis. Curr. Med. Imaging 2021, 17, 1059–1077. [Google Scholar] [CrossRef]
- Su, Y.; Cheng, J.; Cao, G.; Liu, H. How to design a deep neural network for retinal vessel segmentation: An empirical study. Biomed. Signal Process. Control 2022, 77, 103761. [Google Scholar] [CrossRef]
- Dhanagopal, R.; Raj, P.T.V.; Suresh Kumar, R.; Mohan Das, R.; Pradeep, K.; Kwadwo, O.A. An Efficient Retinal Segmentation-Based Deep Learning Framework for Disease Prediction. Wirel. Commun. Mob. Comput. 2022, 2022, 2013558. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, H.; Chen, Z.; Huangliang, K.; Zhang, H. TransUNet plus: Redesigning the skip connection to enhance features in medical image segmentation. Knowl.-Based Syst. 2022, 256, 109859. [Google Scholar] [CrossRef]
- Zhang, H.; Zhong, X.; Li, G.; Liu, W.; Liu, J.; Ji, D.; Li, X.; Wu, J. BCU-Net: Bridging ConvNeXt and U-Net for medical image segmentation. Comput. Biol. Med. 2023, 159, 106960. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In International Conference on Machine Learning; PMLR: New York, NY, USA, 2015. [Google Scholar]
- Glorot, X.; Bordes, A.; Bengio, Y. Deep Sparse Rectifier Neural Networks. In Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics, Fort Lauderdale, FL, USA, 11–13 April 2011; Gordon, G., Dunson, D., Dudík, M., Eds.; PMLR: New York, NY, USA, 2011; Volume 15, pp. 315–323. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Liu, X.; Song, L.; Liu, S.; Zhang, Y. A Review of Deep-Learning-Based Medical Image Segmentation Methods. Sustainability 2021, 13, 1224. [Google Scholar] [CrossRef]
- Kuang, X.; Xu, X.; Fang, L.; Kozegar, E.; Chen, H.; Sun, Y.; Huang, F.; Tan, T. Improved fully convolutional neuron networks on small retinal vessel segmentation using local phase as attention. Front. Med. 2023, 10. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Yang, X.; Cheng, K.T. A Three-Stage Deep Learning Model for Accurate Retinal Vessel Segmentation. IEEE J. Biomed. Health Inform. 2019, 23, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Qin, J.; Lv, L.; Cheng, M.; Li, L.; Xia, D.; Wang, S. MLKCA-Unet: Multiscale large-kernel convolution and attention in Unet for spine MRI segmentation. Optik 2023, 272, 170277. [Google Scholar] [CrossRef]
- Staal, J.; Abramoff, M.; Niemeijer, M.; Viergever, M.; van Ginneken, B. Ridge-based vessel segmentation in color images of the retina. IEEE Trans. Med. Imaging 2004, 23, 501–509. [Google Scholar] [CrossRef]
- Owen, C.G.; Rudnicka, A.R.; Mullen, R.; Barman, S.A.; Monekosso, D.; Whincup, P.H.; Ng, J.; Paterson, C. Measuring Retinal Vessel Tortuosity in 10-Year-Old Children: Validation of the Computer-Assisted Image Analysis of the Retina (CAIAR) Program. Investig. Ophthalmol. Vis. Sci. 2009, 50, 2004–2010. [Google Scholar] [CrossRef]
- Li, Q.; Feng, B.; Xie, L.; Liang, P.; Zhang, H.; Wang, T. A Cross-Modality Learning Approach for Vessel Segmentation in Retinal Images. IEEE Trans. Med. Imaging 2016, 35, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Hoover, A.; Kouznetsova, V.; Goldbaum, M. Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response. IEEE Trans. Med. Imaging 2000, 19, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Charilaou, P.; Battat, R. Machine learning models and over-fitting considerations. World J. Gastroenterol. 2022, 28, 605–607. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Peng, L.; Peng, S.; Xiao, H.; Zhang, Y. Retinal vessel segmentation by using AFNet. Vis. Comput. 2023, 39, 1929–1941. [Google Scholar] [CrossRef]
- Garcea, F.; Serra, A.; Lamberti, F.; Morra, L. Data augmentation for medical imaging: A systematic literature review. Comput. Biol. Med. 2023, 152. [Google Scholar] [CrossRef] [PubMed]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2017, arXiv:1412.6980. [Google Scholar]
- Panigrahi, A.; Chen, Y.; Kuo, C.C.J. Analysis on Gradient Propagation in Batch Normalized Residual Networks. arXiv 2018, arXiv:1812.00342. [Google Scholar]
- Zhang, J.; He, T.; Sra, S.; Jadbabaie, A. Why gradient clipping accelerates training: A theoretical justification for adaptivity. arXiv 2020, arXiv:1905.11881. [Google Scholar]
- Ma, J.; Chen, J.; Ng, M.; Huang, R.; Li, Y.; Li, C.; Yang, X.; Martel, A.L. Loss odyssey in medical image segmentation. Med. Image Anal. 2021, 71, 102035. [Google Scholar] [CrossRef]
- Ye, Y.; Pan, C.; Wu, Y.; Wang, S.; Xia, Y. MFI-Net: Multiscale Feature Interaction Network for Retinal Vessel Segmentation. IEEE J. Biomed. Health Inform. 2022, 26, 4551–4562. [Google Scholar] [CrossRef]
- Yang, D.; Zhao, H.; Yu, K.; Geng, L. NAUNet: Lightweight retinal vessel segmentation network with nested connections and efficient attention. Multimed. Tools Appl. 2023, 82, 25357–25379. [Google Scholar] [CrossRef]
- Rong, Y.; Xiong, Y.; Li, C.; Chen, Y.; Wei, P.; Wei, C.; Fan, Z. Segmentation of retinal vessels in fundus images based on U-Net with self-calibrated convolutions and spatial attention modules. Med. Biol. Eng. Comput. 2023, 61, 1745–1755. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ni, W.; Luo, Y.; Feng, Y.; Song, R.; Wang, X. TUnet-LBF: Retinal fundus image fine segmentation model based on transformer Unet network and LBF. Comput. Biol. Med. 2023, 159, 106937. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.T.; Khan, H.A.; Naveed, K.; Nauman, A.; Gulfam, S.M.; Kim, S.W. LUVS-Net: A Lightweight U-Net Vessel Segmentor for Retinal Vasculature Detection in Fundus Images. Electronics 2023, 12, 1786. [Google Scholar] [CrossRef]
- Wei, X.; Yang, K.; Bzdok, D.; Li, Y. Orientation and Context Entangled Network for Retinal Vessel Segmentation. Expert Syst. Appl. 2023, 217, 119443. [Google Scholar] [CrossRef]
- Ryu, J.; Rehman, M.U.; Nizami, I.F.; Chong, K.T. SegR-Net: A deep learning framework with multi-scale feature fusion for robust retinal vessel segmentation. Comput. Biol. Med. 2023, 163, 107132. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, S.; Naveed, K.; Naqvi, S.S.; Naveed, A.; Khan, T.M. Robust retinal blood vessel segmentation using a patch-based statistical adaptive multi-scale line detector. Digit. Signal Process. 2023, 139, 104075. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).