Integrating FPGA Acceleration in the DNAssim Framework for Faster DNA-Based Data Storage Simulations
Abstract
:1. Introduction
- We have developed a DNA storage simulation (DNAssim) platform to enable a fast full-design exploration of the synthesis and sequencing technologies in the context of storing digital information inside a DNA strand. The tool is entirely built in the Python/C language and features a proprietary Graphical User Interface (GUI). To the best of our knowledge, this is the first framework oriented to the study of DNA-based storage that includes all the steps of the pipeline within a single tool.
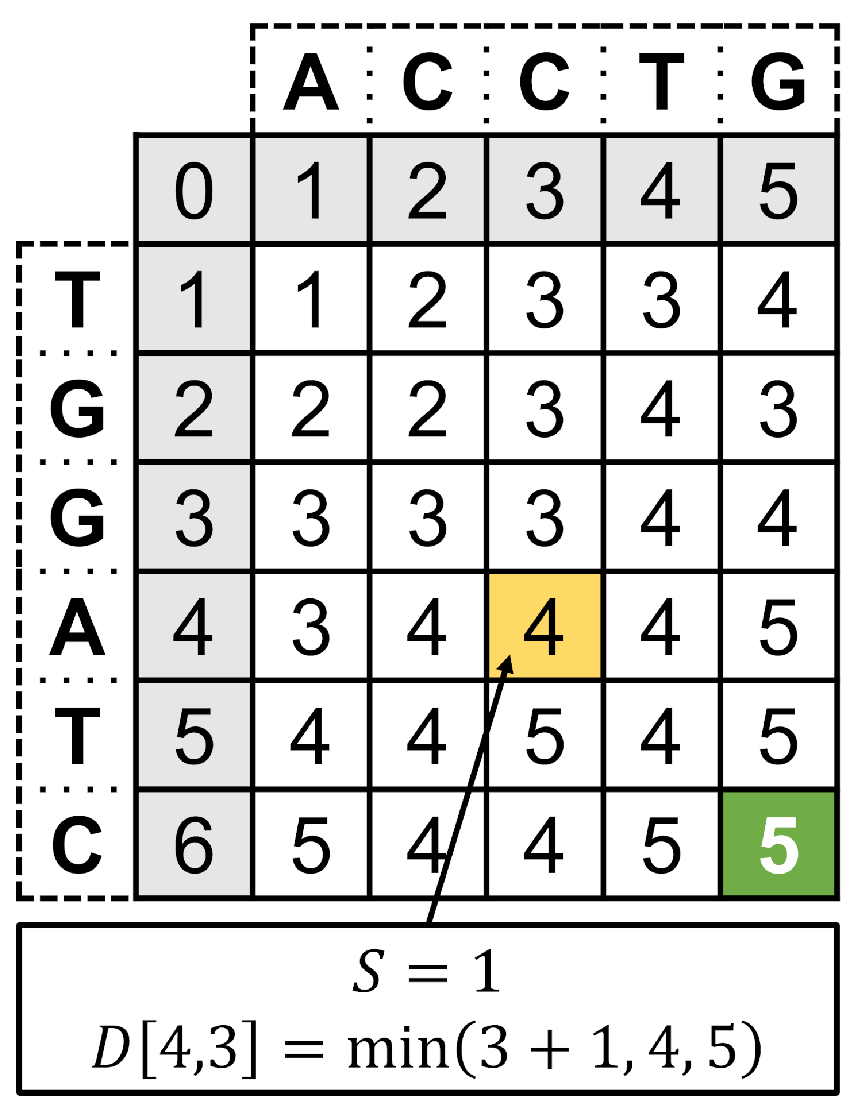
- In the storage pipeline simulation, we identified a possible performance bottleneck in the calculation of the edit distance, which is a similarity metric between DNA strands appearing both in the modeling of the DNA storage noise channel and in the information decoding steps.
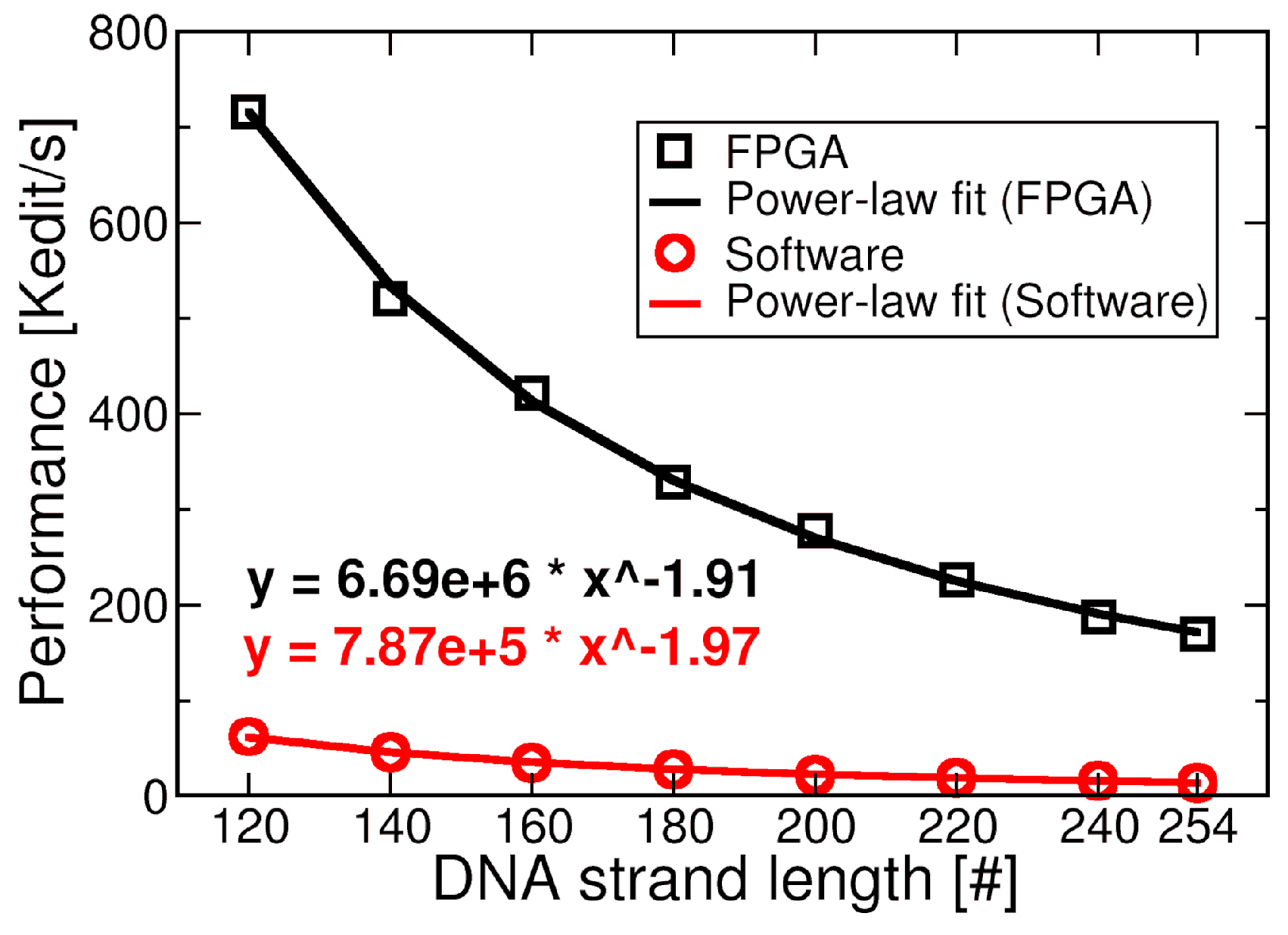
- To this extent, we developed a custom acceleration engine based on a Xilinx VC707 Field Programmable Gate Array (FPGA) that improves edit distance execution with evident advantages in the simulation chain. The accelerator improves the performance with respect to a software counterpart by up to 11 times (700 kedit/s) and consumes up to 7.46 W with a clock frequency of 170 MHz for the computational blocks.
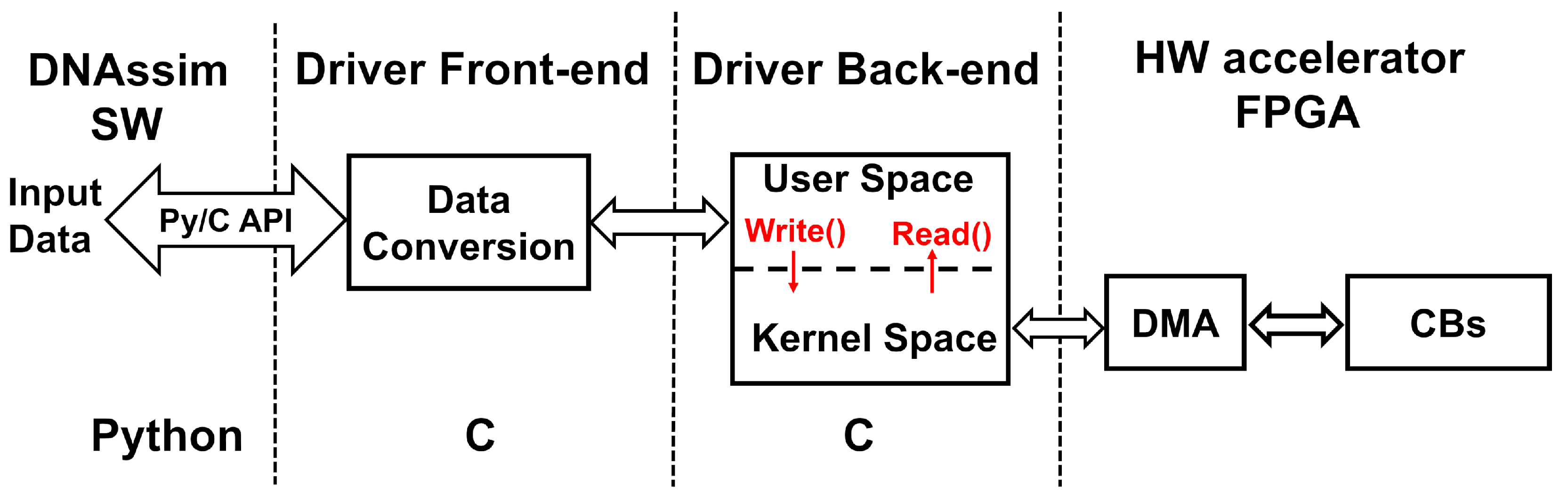
- The accelerator has been integrated with the DNAssim framework by developing a custom driver in the C language that provides data conversion from the software tool and transfers them to the FPGA for subsequent edit distance calculation using the PCIe gen2 protocol [20].
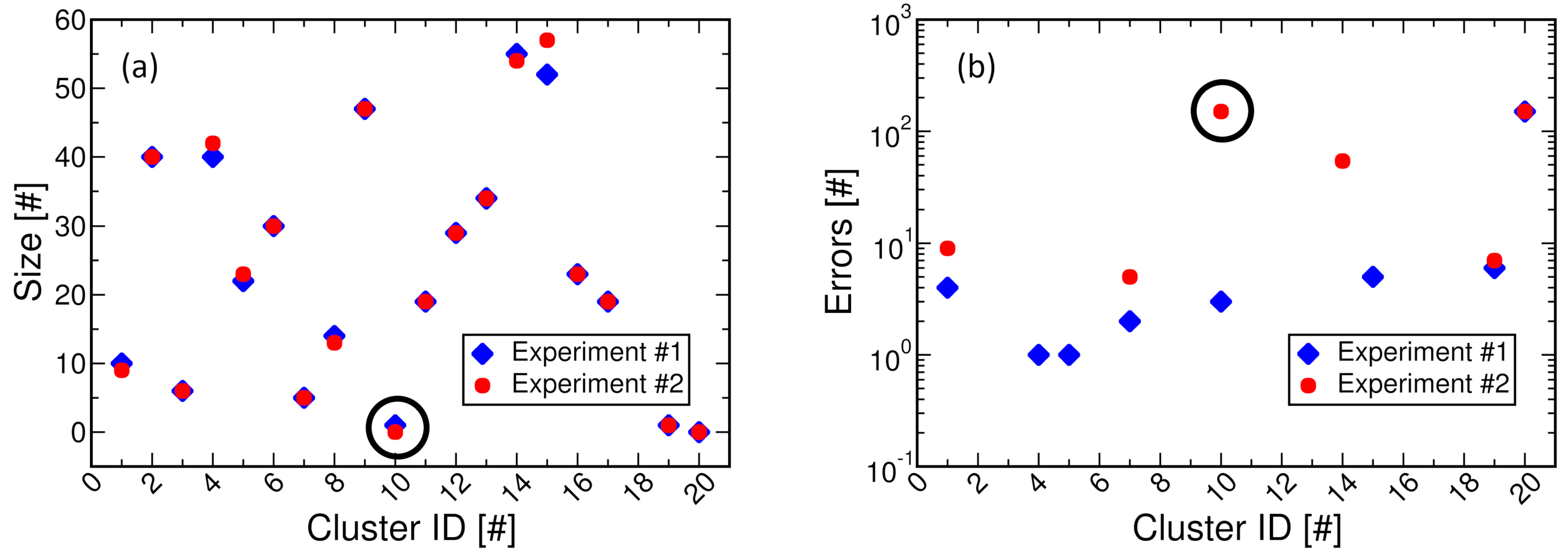
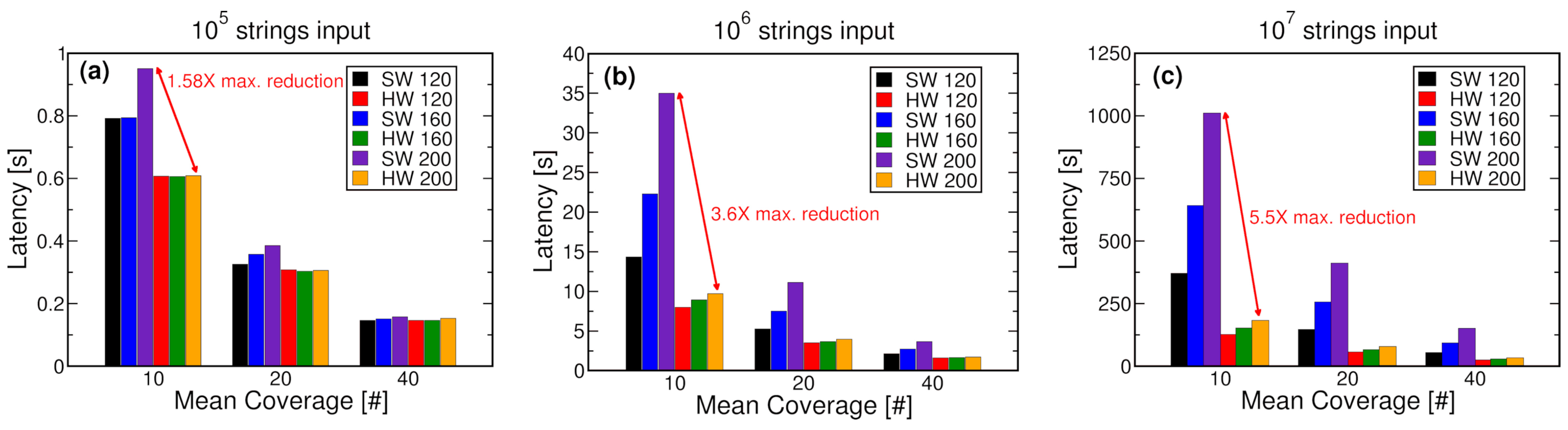
- We have validated the hardware-software co-simulation approach in the clustering operation performed during the DNA storage decoding steps. The experimental results demonstrated a simulation latency reduction of up 5.5 times with respect to a pure software approach. Further, we have projected the simulation speed-ups achievable on real use cases by demonstrating a simulation time reduction of up to 4.2 times when considering the storage of a music file on the DNA.
2. Related Works
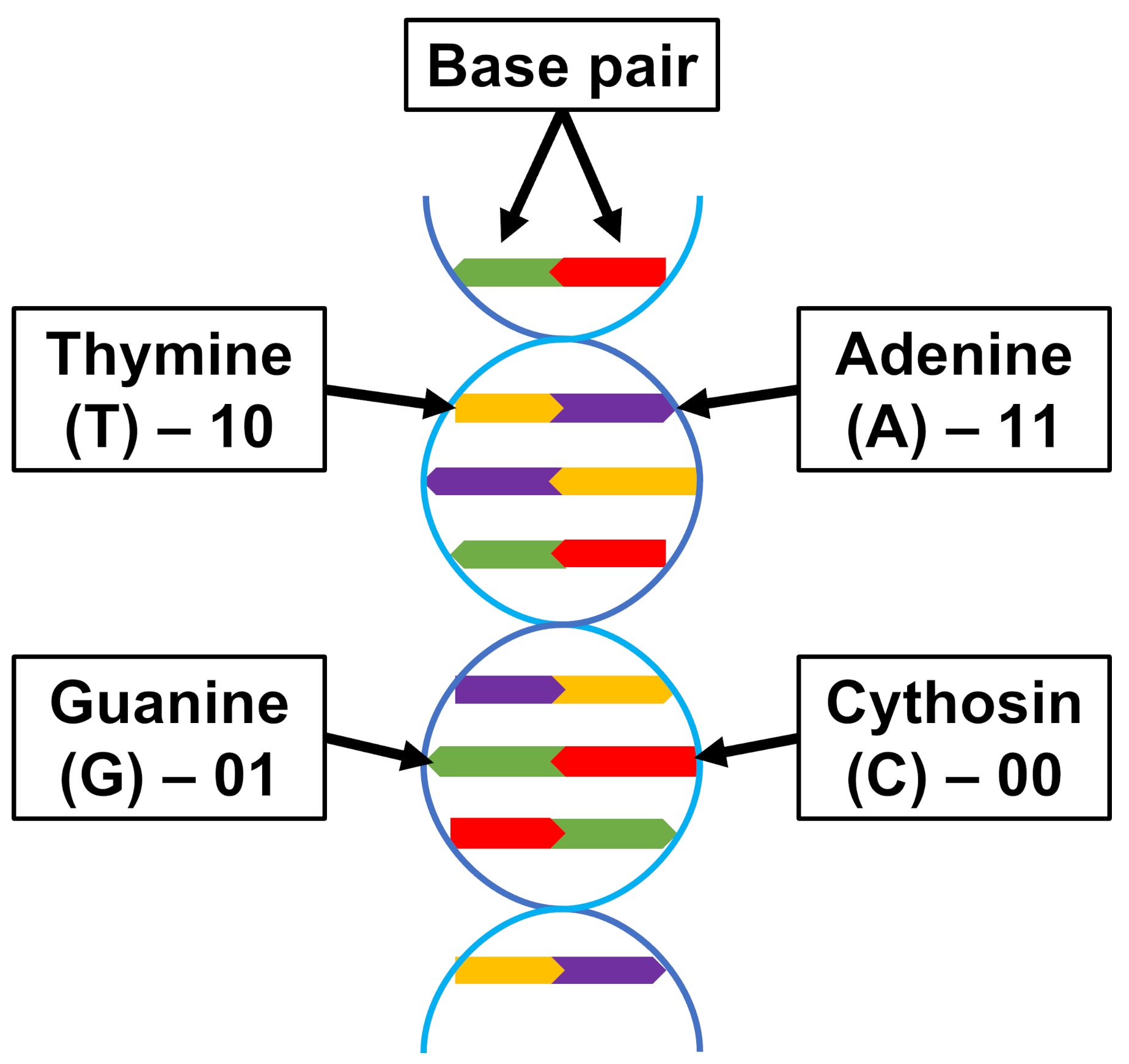
3. The DNA-Based Storage Simulation Engine
3.1. Encoder Blocks
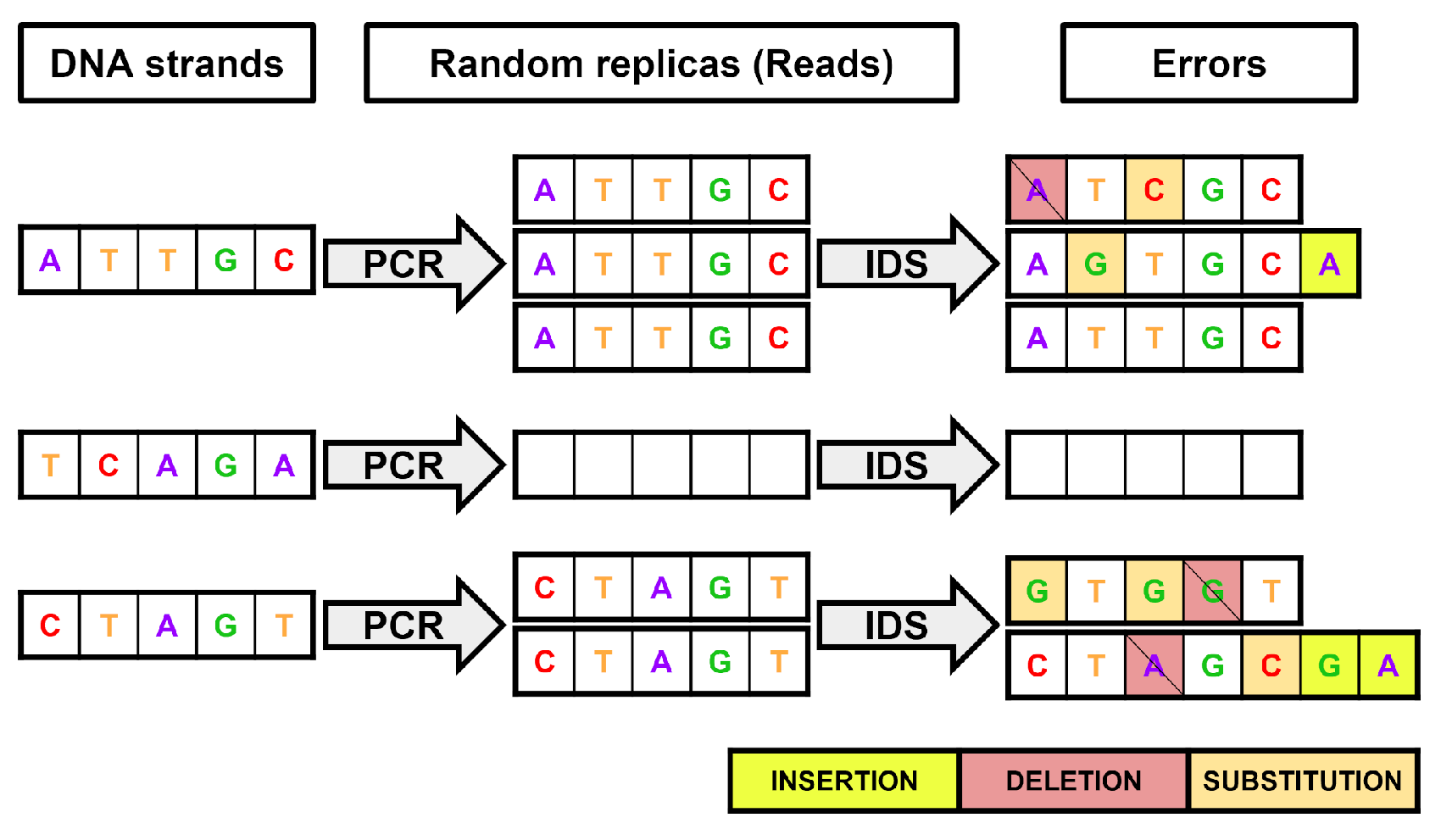
3.2. Noise Model of the Channel
3.3. Decoder Blocks
- Pick a random element (a string) of the cluster and call it x;
- Evaluate which sequence in the enumeration appears first in x;
- Return that sequence followed by the next l character of x after the sequence.
| Algorithm 1 Clustering algorithm |
| Ensure: |
| for all do |
| pick random permutation |
| for all clusters c do |
| pick random element and hash it |
| for all pairs : do |
| if then |
| merge and |
| end if |
| end for |
| end for |
| end for |
3.4. Qualitative Evaluation of DNAssim
4. FPGA Hardware Acceleration of the Edit Distance Computation
4.1. Hardware Design
4.1.1. Implementation of CBs and the Overall Framework
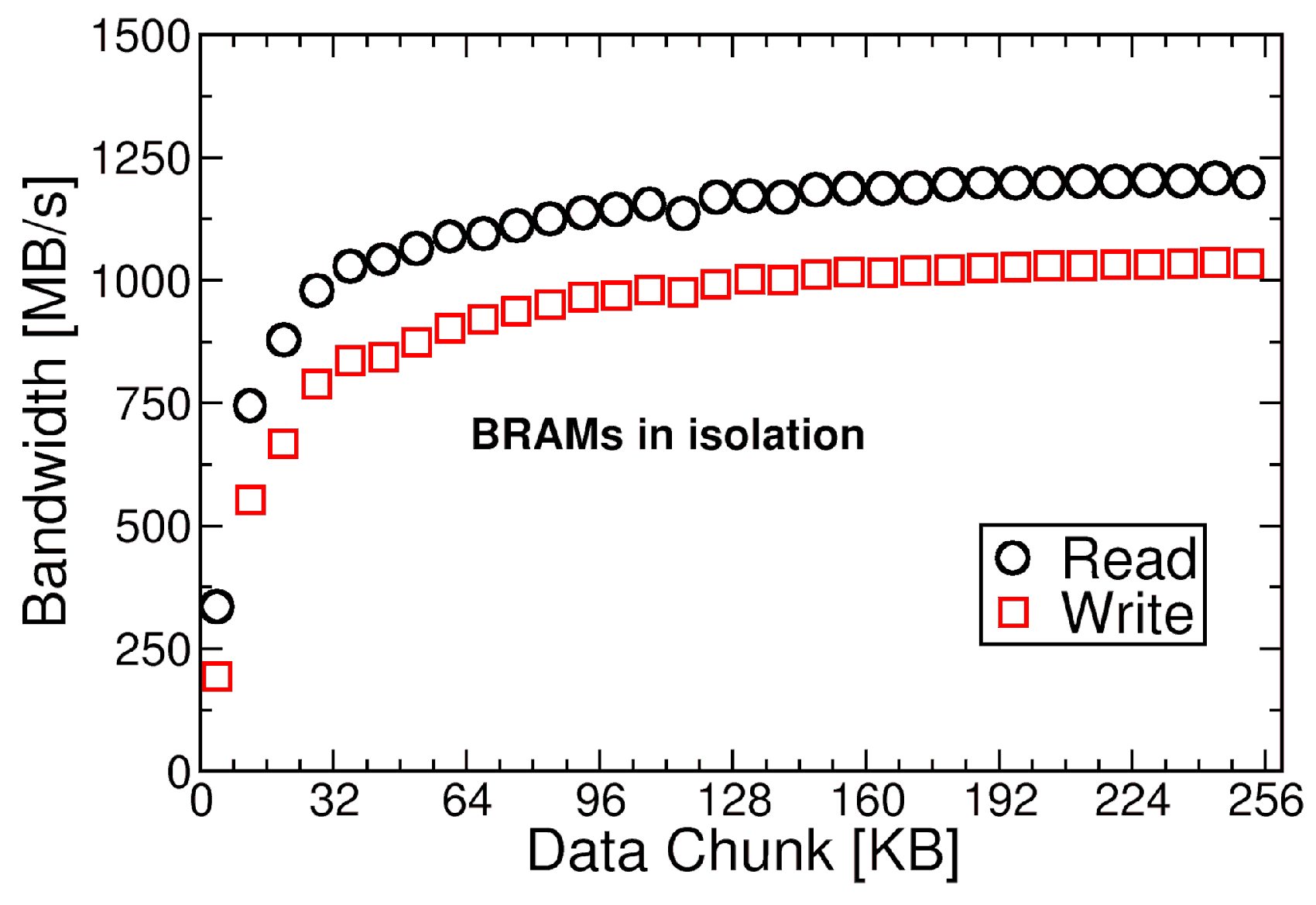
4.1.2. BRAMs Design for Data Transfers
4.1.3. Full Design Results
4.2. Software Driver Design
4.2.1. Front-End
4.2.2. Back-End
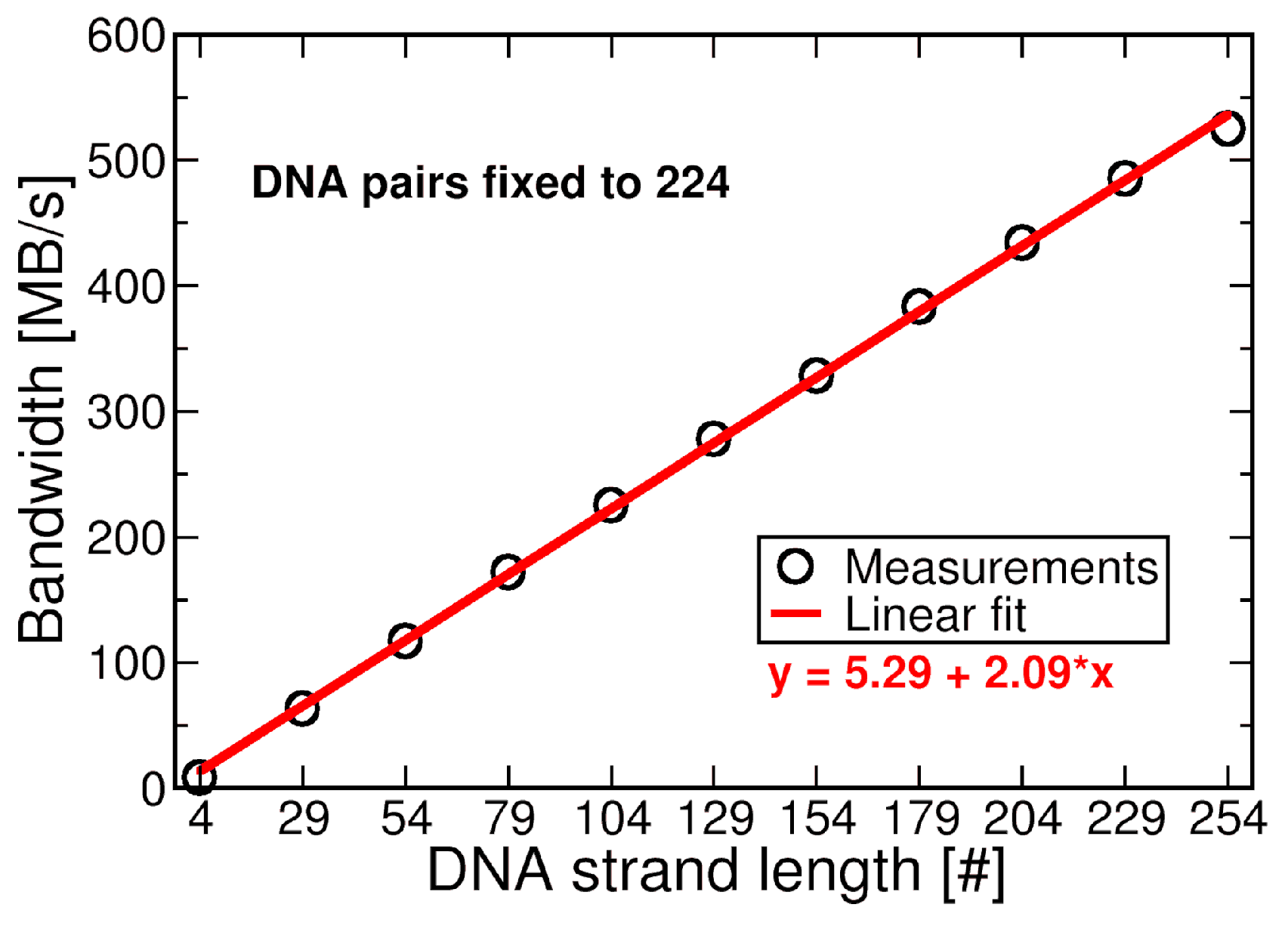
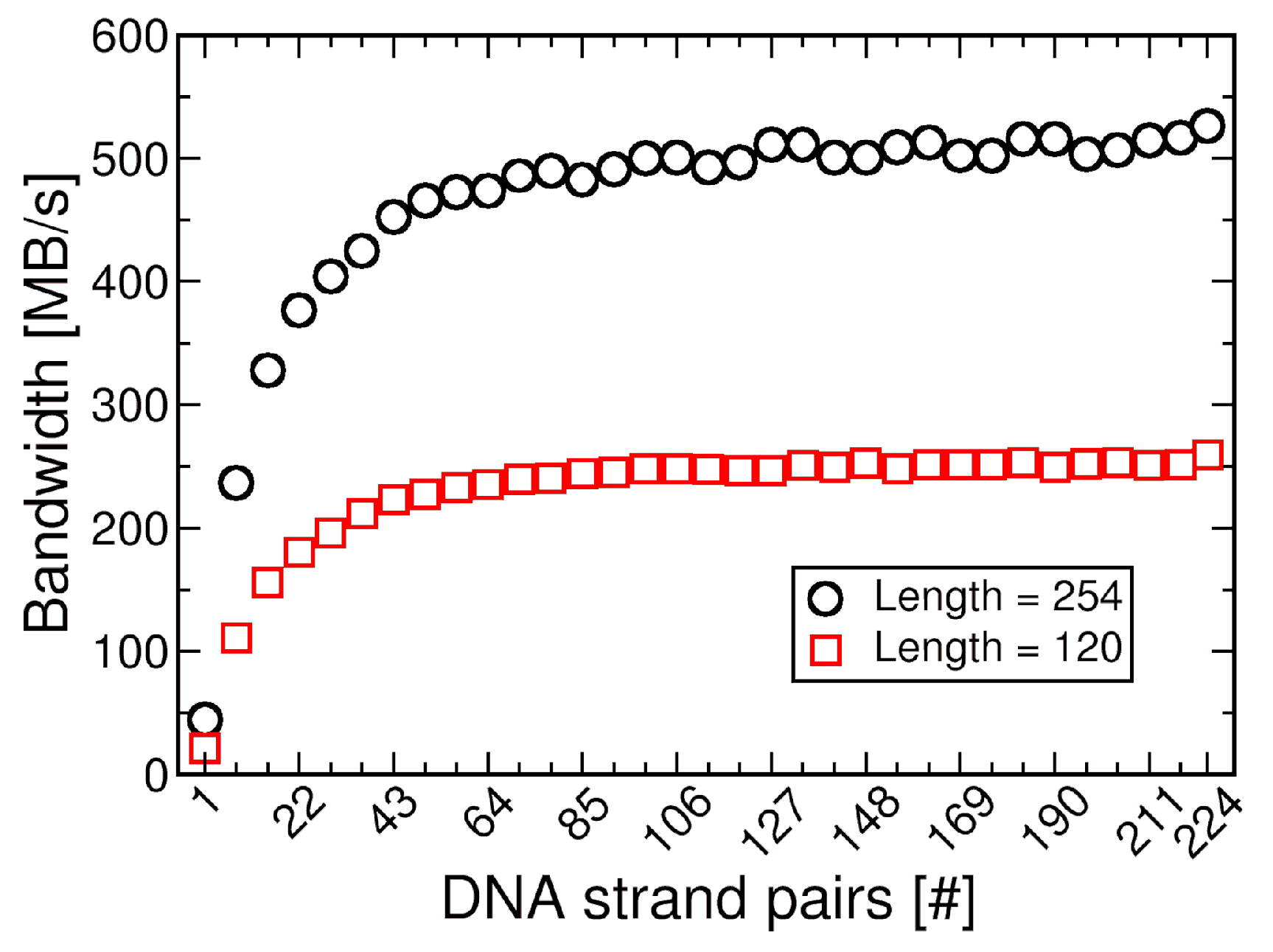
4.2.3. Performance
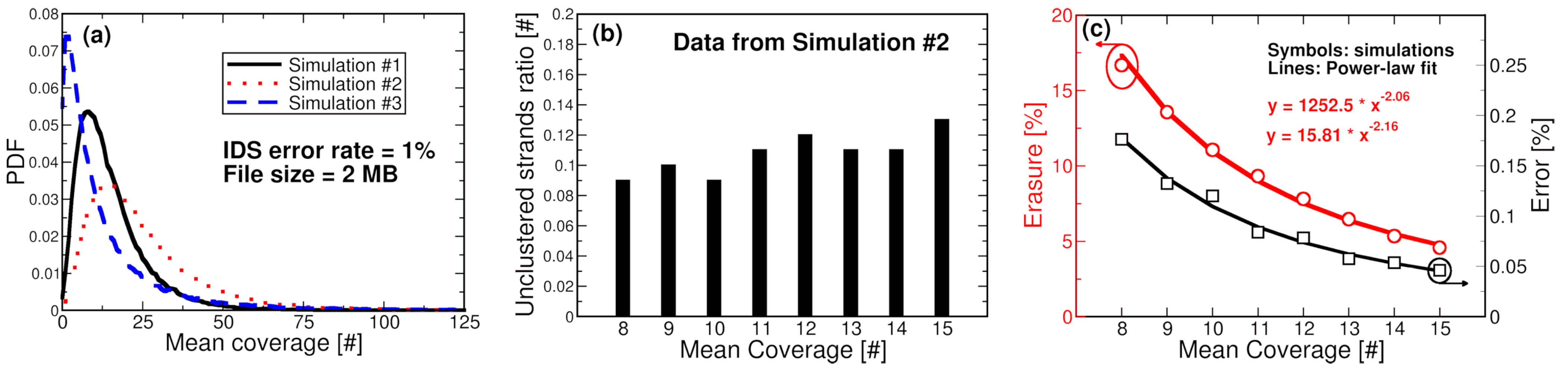
5. Co-Simulation Experiments and Results
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DNA | DeoxyriboNucleic Acid |
| PCR | Polymerase Chain Reaction |
| FPGA | Field Programmable Gate Array |
| IDS | Inserted, Deleted, and transposed/Substituted |
| ECC | Error Correction Codes |
| BRAM | Block Random Access Memory |
References
- Rydning, J.; Reinsel, D. Worldwide Global StorageSphere Forecast, 2021–2025: To Save or Not to Save Data, That Is the Question; Technical Report IDC Doc #US47509621; IDC Corp.: Needham, MA, USA, 2021. [Google Scholar]
- Wieder, P.; Butler, J.M.; Theilmann, W.; Yahyapour, R. Service Level Agreements for Cloud Computing; Springer: New York, NY, USA, 2014. [Google Scholar] [CrossRef]
- DNA Data Storage Alliance. Preserving Our Digital Legacy: An Introduction to DNA Data Storage. Technical Report. 2021. Available online: https://dnastoragealliance.org/dev/wp-content/uploads/2021/06/DNA-Data-Storage-Alliance-An-Introduction-to-DNA-Data-Storage.pdf (accessed on 6 June 2023).
- Alberts, B.; Bray, D.; Lewis, J.; Raff, M.; Roberts, K.; Watson, J. Molecular Biology of the Cell, 4th ed.; Garland: New York, NY, USA, 2002. [Google Scholar]
- Erlich, Y.; Zielinski, D. DNA Fountain enables a robust and efficient storage architecture. Science 2017, 355, 950–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grass, R.; Heckel, R.; Puddu, M.; Paunescu, D.; Stark, W. Robust Chemical Preservation of Digital Information on DNA in Silica with Error-Correcting Codes. Angew. Chem. Int. Ed. 2015, 54, 2552. [Google Scholar] [CrossRef] [PubMed]
- DNA Storage. 2015. Available online: https://www.microsoft.com/en-us/research/project/dna-storage/ (accessed on 15 April 2023).
- Budel, S. Next Generation Sequencing (NGS) Market Assessment Trends (2018–2024); Technical Report; DeciBio: Los Angeles, CA, USA, 2021. [Google Scholar]
- Brown, K. A $100 Genome within Reach, Illumina CEO Asks If World Is Ready. 2019. Available online: https://www.bloomberg.com/news/articles/2019-02-27/a-100-genome-within-reach-illumina-ceo-asks-if-world-is-ready (accessed on 15 April 2023).
- Genscript. Gene Synthesis & DNA Synthesis Service. 2023. Available online: https://www.genscript.com/gene_synthesis.html?src=google&gclid=Cj0KCQjwyLGjBhDKARIsAFRNgW_Y6C7bL0pr-U_MZA_2tmShoNPCZWmjEZuLPCm4OjBff-LARSzPE3oaAu3BEALw_wcB (accessed on 24 April 2023).
- Saiki, R.; Gelfand, D.; Stoffel, S.; Scharf, S.; Higuchi, R.; Horn, G.; Mullis, K.; Erlich, H. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 1988, 239, 487–491. [Google Scholar] [CrossRef] [Green Version]
- Chandak, S.; Tatwawadi, K.; Lau, B.; Mardia, J.; Kubit, M.; Neu, J.; Griffin, P.; Wootters, M.; Weissman, T.; Ji, H. Improved Read/Write Cost Tradeoff in DNA-Based Data Storage Using LDPC Codes. In Proceedings of the 57th Annual Allerton Conference on Communication, Control, and Computing (Allerton), Monticello, IL, USA, 24–27 September 2019; pp. 147–156. [Google Scholar] [CrossRef]
- Mitzenmacher, M. A survey of results for deletion channels and related synchronization channels. Probab. Surv. 2009, 6, 1–33. [Google Scholar] [CrossRef]
- Church, G.M.; Gao, Y.; Kosuri, S. Next-generation digital information storage in DNA. Science 2012, 337, 1628. [Google Scholar] [CrossRef] [Green Version]
- Goldman, N.; Bertone, P.; Chen, S.; Dessimoz, C.; LeProust, E.M.; Sipos, B.; Birney, E. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature 2013, 494, 77–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blawat, M.; Gaedke, K.; Hütter, I.; Chen, X.M.; Turczyk, B.; Inverso, S.; Pruitt, B.W.; Church, G.M. Forward Error Correction for DNA Data Storage. Procedia Comput. Sci. 2016, 80, 1011–1022. [Google Scholar] [CrossRef] [Green Version]
- Tabatabaei Yazdi, S.M.H.; Gabrys, R.; Milenkovic, O. Portable and Error-Free DNA-Based Data Storage. Sci. Rep. 2017, 7, 5011. [Google Scholar] [CrossRef] [Green Version]
- Organick, L.; Ang, S.; Chen, Y.J.; Lopez, R.; Yekhanin, S.; Makarychev, K.; Racz, M.; Kamath, G.; Gopalan, P.; Nguyen, B.; et al. Random access in large-scale DNA data storage. Nat. Biotechnol. 2018, 36, 242–248. [Google Scholar] [CrossRef]
- Heckel, R.; Mikutis, G.; Grass, R.N. A Characterization of the DNA Data Storage Channel. Sci. Rep. 2019, 9, 9663. [Google Scholar] [CrossRef] [Green Version]
- AXI Memory Mapped to PCI Express (PCIe) Gen2 v2.9. 2021. Available online: https://docs.xilinx.com/v/u/en-US/pg055-axi-bridge-pcie/ (accessed on 18 April 2023).
- Marelli, A.; Chiozzi, T.; Zuolo, L.; Battistini, N.; Lanzoni, G.; Olivo, P.; Zambelli, C.; Micheloni, R. DNAssim: A Full System Simulator for DNA Storage. In Proceedings of the Flash Memory Summit, Santa Clara, CA, USA, 8–10 August 2022. [Google Scholar]
- Marelli, A.; Chiozzi, T.; Zuolo, L.; Battistini, N.; Olivo, P.; Zambelli, C.; Micheloni, R. DNAssim: A Full System Simulator for DNA Storage. In Proceedings of the Storage Developer Conference, Fremont, CA, USA, 12–15 September 2022. [Google Scholar]
- Rashtchian, C.; Makarychev, K.; Racz, M.; Ang, S.; Jevdjic, D.; Yekhanin, S.; Ceze, L.; Strauss, K. Clustering Billions of Reads for DNA Data Storage. In Proceedings of the Advances in Neural Information Processing Systems 30; MIT Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Whitwam, R. Microsoft Automates DNA-Based Data Storage. 2019. Available online: https://www.extremetech.com/extreme/288240-microsoft-automates-dna-based-data-storage (accessed on 15 April 2023).
- Lassmann, T.; Frings, O.; Sonnhammer, E. Kalign2: High-performance Multiple Alignment of Protein and Nucleotide Sequences Allowing External Features. Nucleic Acids Res. 2009, 37, 858–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivasavaradhan, S.R.; Gopi, S.; Pfister, H.D.; Yekhanin, S. Trellis BMA: Coded Trace Reconstruction on IDS Channels for DNA Storage. In Proceedings of the 2021 IEEE International Symposium on Information Theory (ISIT), Melbourne, Australia, 12–20 July 2021; pp. 2453–2458. [Google Scholar] [CrossRef]
- Zuolo, L.; Zambelli, C.; Marelli, A.; Micheloni, R.; Olivo, P. LDPC Soft Decoding with Improved Performance in 1X-2X MLC and TLC NAND Flash-Based Solid State Drives. IEEE Trans. Emerg. Top. Comput. 2019, 7, 507–515. [Google Scholar] [CrossRef]
- Zuolo, L.; Zambelli, C.; Micheloni, R.; Indaco, M.; Carlo, S.D.; Prinetto, P.; Bertozzi, D.; Olivo, P. SSDExplorer: A Virtual Platform for Performance/Reliability-Oriented Fine-Grained Design Space Exploration of Solid State Drives. IEEE Trans. Comput. Aided Des. Integr. Circuits Syst. 2015, 34, 1627–1638. [Google Scholar] [CrossRef] [Green Version]
- Caffarena, G.; Pedreira, C.; Carreras, C.; Bojanic, S.; Nieto-Taladriz, O. FPGA Acceleration for DNA sequence alignment. J. Circuits Syst. Comput. 2007, 16, 245–266. [Google Scholar] [CrossRef]
- Kent, K.; Proudfoot, R.; Zhao, Y. Parameter-Specific FPGA Implementation of Edit-Distance Calculation. In Proceedings of the Seventeenth IEEE International Workshop on Rapid System Prototyping (RSP’06), Chania, Greece, 14–16 June 2006; pp. 209–215. [Google Scholar] [CrossRef]
- Dydel, S.; Bała, P. Large Scale Protein Sequence Alignment Using FPGA Reprogrammable Logic Devices. In Proceedings of the Field Programmable Logic and Application; Becker, J., Platzner, M., Vernalde, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2004; pp. 23–32. [Google Scholar]
- Castells-Rufas, D.; Marco-Sola, S.; Moure, J.C.; Aguado, Q.; Espinosa, A. FPGA Acceleration of Pre-Alignment Filters for Short Read Mapping with HLS. IEEE Access 2022, 10, 22079–22100. [Google Scholar] [CrossRef]
- Marchisio, A.; Teodonio, F.; Rizzi, A.; Shafique, M. ISMatch: A real-time hardware accelerator for inexact string matching of DNA sequences on FPGA. Microprocess. Microsystems 2023, 97, 104763. [Google Scholar] [CrossRef]
- Cai, K.; Chee, Y.M.; Gabrys, R.; Kiah, H.M.; Nguyen, T.T. Correcting a Single Indel/Edit for DNA-Based Data Storage: Linear-Time Encoders and Order-Optimality. IEEE Trans. Inf. Theory 2021, 67, 3438–3451. [Google Scholar] [CrossRef]
- Leung, K.; Welch, L. Erasure decoding in burst-error channels. IEEE Trans. Inf. Theory 1981, 27, 160–167. [Google Scholar] [CrossRef]
- Skiena, S.S. Hashing and Randomized Algorithms. In The Algorithm Design Manual; Springer: Cham, Switzerlands, 2020; pp. 171–195. [Google Scholar] [CrossRef]
- Shomorony, I.; Heckel, R. DNA-Based Storage: Models and Fundamental Limits. IEEE Trans. Inf. Theory 2021, 67, 3675–3689. [Google Scholar] [CrossRef]
- Mao, W.; Diggavi, S.N.; Kannan, S. Models and Information-Theoretic Bounds for Nanopore Sequencing. IEEE Trans. Inf. Theory 2018, 64, 3216–3236. [Google Scholar] [CrossRef]
- Berger, B.; Waterman, M.S.; Yu, Y.W. Levenshtein Distance, Sequence Comparison and Biological Database Search. IEEE Trans. Inf. Theory 2021, 67, 3287–3294. [Google Scholar] [CrossRef] [PubMed]
- Navarro, G. A Guided Tour to Approximate String Matching. ACM Comput. Surv. 2001, 33, 31–88. [Google Scholar] [CrossRef]
- AMBA AXI4 Protocol. 2019. Available online: https://developer.arm.com/products/architecture/system-architectures/amba/amba-4 (accessed on 15 April 2023).
- Xilinx Integrated Logic Analyzer (ILA) v2.0 IP-Core. 2012. Available online: https://docs.xilinx.com/v/u/en-US/ds875-ila (accessed on 15 April 2023).
- Organick, L.; Ang, S.D.; Chen, Y.J.; Lopez, R.; Yekhanin, S.; Makarychev, K.; Racz, M.Z.; Kamath, G.; Gopalan, P.; Nguyen, B.; et al. Scaling up DNA data storage and random access retrieval. bioRxiv 2017. [Google Scholar] [CrossRef] [Green Version]





| Name | FF | LUT |
|---|---|---|
| Expression | 0 | 5915 |
| Instance | 135 | 118 |
| Memory | 272 | 96 |
| Multiplexer | - | 1849 |
| Register | 4266 | - |
| Total | 4633 (0.76%) | 7978 (2.62%) |
| Available | 607,200 | 303,600 |
| Site Type | Used | Available | Usage % |
|---|---|---|---|
| Slice LUTs | 196,785 | 303,600 | 64.82 |
| LUT as Logic | 174,277 | 303,600 | 57.40 |
| LUT as Memory | 22,508 | 130,800 | 17.21 |
| LUT as distrib. RAM | 21,304 | - | - |
| LUT as Shift Reg. | 1204 | - | - |
| Slice Registers | 237,117 | 607,200 | 39.05 |
| Register as FF | 237,177 | 607,200 | 39.05 |
| Register as Latch | 0 | 607,200 | 0.00 |
| F7 Muxes | 5065 | 151,800 | 3.34 |
| F8 Muxes | 1404 | 75,900 | 1.85 |
| Dataset | # Reads (Closest) | Avg. Strand Length (Closest) | Description | Speed-Up |
|---|---|---|---|---|
| 3.1M | 3,103,511 (10) | 150 (160) | Movie file | 2.49× |
| 13.2M | 13,256,431 (10) | 150 (160) | Music file | 4.2× |
| 12M | 11,973,538 (10) | 110 (120) | Text file | 2.91× |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marelli, A.; Chiozzi, T.; Battistini, N.; Zuolo, L.; Micheloni, R.; Zambelli, C. Integrating FPGA Acceleration in the DNAssim Framework for Faster DNA-Based Data Storage Simulations. Electronics 2023, 12, 2621. https://doi.org/10.3390/electronics12122621
Marelli A, Chiozzi T, Battistini N, Zuolo L, Micheloni R, Zambelli C. Integrating FPGA Acceleration in the DNAssim Framework for Faster DNA-Based Data Storage Simulations. Electronics. 2023; 12(12):2621. https://doi.org/10.3390/electronics12122621
Chicago/Turabian StyleMarelli, Alessia, Thomas Chiozzi, Nicholas Battistini, Lorenzo Zuolo, Rino Micheloni, and Cristian Zambelli. 2023. "Integrating FPGA Acceleration in the DNAssim Framework for Faster DNA-Based Data Storage Simulations" Electronics 12, no. 12: 2621. https://doi.org/10.3390/electronics12122621
APA StyleMarelli, A., Chiozzi, T., Battistini, N., Zuolo, L., Micheloni, R., & Zambelli, C. (2023). Integrating FPGA Acceleration in the DNAssim Framework for Faster DNA-Based Data Storage Simulations. Electronics, 12(12), 2621. https://doi.org/10.3390/electronics12122621








