1. Introduction
The bare minimum list of essential smart healthcare services includes emergency, inpatient, hospital, medical care, outpatient, and preventative medical services [
1,
2]. A data leak can harm both doctors’ and their patients’ reputations. Cyberattack risk is decreased by enhancing the security of IT systems that store and process medical records. To preserve medical information, laws establish security standards and place protections around patient data and healthcare facilities. Regarding data storage, “on-premise” refers to the healthcare provider’s data center [
3]. Medical professionals use Artificial Intelligence (AI) (e.g., Machine Learning (ML) and Deep Learning (DL)) to assist with patient care and clinical data management [
4]. DL is progressively integrated into advanced systems with notable clinical implications. Some of the most promising applications involve patient-facing innovations and surprisingly mature approaches to enhancing the user experience of health IT [
5]. Among these, Convolutional Neural Networks (CNNs), a category of DL, demonstrate remarkable proficiency in analyzing images, such as X-rays or findings from MRI scans [
6]. CNNs were intentionally designed by computer scientists at Stanford University to excel in image processing, with the goal of enabling them to operate more efficiently and manage larger images. Consequently, certain CNNs surpass human diagnosticians in accurately identifying crucial details within diagnostic imaging assessments [
7]. Federated Learning (FL) enables non-affiliated hospitals to participate without sharing their rich datasets, alleviating the need to co-locate all the data in a central location and enabling independent hospitals to gain benefits from them [
8]. Significant problems, such as data security, privacy, and access to diverse data, are solved by this method [
9,
10].
FL involves the collective sharing of data among multiple individuals to collaboratively train a singular DL model and progressively enhance its capabilities; it resembles a collaborative group presentation or report [
11]. Each participant receives the model from a cloud data center, often starting with a foundational model that has previously undergone training. It is a good option for private information belonging to patients, persons, businesses, or other sectors that must adhere to strict privacy regulations [
12]. FL offers an alternative to modeling problems by allowing numerous edge devices or organizations to train a global model using various local data. FL mainly or partially supports data privacy and security challenges [
13,
14]. However, there is a conflict when we discuss ML and privacy. Indeed, to perform well, ML in general—and DL models in particular—require access to massive datasets. Unfortunately, due to privacy issues and liability risks, these data are frequently maintained by many companies. Privacy, data ownership, legal, and technical issues make it challenging to acquire most data, especially in the healthcare industry.
Furthermore, in the healthcare industry, data management strategies are severely impacted by international regulations, such as the Health Insurance Portability and Accountability Act (HIPAA) in the United States and the General Data Protection Regulation (GDPR) in the European Union [
15,
16]. Data collection from clients in bulk without a clear service goal is no longer an issue. The GDPR provides the legal framework to protect personal data in the European Union. The GDPR increases corporate accountability by imposing additional requirements on service providers concerning data handling, specifically emphasizing controlling its centralization. Privacy protection has assumed paramount importance at the heart of data processing now more than ever [
17]. These challenges pose a serious concern for organizations and data enthusiasts dealing in the healthcare industry to build AI healthcare applications. In other words, one must disclose data to utilize these full diagnostics.
A distributed ML approach called FL, which Google first unveiled in 2017 [
18], enables cross-organization collaboration for training DL models without disclosing their datasets for cases for which training data are to be maintained locally by device users (nodes) as opposed to a centralized data center; FL emerges as a compelling ML approach [
19]. These nodes use their data to compute, and then they update a global model. Since these nodes can be located in various geographical regions subjected to different regulations and with different data generation and preparation techniques, the data distribution patterns across these nodes can have a high level of variance. In a heterogeneous multi-node environment, some clients may have more or different data than others. Therefore, identifying a data sample that generalizes the entire data distribution is an unlikely event. Two primary features that distinguish federated optimization problems from their distributed counterparts are highlighted in this context. First, the data used by each user for training are non-identically and independently distributed (Non-IID), eliminating the existence of a consolidated representation of the entire population’s dataset. Second, data distribution is unbalanced, with varying amounts of data held by each user compared to others. Leveraging the benefits of Federated Learning (FL), we have employed FL-based techniques to address a compassionate issue in the healthcare sector: the coronavirus disease (COVID-19). This infectious ailment, caused by the SARS-CoV-2 virus, was initially reported in Wuhan, China, in December 2019 [
20]. The outbreak spread rapidly throughout China and most of the world’s nations. Stopping the virus is extremely difficult given the pandemic’s rapid escalation (thousands of deaths and hundreds of thousands of infections). There are currently numerous diagnostic techniques available for the identification of coronavirus.
Chest X-ray imaging and CT scans, which are widely utilized [
21,
22], play a pivotal role. The lung damage caused by COVID-19, which targets respiratory tract epithelial cells, is assessed through chest X-ray images [
23]. Furthermore, the availability of X-ray imaging equipment globally and its heightened diagnostic accuracy confers a notable advantage in COVID-19 diagnosis compared to specialized testing kits. AI-based models for processing chest X-ray images offer an expedient and cost-effective alternative for COVID-19 screening, prompting substantial interest in the scientific community to develop ML-based strategies for outbreak prediction and diagnosis [
24]. This study’s objective—the cornerstone of the article’s distinctive concepts—is to formulate and assess an FL-based system for identifying COVID-19 through chest X-ray images. A pioneering effort, this research employs FL techniques to detect COVID-19 in chest X-rays proficiently. The principal contributions of this research encompass:
This research proposes a collaborative and decentralized system specifically designed to empower clinicians with valuable insights for diagnosing COVID-19 while ensuring stringent safeguards are in place for patient data privacy.
The study introduces a robust, decentralized fog-based Federated Learning model. This model is unique in its ability to effectively handle dispersed and unevenly distributed data, yielding results that are on par with those obtained from traditional centralized ML approaches.
To ensure clarity and facilitate future research, the mathematical equations and algorithms used in this study are provided and explained in detail. These resources serve as a valuable tool for readers who wish to further understand or build upon this work.
A comprehensive analysis of the model’s performance has been conducted, demonstrating high accuracy and precision. Further examination of the model’s performance reveals equally impressive recall and F1-scores. Additionally, a detailed confusion matrix is provided, offering an in-depth understanding of the model’s performance across different scenarios. This demonstrates the model’s robustness and reliability, underscoring the effectiveness of the proposed methodology.
The subsequent sections of the paper are structured as follows:
Section 2 provides an overview of the current state-of-the-art, delving into the latest advancements. The proposed methodology is expounded upon in
Section 3, alongside exploring the evaluation parameters employed.
Section 4 furnishes details about the dataset, experimental procedures, and the outcomes garnered from this study. Lastly, the paper concludes in
Section 5 by summarizing key findings and delineating avenues for future research.
2. Literature Review
This section discusses different FL-based models for detecting COVID-19.
Sadly, the dangerous COVID-19 virus caused a pandemic that spread worldwide. Researchers, scientists, medical experts, and executives worldwide face a significant risk if it requires future treatment. In a recent study by Kandati et al. [
23] a comprehensive strategy is recommended amalgamating Federated Learning (FL) with a Particle Swarm Optimization algorithm (PSO). This fusion aims to accelerate the government’s responsiveness in addressing COVID-19-induced chest lesions. The Federated Particle Swarm Optimization method’s efficacy is evaluated using a diverse dataset of images featuring chest lesions associated with COVID-19 infections alongside pneumonia cases from Kaggle’s repository of chest X-ray images. In another innovative undertaking outlined in Bian et al.’s work [
20], the focus is on enhancing the efficiency and precision of global models during training through the incorporation of pre-trained models (PTMs) within the ambit of Federated Learning (FL). This endeavor seeks to curtail computational and communication demands, ultimately fostering improved outcomes. The user discusses introducing a secure aggregation protocol that utilizes homomorphic encryption and differential privacy. The findings in reference [
21] show better performance than several other strategies already in use. This strategy is suitable and efficient for classifying COVID-19 chest X-rays. Reference [
25] demonstrated that FL could aggregate COVID-19 information from many participating centers while respecting patient privacy. Recently, it has been demonstrated that other fields, such as cloud computing, can benefit from FL. The authors in [
24] proposed a method for identifying CT scans of COVID-19 patients utilizing data from several hospitals to create a precise collaborative model.
The COVID-19 epidemic is one of the significant medical disasters that the entire world is undergoing. Reference [
26] presents a collaborative FL model that enables COVID-19 detection screening from CXR (chest X-ray) pictures by numerous medical institutions. COVID-19 was classified as a pandemic by the World Health Organization in March 2020. To slow the pandemic’s spread, practical testing is essential. In this regard, ref. [
27] examines recent issues using DL and FL approaches for COVID-19 detection, focusing on that. One of the biggest challenges in detecting COVID-19 is the virus’s quick spread and the need for valid testing models. For clinicians, this issue continues to be the most significant burden. The study presented in [
28] examined reducing medical imaging analytic techniques in prediction, e-treatment, and data transfer to identify diseases as early as feasible. The outcomes reveal that the proposed approach did best in accuracy and precision. In [
29], a proposed model focuses on COVID-19 data and creates a more accurate COVID-19 diagnosis model based on patient symptoms. The data are then reanalyzed using ML, and a computational model for determining if a person has COVID-19 based only on clinical data is be created. Taking advantage of FL’s quick, centrally managed experiment start and enhanced data traceability and assessing the impact of algorithmic adjustments, ref. [
3] used FL to improve the prediction accuracy of the websites of testing sets.
The image classification job examines an algorithm’s capacity to identify the image’s constituent elements without necessarily locating them. This study addresses the establishment of a standardized dataset and the ensuing possibilities in object recognition that emerge as a consequence. In a related work by Dou et al. [
14], the authors detailed the extensive data collection procedure of ILSVRC (ImageNet Large Scale Visual Recognition Challenge), presented a summary, and analyzed data and failure modes pertinent to these algorithms. The challenges of procuring comprehensive ground truth annotations are discussed. At the same time, noteworthy strides in categorical object recognition are underscored. “Object recognition” is employed inclusively to encompass both image classification and object identification. Similarly, in [
10], the authors aim to create a reliable, transferable model to help with patient triage. They predicted that it would surpass regional models and more truly describe healthcare systems. On the test data for each customer, they compared the trained local models with the global FL model.
Due to substantial uncertainty and insufficient availability of requisite data, standard models have exhibited restricted accuracy in long-term prediction. Several attempts to tackle this problem have been made in the literature; however, optimizing current models’ essential applicability and robustness abilities is still important. To predict the detection of the COVID-19 outbreak, the authors in [
3] compare ML and computing models. To gain knowledge about the probable progression and impact of infections, having access to accurate outbreak estimation methods is essential. They examine and contrast ML and ML methods to forecast the COVID-19 outbreak.
3. Methodology
This section outlines the methodology for identifying COVID-19 chest X-ray images while excluding non-infected instances. Following a formal introduction, we delve into a comprehensive overview of our proposed approach: including the architecture of the ML model integrated with FL, the model training process at the client end, and the mechanism for aggregating trained models on the server.
3.1. Formal Description of the Proposed Framework
The proposed framework is tailored for classifying chest X-ray images into infected and non-infected categories. The foundation of this approach is the FedAvg algorithm, which facilitates efficient distributed training while safeguarding data privacy. Let N denote the total number of participating clients in the FL process.
Let be the global model parameters, and represents the local model parameters of client i ().
- 1.
Initialization: The central server initiates the global model parameters and disseminates them to a subset of clients , where k signifies the communication round index.
- 2.
Local Training: Each client, denoted as
i, undertakes local training utilizing its own dataset. The objective of this training is to minimize the local loss,
, which is a function of the model parameters. In this context,
represents the current model parameters for client
i. The process of minimizing the local loss is represented mathematically in Equation (
1) as follows:
In the above equation, represents the updated model parameters for client i after the local training. These are obtained by finding the argument that minimizes the local loss function .
- 3.
Model Update: Subsequent to local training, each client i transmits its updated model parameters back to the central server.
- 4.
Aggregation: The central server aggregates the received local model parameters via weighted averaging, as depicted in Equation (
2):
3.2. Deep CNN Model
The classification model accepts an X-ray image
X as input and generates the probability
of being COVID-19 infected. Let
denote the CNN model’s mapping function parameterized by
, as expressed in Equation (
3):
3.3. Overall Model Training Objective
Define the combined loss function using Equation (
4):
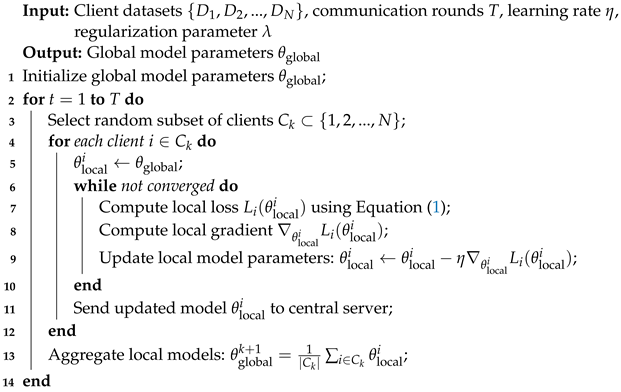
Algorithm 1 starts with global model parameter initialization. A random subset of clients is selected over a set number of communication rounds. For each client, local training is executed iteratively until convergence, adjusting their local model parameters using stochastic gradient descent. The updated local models are aggregated at the central server through weighted averaging. The deep Convolutional Neural Network (CNN) model processes X-ray images, and post-processing techniques, such as lung segmentation and data augmentation, enhance robustness. The overall training objective involves a combined loss function incorporating local and CNN classification loss. Implementation details, results, and conclusions provide further insights into the methodology’s effectiveness.
| Algorithm 1: Federated Learning for COVID-19 classification. |
![Electronics 12 04074 i001]() |
3.4. Proposed Framework Overview
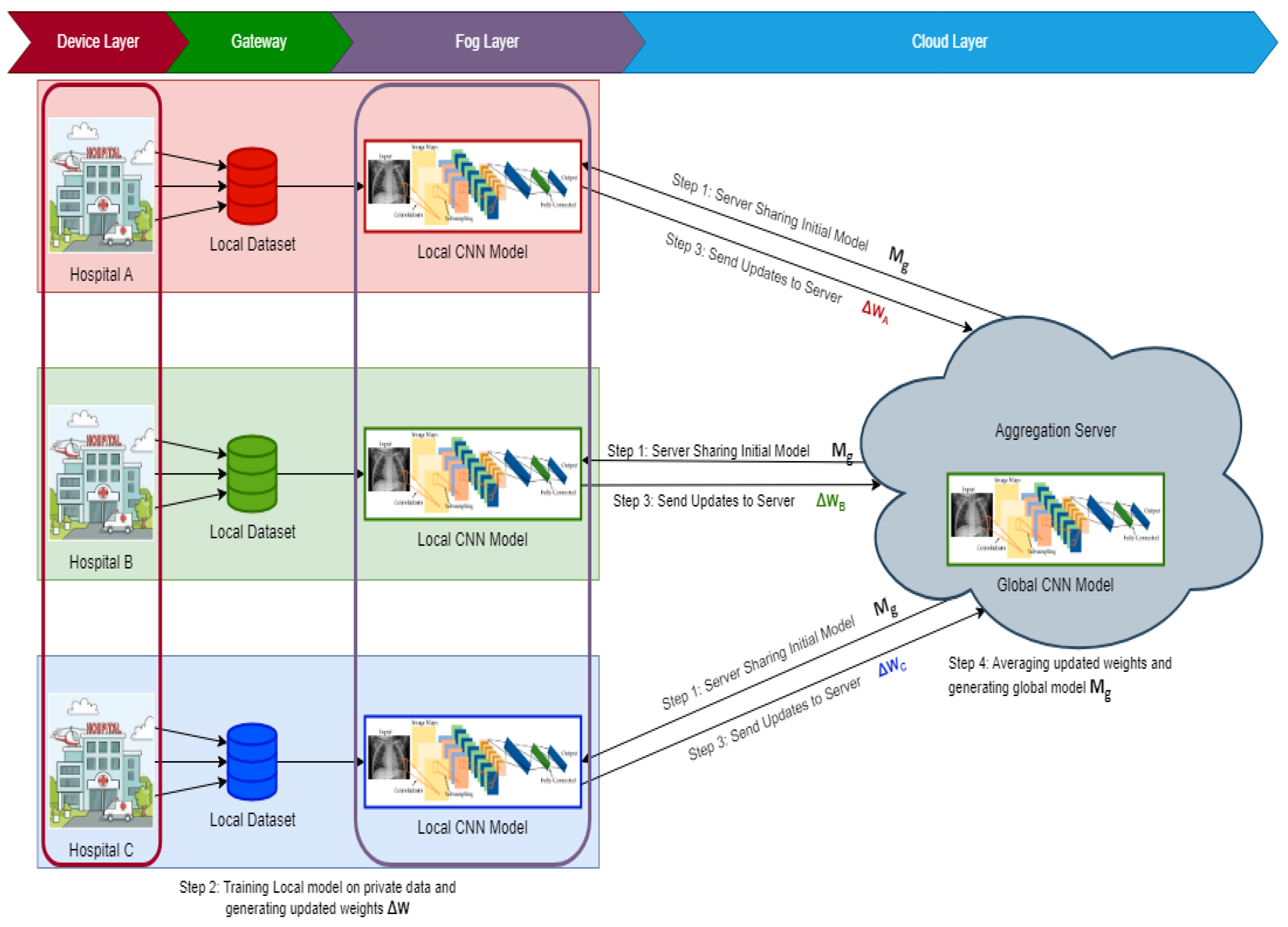
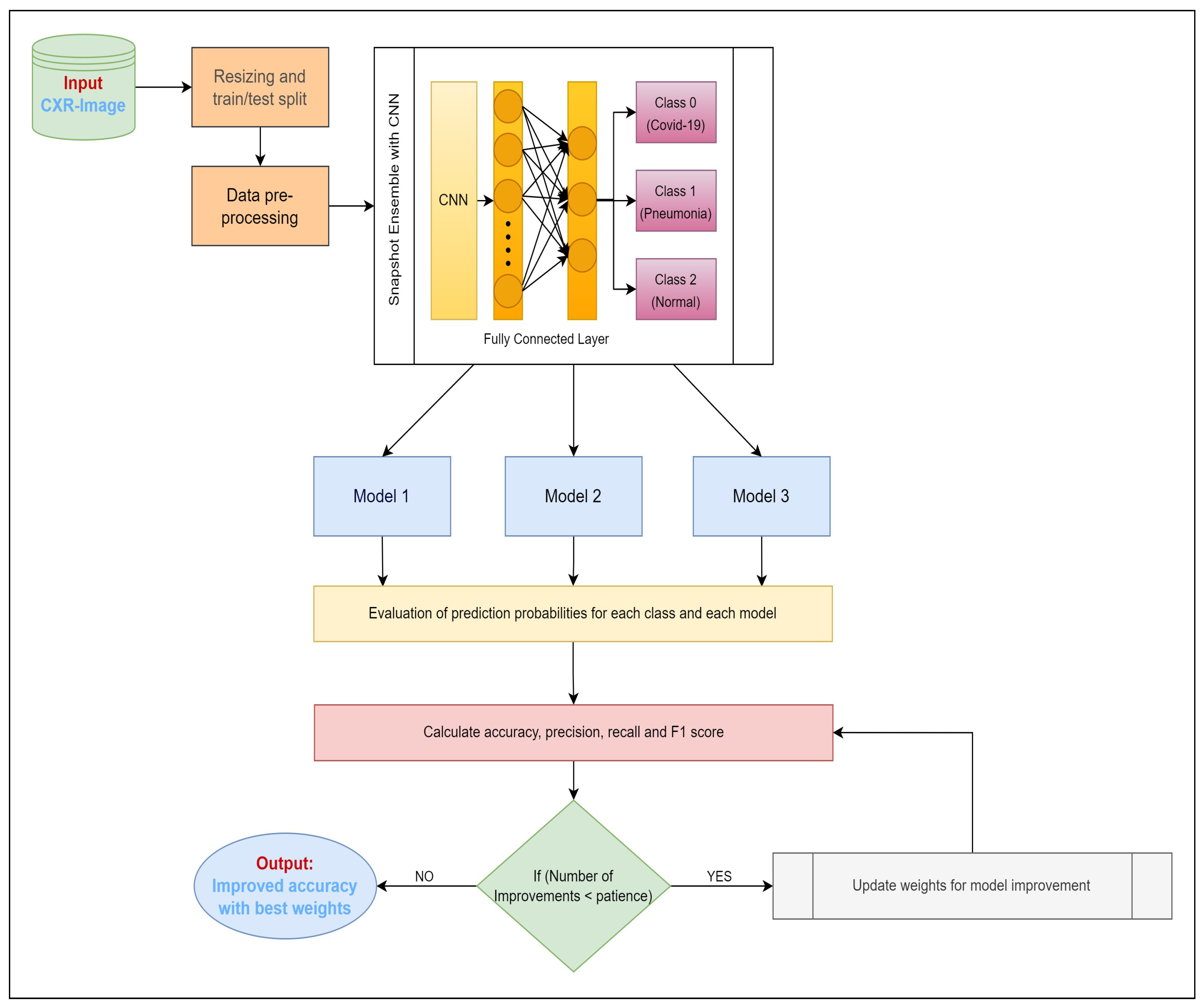
To address the task of differentiating images capturing distinct health conditions, our approach revolves around exploring an FL structure rooted in a client–server setup (depicted in
Figure 1). This architecture is underpinned by the FedAvg algorithm, which is renowned for its pivotal role. FedAvg operates as a communication-efficient mechanism within distributed training, seamlessly integrating with localized models. In this context, localized models maintain their data locally to uphold privacy, while a central server facilitates communication between these localized counterparts. This framework hinges upon a central parameter server tasked with orchestrating client alterations and overseeing a standardized global model. Through collaborative efforts, clientele leverage their distinctive datasets to formulate a robust model. Our proposition entails the design of an intricate CNN model primed to undertake tasks encompassing classification and feature extraction, particularly within the realm of chest imagery analysis. The model’s capacity to gauge the likelihood of infection is grounded in scrutinizing input images. Elaborated specifications regarding this CNN model’s architecture are presented in
Section 3.5. The learning phase of this CNN model encompasses multiple communication cycles between clients and the central server. Before the commencement of training sessions, the CNN model is initialized with random base weights. We assume that each privately stored local X-ray image is accessible to clients. Each communication cycle comprises four distinct steps:
- Step 1:
A global central architecture is maintained on the central aggregation server, which is initially endowed with weights. These weights are then broadcast to a subset of client devices selected at random (e.g., hospitals).
- Step 2:
Subsequent to receiving the initial parameters, each client embarks on training using a subset of its data via mini-batch stochastic gradient descent (SGD) over multiple epochs. This endeavor aims to minimize local gradients. Notably, the convergence of each local model transpires at varying epoch counts. Clients iteratively refine their models by minimizing classification loss via categorical cross-entropy.
- Step 3:
Users stationed at the client nodes update the server upon completion of their respective local training iterations (involving SGD across epochs on local data points).
- Step 4:
After collecting data from all participating nodes and calculating an average model, the central server updates the global model parameters.
A single Federated Learning (FL) round for our CNN model entails these four phases. This sequence of actions is then repeated iteratively (across rounds). The server dispatches the updated global model parameters from the preceding round during each new round. Furthermore, if an abundant number of clients are available, the subset of participating clients can be modified between rounds.
3.5. Preprocessing, Model Training, and Postprocessing
The Hounsfield units for each volume cut for preprocessing before their values show at [1.0, 1.0]. Experimental observations show that standardizing the data to have a mean of zero and unit variance and using individual volume statistics rather than global dataset statistics helps improve the model’s generality. Furthermore, using three neighboring slices as input for convolutional neural models after normalizing the data yields better results. Every local client used the dataset to improve his or her model for one epoch during each round of FL. All local clients used the Adam optimizer, which had a learning rate of
, 0.9, 0.999, and
for beta1, beta2, and epsilon. Following the analysis of the biases of the dataset, we observed that the classification algorithms are mainly based on the pixels at the edge of the image to make their decision. Therefore, we aim to remove these edges and focus only on the lungs with the information needed for the classification. This required using a U-Net neural network pretrained on CXR images and developed especially for lung segmentation. After segmentation, the images were cropped around the lungs with a margin of 10 pixels. The homogenized and cropped images were then used to build our new dataset for training and testing the DL model [
30].
A variety of data augmentation techniques, such as random horizontal and vertical flips (with a probability of 50%), lucky clockwise rotations (with an amplitude ranging from 0.1 to 0.1), random horizontal and vertical translations (with a range of 0.1 to 0.1 of the input image’s length or width), and random shear and scaling were applied to the training data (with a range of 0.1 to 0.1). Different Python libraries were used for data preprocessing before giving the data to the model for training. In this research work, we also utilized some of those libraries. Panda and Numpy libraries were used for data cleaning. Panda, Numpy, and Matplotlib libraries were used for data processing. SKlearn was also used for data augmentation purposes. The Numpy and TensorFlow programming frameworks were used in order to put these strategies into action. The Keras library facilitated accessing different layers of models for classification. A relatively small piece of the training data was extracted at each process step to assess the model’s performance. If the global performance of the model on the local test dataset did not improve for five federated rounds in a row, we considered the training to have collapsed and the FL to have come to an end. The deep CNNs were built using a single NVIDIA TitanXp GPU (graphics processing unit).
During the post-processing phase, we used non-maximum suppression30, a method prevalent in image processing, to separate overlapping elements. We separated an overlapping series of frames using non-maximum suppression to create bounding boxes with the greatest anticipated probability. For more accuracy, we eliminated any bounding boxes in an image with a chance lower than a certain threshold, which was determined by looking at a collection of projected bounding boxes. Out of the remaining bounding boxes, we chose the one with the greatest probability and eliminated those with an Intersection over Union (IoU) value larger than 0.5. In addition, we used an open-source AI model for lung segmentation31 to eliminate any false-positive detections that occurred outside of the lung area.
3.6. Model Architecture
This research suggests a collaborative and decentralized architecture for chest X-ray imaging-based COVID-19 screening. We aim to demonstrate that a deep CNN model can perform federated learning to benefit from rich private data interchange while upholding privacy. The specific design of the CNN is not the focus of this work because many different architectural solutions could only slightly improve or worsen the overall performance. The information exchange flow between the central server and federated clients is shown in
Figure 2. The parts that follow give information on these two parties as discussed subsequently:
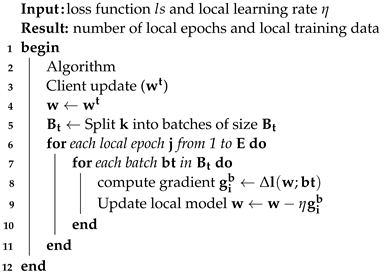
3.6.1. Client-Side Model Update
On the client side, training is carried out since every federated client has access to the same dataset and has the computing capacity to carry out mini-batch Stochastic Gradient Descent (SGD). Clients whose CNN architecture and loss functions are the same are ignored. Algorithm 2 provides the steps used in training. At the beginning of the round, a copy of the global model is sent to each client’s computer so that it may be initialized. Once the client has finished the same number of iterations as there are local epochs, they compute a gradient update to generate the new model, which is then shared with the aggregation server. During this training process, the local data for each customer are treated as private information and are never discussed with anyone else. This describes the procedure done on the client side. The client side can be stated as local algorithms working on Hospitals A, B, and C with local datasets. On the client side, the model is trained, and weights are sent to the global model. During training, dataset Pk is divided into equal sizes of batch B. For each batch B, the gradient is computed, and the weight of the local model is updated.
| Algorithm 2: Client-side algorithm. |
![Electronics 12 04074 i002]() |
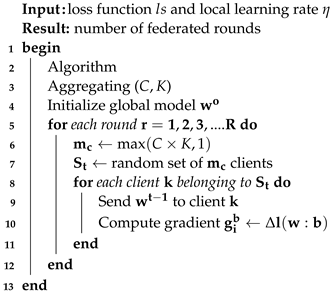
3.6.2. Server-Side Model Aggregation
An aggregation server is responsible for managing the global model. This server also ensures that each participating client receives the initial model and tracks the progression of the training. The server receives synchronized updates from all clients at each federated round and utilizes these changes to generate a new model with updated parameters following Equation (
2). Algorithm 3 describes the procedure done on the server side. The server side can be stated as a global algorithm working on Hospitals A, B, and C with the global dataset. The model is trained on the server side, and weights are sent to the local models. During training, weights are updated after each round t, i.e., 1, 2, 3, and so on. At the end, the gradient value for the global model is computed.
| Algorithm 3: Server-side algorithm. |
![Electronics 12 04074 i003]() |
4. Findings and Discussion
This section presents an evaluation, and the outcomes of our suggested paradigm are described. The performance of our model was evaluated by simulation. The experimentation setup used to evaluate and train the model is also elaborated on here. Results show that our model is better regarding accuracy, precision, and recall.
4.1. Description of the Dataset
The dataset used in this study comprises 2473 X-ray images from the COVID-19 CXR dataset. This dataset is sourced from the COVID-19 Radiography Database, available on Kaggle. It includes 1345 viral pneumonia images, 219 COVID-19 images, and 1341 regular CXR images. The detailed dataset composition is presented in the following table.
4.2. COVID-19 Chest X-ray Database
A collaborative effort between researchers from the University of Qatar, Doha, Qatar; Dhaka University, Bangladesh; Pakistan; and Malaysia resulted in a chest X-ray image dataset. This dataset includes positive samples of COVID-19, regular X-rays, and viral and bacterial pneumonia images. The dataset was released in multiple phases. The initial release contained 219 COVID-19, 1341 regular, and 1345 viral chest X-ray images. The COVID-19 class has since been expanded to 1200 CXR images. In a subsequent update, the dataset was expanded further with the addition of 3616 COVID-19-positive cases, 10,192 normal cases, 6012 lung opacity non-COVID infections, and 1345 viral influenza images. Regular updates are performed to incorporate new X-ray images of COVID-19 pneumonia cases as they become available. The details of datasets are provided in
Table 1, and the dataset distribution is outlined in
Table 2.
4.3. Experimentation Setup
In opposition to previous models built using TensorFlow version 1.8, the suggested model was established with open-sourced TensorFlow federation version 2.1.0, with Keras providing the backend in both cases. The use of TensorFlow 1.8 for previous models was primarily based on the existing infrastructure and legacy models already built using that version. However, TensorFlow 2.1.0, which we used for our proposed model, offers several improvements over the older versions. It brings tighter Keras integration, easier model exporting for deployment, and enhanced performance optimizations. These features aligned well with the requirements of our proposed model, prompting us to transition to the newer version. Different Python libraries were used for data preprocessing before giving the data to the model for training. In this research work, we also utilized some of those libraries. Panda and Numpy libraries were used for data cleaning. Panda, Numpy, and Matplotlib libraries were used for data processing. SKlearn was also used for data augmentation purposes. The Numpy and TensorFlow programming frameworks were used in order to put these strategies into action. The Keras library facilitated accessing different layers of models for classification. The experiment was run on a PC desktop with an Intel i5 6th generation CPU, an NVIDIA Titan GPU, 16 GB RAM, and a 3.5 GHz speed control. Each dataset was split into three portions: 70% for instruction, 20% for evaluation, and 10% for validation. The proposed system was trained using TensorFlow federation version 2.1.0, and the other classic model was trained using TensorFlow version 1.8; both used 70% of the three databases: 23,804, 31,501, and 2443 pictures. Python 3.9.1 was used to create these models.
Table 3 summarizes the parameters and their values used in this experiment.
4.4. Evaluation Metrics
In this part, the discussion of the evaluation metrics is given and offers a thorough analysis of the outcomes. The classified accuracy uses statistics for evaluating the classification of the model. It is calculated by dividing the total number of examples (images) in the database under examination by the set of instances (images) that were correctly categorized. The numerical equivalent is:
Here, stands for true positive, and the number of medical images properly defined as true is counted. stands for false positive and is a count of medical photos wrongly defined as false. stands for true negative and is the count of correct medical images identified as true, and stands for false negative. It is the count of medical images incorrectly identified as false.
In image classification systems, recall and precision are used for performance measurement. The proportion of correct positive images to all classified images is known as precision. The formula for precision (
) is:
Along with precision (
), performance measurement is accomplished by
Recall. The proportion of correctly categorized images to all images in the database is known as recall. It can be summed up as follows in formula form:
The Harmonic mean of the recall and precision is the
F-score; a greater number indicates that the system has superior predictive capacity. The performance of systems cannot be judged just based on precision or recall. The
F-score can be calculated as follows:
4.5. Accuracy
Figure 3 explains the COVID-19 image classification by the two different models, i.e., the local and the global model. The accuracies of both software configurations varies for different hospital models. As the figure explains, the accuracy of the local model was moderate throughout all hospital models. In contrast, the FL model surpassed the local model in all three hospitals in patient detection regarding accuracy and precision. The figure also shows that the accuracy of the FL model constantly increased in the respective hospitals, and the local model has the lowest accuracy in Model B hospitals.
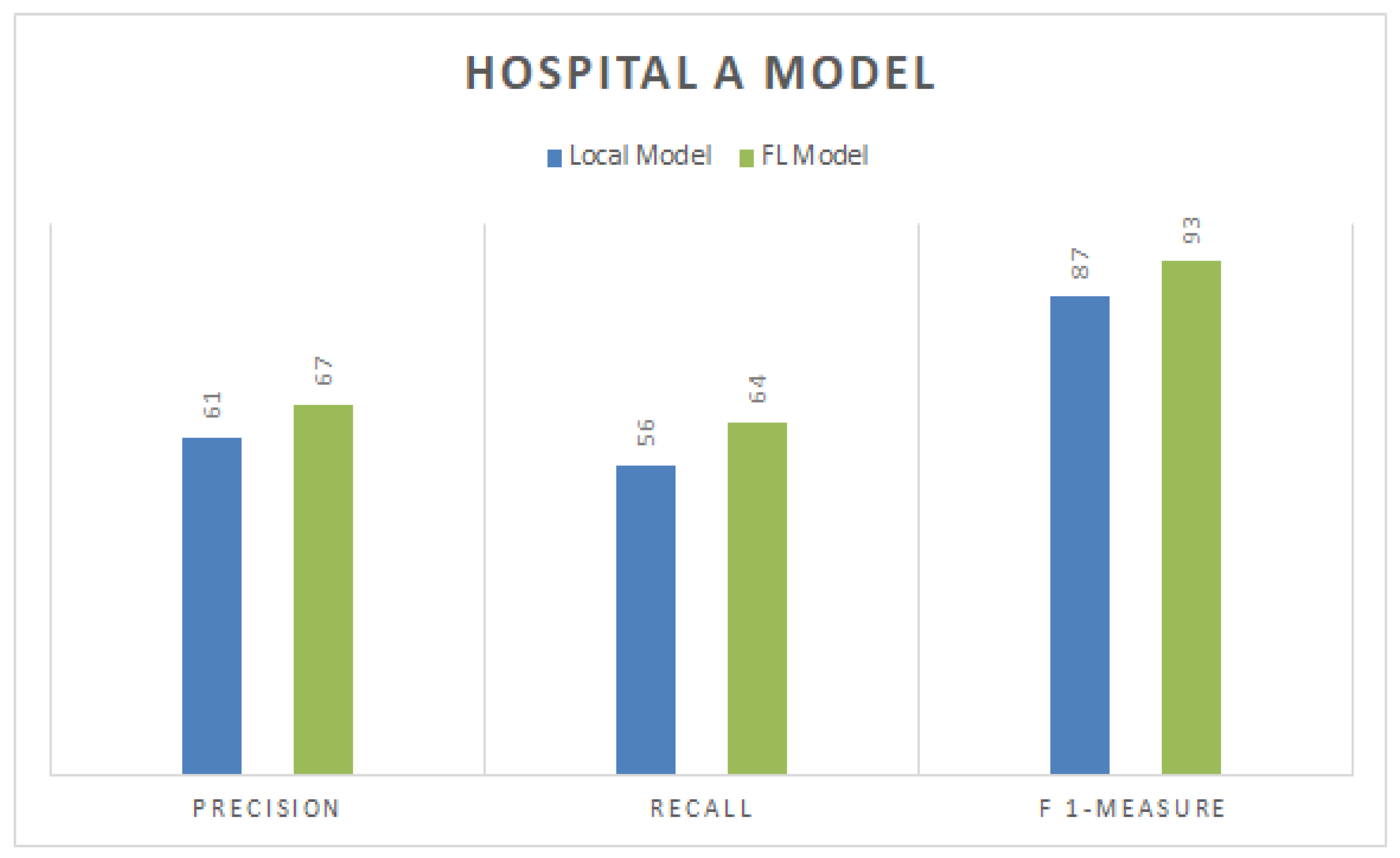
4.6. Precision, Recall, and F1-Measure for Hospital A
Figure 4 represents the precision, recall, and F1-measure values for the local and FL-based models for a particular dataset of Hospital A. The figure shows that the FL-based model scored the best precision, recall, and F1-score, with respective values of 67%, 64%, and 93%. At the same time, other CNN-based local models gained 61%, 56%, and 87% precision, recall, and F1-measure scores, respectively. Models that are based locally often necessitate longer training periods compared to those that employ FL. This is primarily due to the distributed nature of local models, where the data and the computational resources are scattered across multiple devices or locations. This distribution can complicate the process and extend the time required for model training. On the other hand, Federated-Learning-based models have proven to be more efficient. FL allows a model to be trained across multiple decentralized devices or servers, which hold local data samples without exchanging them. It allows for faster, more efficient training because it eliminates the need for data centralization and, therefore, speeds up the entire process. Comparatively, when it comes to efficiency and speed, FL-based models outperform local CNN-based models. The latter often rely on local resources and data, which can limit their performance and extend their training time. Hence, strategies that leverage FL are generally considered superior to those based on a local CNN.
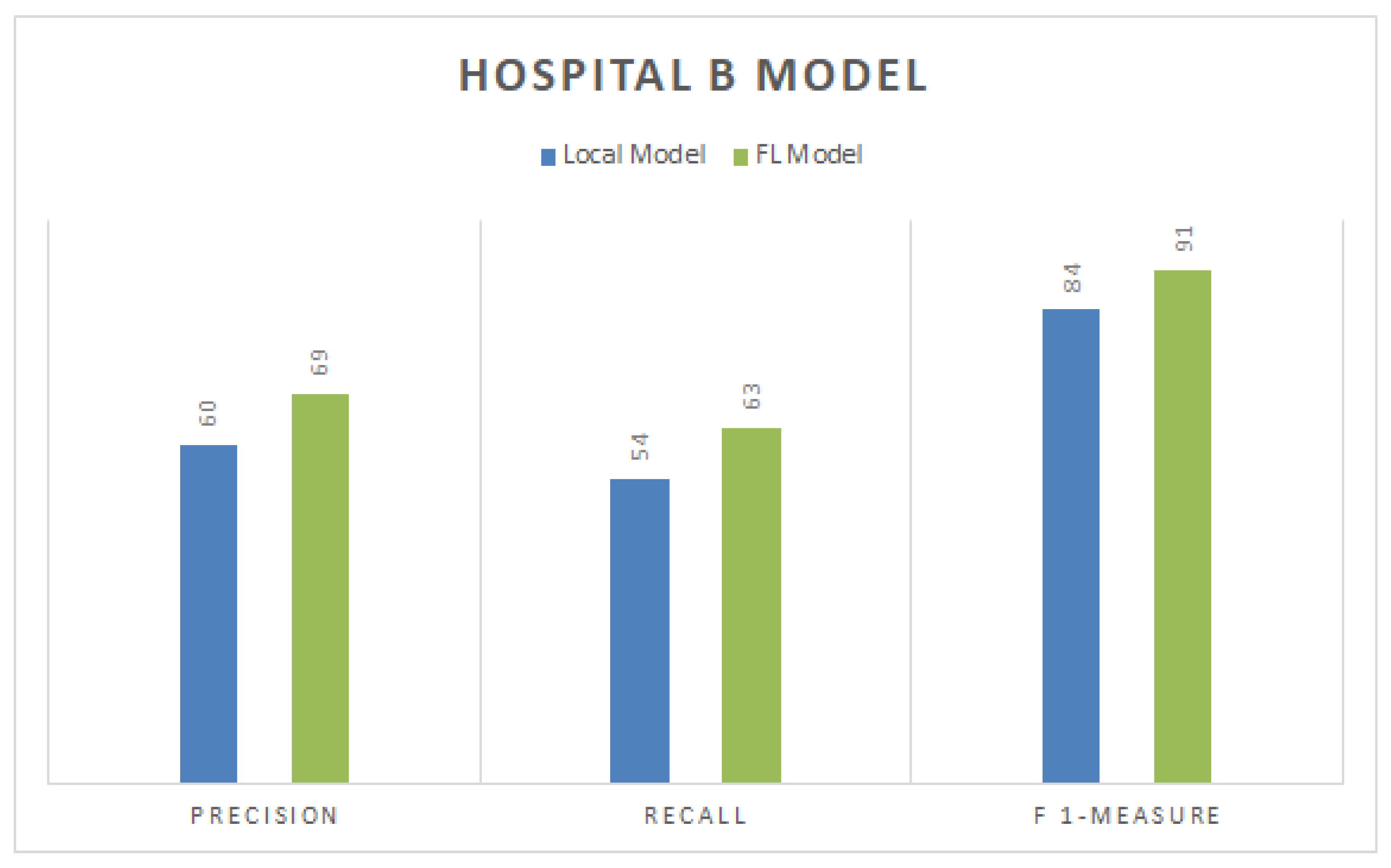
4.7. Precision, Recall, and F1-Measure for Hospital B
Figure 5 represents the precision, recall, and F1-measure values for the local and FL-based models for a particular dataset of Hospital B. The Figure shows that the FL-based model scored the best precision, recall, and F1-score, with respective values of 69%, 63%, and 91%. In contrast, other CNN-based local models gained 60%, 54%, and 84% precision, recall, and F1-measure scores, respectively. Local models also require more time for training than FL-based models due to their distributed nature. This demonstrates that approaches based on FL are more efficient than those based on a local CNN.
4.8. Precision, Recall, and F1-Measure for Hospital C
Figure 6 represents the precision, recall, and F1-measure values for the local and FL-based models for a particular dataset of Hospital C. The figure shows that the FL-based model scored the best precision, recall, and F1-score, with respective values of 71%, 66%, and 94%. In comparison, other CNN-based local models gained 62%, 57%, and 88% precision, recall, and F1-measure scores, respectively. Local models also require more time for training than FL-based models due to their distributed nature. This demonstrates that approaches based on FL are more efficient than those based on a local CNN.
4.9. Confusion Matrix
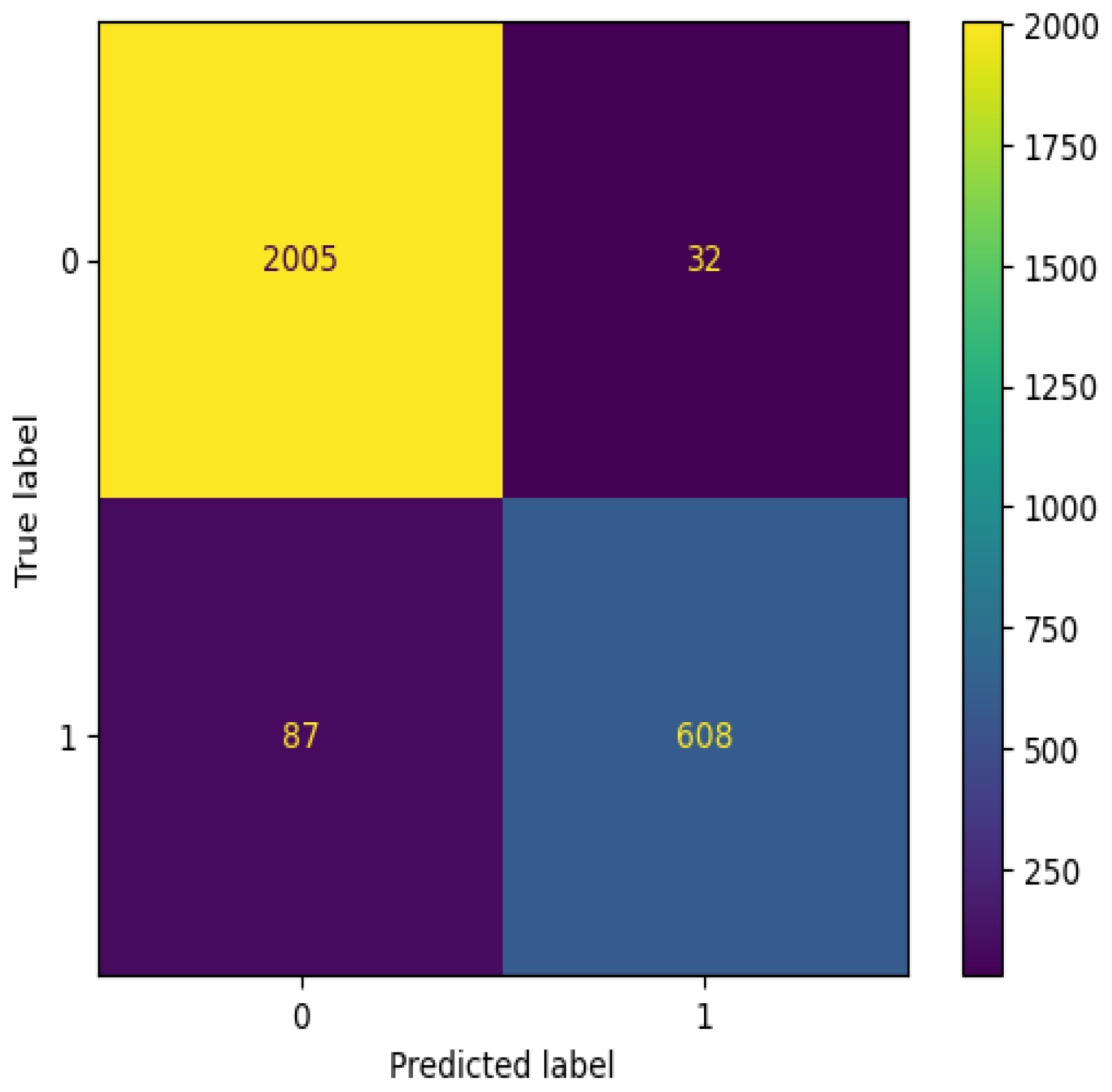
The confusion matrix for the training phase is shown in
Figure 7. The results show that there were 10,946 true positives and 7860 true negatives. On the other hand, there were 291 false positives and 624 false negatives. In contrast, the confusion matrix for the validation phase is shown in
Figure 8. The results show that there were a total of 608 true positives and 2005 true negatives. On the other hand, there were 32 false positives and 87 false negatives. The analysis of our confusion matrix results reveals notable percentage differences between the training and testing (validation) phases, shedding light on how our COVID-19 screening model using chest X-ray images generalizes to unseen data. In the training phase, we achieved a substantial count of 10,946 true positives, indicating the model’s proficiency in correctly identifying COVID-19 cases. However, during the testing phase, the true positive count dropped significantly to 608, representing a percentage difference of approximately 94.44%. This decline suggests a challenge in the model’s ability to generalize effectively to new data, particularly in identifying COVID-19 cases in real-world scenarios.
Conversely, there was a noticeable decrease in false positives during testing, with a percentage difference of around 89.02%. This is a positive development, as it indicates a lower rate of false alarms when applying the model to unseen data. Additionally, the false negatives decreased by approximately 86.10% during testing, implying a reduction in missed diagnoses, which is crucial for patient care. The most pronounced percentage difference is in the true positive rate, underscoring the need for further optimization to enhance the model’s performance on unseen data. These findings highlight the critical importance of rigorous model evaluation and validation, as they offer insights into the model’s generalization capabilities and potential areas for improvement. Future work should focus on fine-tuning the model to achieve more consistent performance between the training and testing phases, ultimately increasing its reliability in real-world healthcare applications.
4.10. Discussion
This research shows that it is possible to aggregate COVID-19 data across detailed listings while respecting patient privacy using FL. During COVID-19, when no time is available to set up complex material around institutions and countries, this decentralized training strategy is a crucial scaling enabler of AI-based technologies. The application of FL has recently been shown in different industries, such as edge computing for digital equipment. Still, diagnostic imaging is getting more complicated. It involves common challenges, such as data with high dimensionality and with improper cohort sizes, that exert untested influences on the current FL methods.
This is the first study showing FL’s feasibility and efficiency for COVID-19 image processing. In this field, teamwork is vital during crises around the world. According to our experimental findings, FL outperformed single-site models and their ensembles regarding generalization ability. This is an example of successful decentralized optimization using various training with multidimensional data. In a solid evidence study, we demonstrate the viability of a convolution neural federated DL system for identifying chest CT problems in COVID-19 disease patients. Significantly, the AI model developed using cohorts of Hong Kong performed well on outer, hidden, independent datasets gathered by institutions in Europe and Asia in addition to internal testing cases.
These international centers used different scanner brands, imaging protocols, and groups of patients, and the study participants’ COVID-19 pneumonia severity varied. The diversity of the data in this comprehensive study shows that it is possible to create reliable and generalized AI techniques for battling COVID-19 through image processing in diverse healthcare situations. Our model may allow proper use in reality as it usually requires 40 ms to assess one CT volume. The unreliability of AI techniques was expected to be improved by multicenter training, which has been proven in various medical rummaging scenarios 25–27. The Individual-2-model trained on 4146 photos of CT slices outperformed the two single-site systems trained on 958 and 660 data points, respectively, in our studies on all three external testing sites. As a result, it was discovered that more extensive training databases could enhance the model’s performance on new datasets.
Furthermore, merging all three main training sites might improve the test accuracy, even if two areas contributed fewer cases. This indicates that larger data scales and the diversity of richer data through imaging scanners and protocols are also crucial for lowering model error and enhancing generalizability. In this view, a collaboration between various clinical institutions is a necessary step in creating AI for broader adoption, particularly in the case of the COVID-19 pandemic, when global cooperation is vital. Despite enrolling patients from seven clinical centers in various regions, each participating institution only had a modest amount of patients. There was an imbalance between the locations due to the pandemic performance management system receiving more cases than other hospitals. This reflected the real-world circumstance that most single sites had difficulty finding enough COVID-19 patients to train their internal AI systems.
Therefore, multicenter research with collaborative data merging efforts is crucial and helpful to handle the COVID-19 long-tail dispersion. More patients and institutions will be involved in FL in upcoming work. However, it is essential to note that the current solid evidence study’s tiny patient population cannot prevent the creation of a DL model due to a sizable number of lesions identified by the CT volumes. The German cohort’s model extended less effectively compared to those of other foreign affiliates. The patient populations’ varied ethnicities could be one factor. The cohort of hand annotations is not directly compatible through training examples due to the diverse lesion annotation processes used at various clinical sites, known in computer vision as idea shift. For instance, the training data noted floor occlusion and consolidating.
In contrast, the German cohort had a small number of pleural effusion lesions—atypical in COVID-19—that contrasted only slightly with healthy lung tissue. There were 15 cases with mild lesions (difficult to visualize in the window of the lung), 5 cases with a diffuse lesion that was inappropriate for detection processes that anticipate lesion bounding boxes, and 1 case was disqualified due to no CT findings. Image characteristic of individuals eliminated from the German cohort are shown in
Figure 3. This allowed the model to maintain 35 examples while achieving an AUC of 88.15% (95% CI 86.38–89.91). The model obtained an AUC of 77.15% (95% CI 72.84–81.47) when tested on all 56 cases of the German data. The tool for CT abnormality detection is meant to be applicable in conjunction with the typical visual assessment performed by qualified radiologists. By providing quantifiable data during clinical decision making, the AI tool helps the expert. The loop expert also serves as a buffer against incorrect predictions, such as false positives in the scanning process, that are not worrisome.
It is crucial to remember a shift in concepts, such as that seen in the cohort of Germans, will prevent upcoming multinational trials that employ FL if each site follows a set protocol. Following a set protocol was not feasible due to the random design of the study, which comprised data from different websites and each cohort’s independent collection of data. Despite these drawbacks, our multinational verification highlights the practical challenges of conducting such research while demonstrating our strategy’s potential. In conclusion, the CNN-based AI model is efficient for detecting CT anomalies of COVID-19 disease patients after being trained using the confidential FL model. The work of AI in delivering less cost and scalable methods for the burden of lesion estimation for enhanced treatment of clinical diseases is demonstrated by the overall unreliability of regional and worldwide cohorts as external, which is beneficial by adding various datasets.
Despite the significant contributions made with the fog-based FL model introduced in this study, there are a few constraints to consider. Primarily, the model’s effectiveness is heavily reliant on both the quality and quantity of data provided by the participating nodes. If the data are insufficient or skewed, this could impact the model’s accuracy. Another constraint lies in the decentralized nature of the FL model itself. While this decentralization offers enhanced data privacy, it also introduces complexity in terms of coordination and synchronization among nodes. These complexities could result in increased computational time and resource usage. It is also worth noting that the model’s performance may vary based on different datasets and real-world scenarios, which further underlines the importance of robust and diverse data sources. Future research should aim to address these limitations to further improve the performance and applicability of the FL model.