BTENet: Back-Fat Thickness Estimation Network for Automated Grading of the Korean Commercial Pig
Abstract
:1. Introduction
2. Materials and Methods
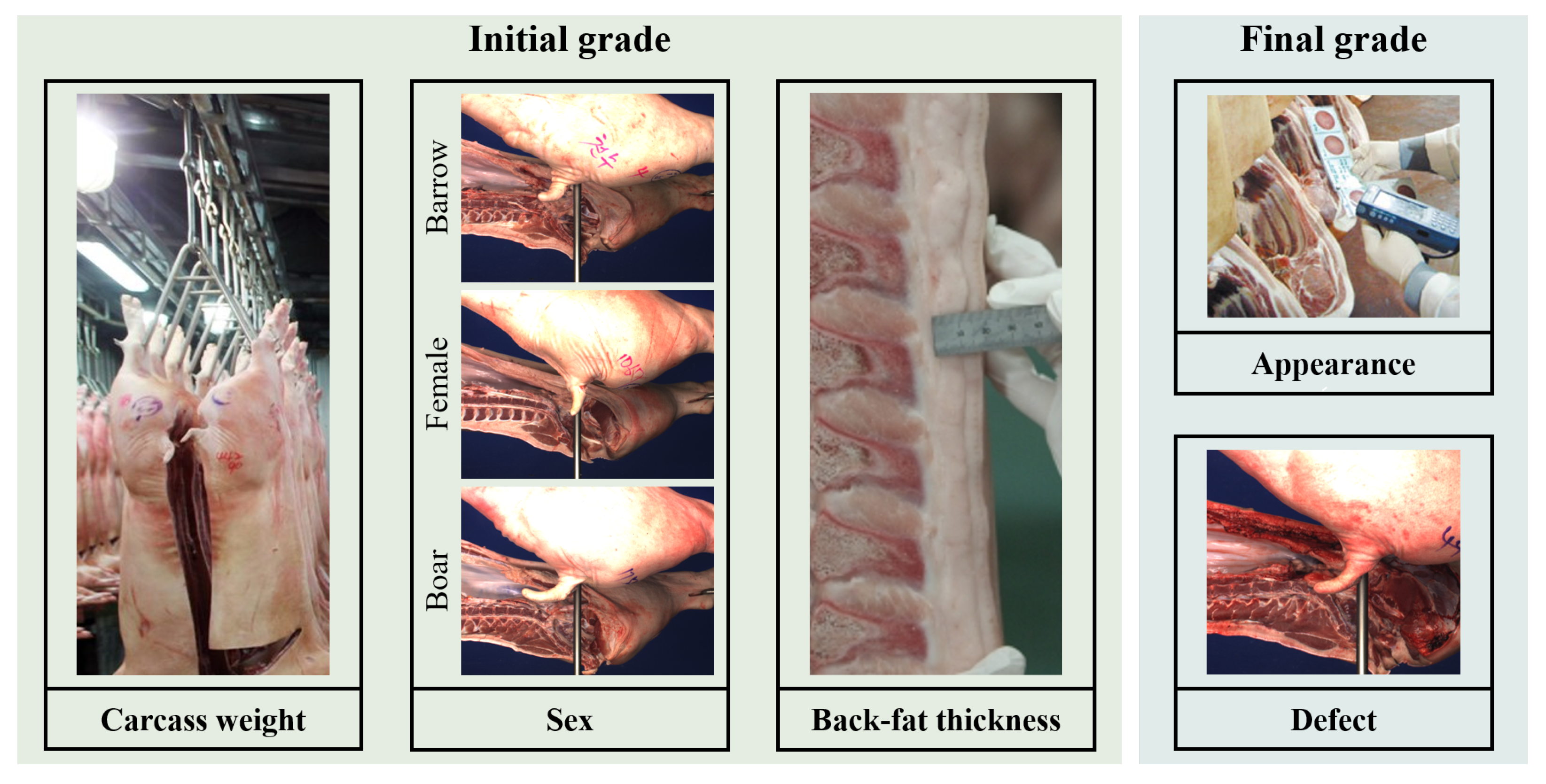
2.1. Dataset
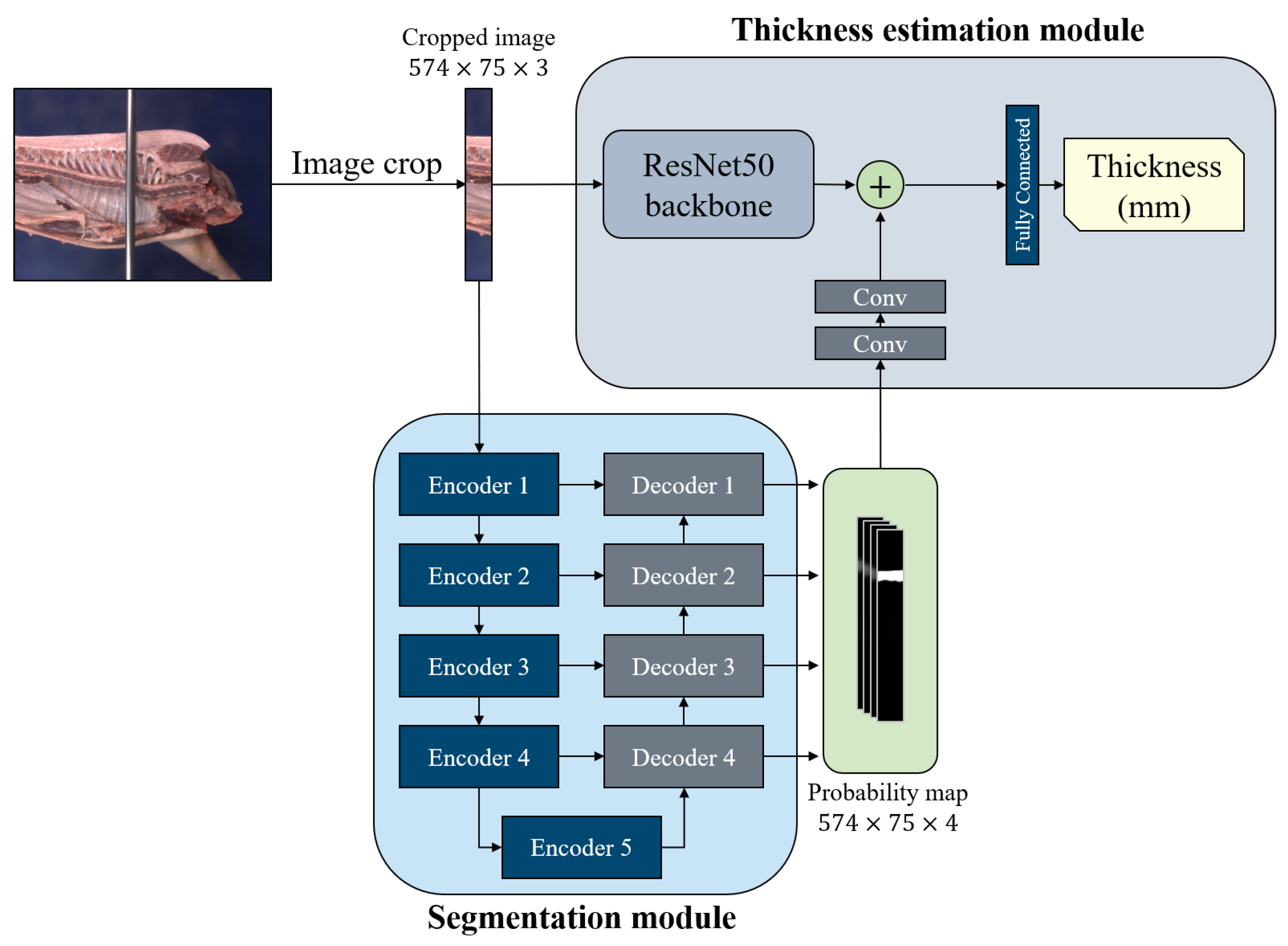
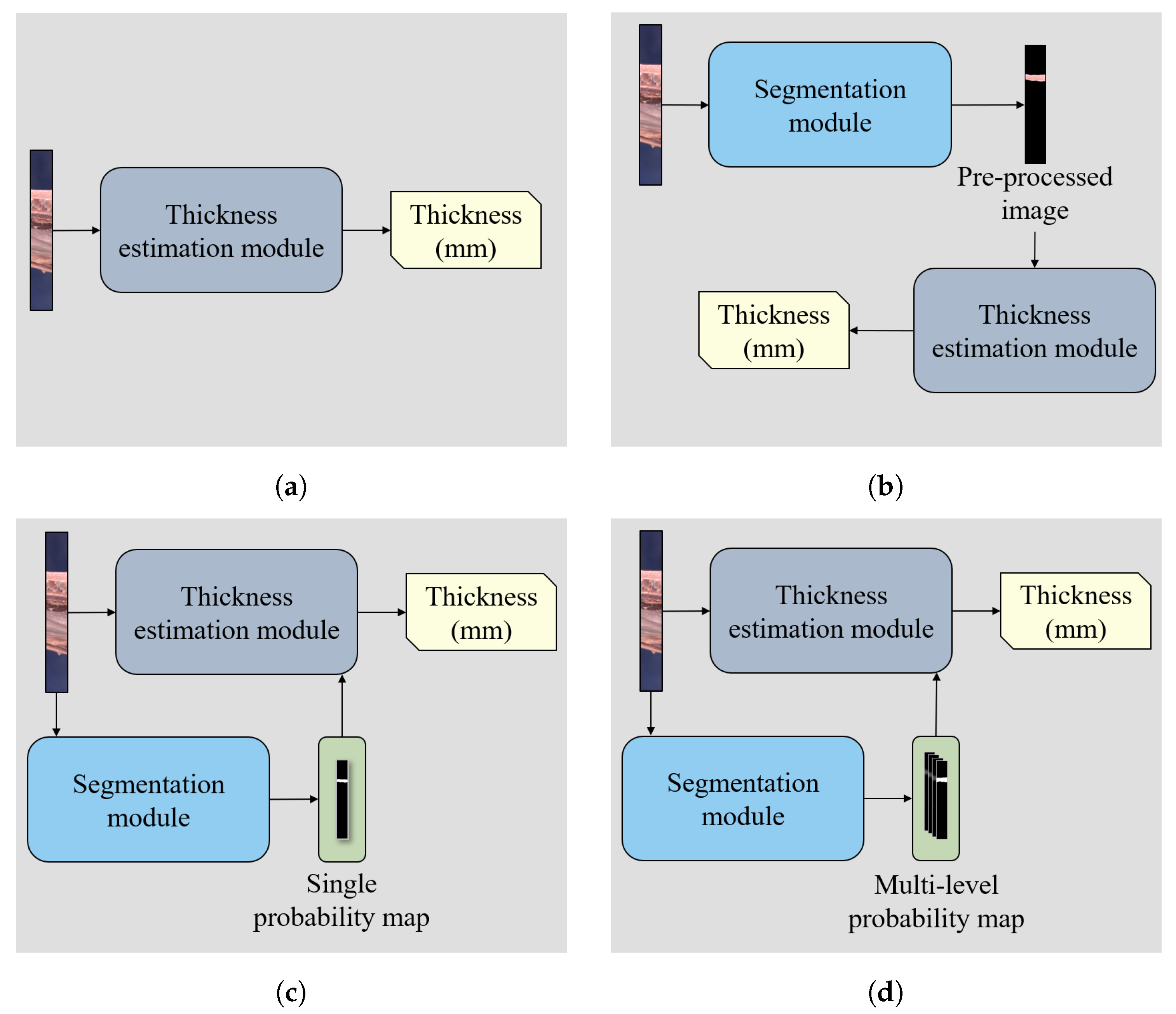
2.2. Back-Fat Thickness Estimation Network
- Segmentation module: To construct the segmentation module, we employed the encoder–decoder structure in U-Net [15]. Specifically, the segmentation module consists of five encoder blocks and four decoder blocks. Each encoder block has two convolution layers with ReLU activation. Furthermore, except for the last encoder block, each encoder performed max pooling operation with stride 2 to extract a high-level context for segmentation. Each decoder block takes two inputs: the output of the previous decoder block and the intermediate feature of the encoder. This makes the first input have the same size as the second input through the upsampling operation, and then concatenates the upsampled first input and the second input along the channel dimension. Then, the decoder block produces an output through two convolution layers with ReLU activation and upsampling operation. Four decoder blocks sequentially predict a probability map of the back-fat area. To transfer information on the back-fat area to the thickness estimation module, the segmentation module yielded multi-scale segmentation masks. The output of each decoder block was transformed to the probability map ranging from 0 to 1 through one convolution layer with sigmoid function. A total of four segmentation masks was interpolated to the size of the input cropped image and concatenated by its channel dimension. To this end, the concatenated probability map was transferred to the thickness estimation module. With the multi-level probability map, the thickness estimation module can extract a feature of back-fat area in various resolutions. For training BTENet and evaluating its segmentation performance, we only use the probability map of the last decoder block as the final segmentation mask.
- Thickness estimation module: The thickness estimation module takes two inputs: the cropped image and multi-level probability map from the segmentation module. It extracts cropped image features throughout backbone networks. Here, we use ResNet50 [7] to the backbone networks. The probability map is transformed to the back-fat area features through two convolution layers with ReLU activation and a global average pooling. Both the cropped image features and back-fat area features are concatenated and transferred to the final fully connected layer, which yields a scalar corresponding to the back-fat thickness of the input image.
2.3. Implementation Details
3. Experimental Results
3.1. Quantitative Results
3.2. Qualitative Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oh, S.H.; Whitley, N.C. Pork production in China, Japan and South Korea. Asian-Australas. J. Anim. Sci. 2011, 24, 1629–1636. [Google Scholar] [CrossRef]
- Korean Statistical Information Service. Livestock Statistics Survey. Available online: https://kosis.kr/ (accessed on 6 December 2021).
- Kim, J.; Han, H.D.; Lee, W.Y.; Wakholi, C.; Lee, J.; Jeong, Y.B.; Bae, J.H.; Cho, B.K. Economic analysis of the use of VCS2000 for pork carcass meat yield grading in Korea. Animals 2021, 11, 1297. [Google Scholar] [CrossRef] [PubMed]
- Brøndum, J.; Egebo, M.; Agerskov, C.; Busk, H. On-line pork carcass grading with the autofom ultrasound system. J. Anim. Sci. 1998, 76, 1859–1868. [Google Scholar] [CrossRef] [PubMed]
- Carabús, A.; Sainz, R.D.; Oltjen, J.W.; Gispert, M.; Font-i Furnols, M. Predicting fat, lean and the weights of primal cuts for growing pigs of different genotypes and sexes using computed tomography. J. Anim. Sci. 2015, 93, 1388–1397. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Leng, S.; Ma, W.; Xu, Q.; Xie, X. Method for detection of the lean meat ratio in pork carcass based on fiber optic sensor. MATEC Web. Conf. 2017, 95, 14009. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 30 June 2016; pp. 770–778. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019; pp. 6105–6114. [Google Scholar]
- Tschandl, P.; Sinz, C.; Kittler, H. Domain-specific classification-pretrained fully convolutional network encoders for skin lesion segmentation. Comput. Biol. Med. 2019, 104, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Alakwaa, W.; Nassef, M.; Badr, A. Lung cancer detection and classification with 3D convolutional neural network (3D-CNN). Lung Cancer 2017, 8, 409. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, A.F.; Dórea, J.R.; Valente, B.D.; Fitzgerald, R.; Herring, W.; Rosa, G.J. Comparison of data analytics strategies in computer vision systems to predict pig body composition traits from 3D images. J. Anim. Sci. 2020, 98, skaa250. [Google Scholar] [CrossRef] [PubMed]
- Kvam, J.; Kongsro, J. In vivo prediction of intramuscular fat using ultrasound and deep learning. Comput. Electron. Agric. 2017, 142, 521–523. [Google Scholar] [CrossRef]
- Lohumi, S.; Wakholi, C.; Baek, J.H.; Do Kim, B.; Kang, S.J.; Kim, H.S.; Yun, Y.K.; Lee, W.Y.; Yoon, S.H.; Cho, B.K. Nondestructive estimation of lean meat yield of South Korean pig carcasses using machine vision technique. Korean J. Food Sci. Anim. Resour. 2018, 38, 1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2015; pp. 234–241. [Google Scholar]
- Kingma, D.P.; Ba, J.L. Adam: A method for stochastic gradient descent. In Proceedings of the ICLR: International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015; pp. 1–15. [Google Scholar]
- Dai, Z.; Liu, H.; Le, Q.; Tan, M. Coatnet: Marrying convolution and attention for all data sizes. Adv. Neural Inf. Process. Syst. 2021, 34, 3965–3977. [Google Scholar]




| Statistic | Train | Validation | Test | Total |
|---|---|---|---|---|
| Mean | 21.39 | 21.41 | 21.19 | 21.36 |
| Std | 4.47 | 4.42 | 4.26 | 4.42 |
| N | 2269 | 756 | 757 | 3782 |
| Model | Segmentation | Thickness Estimation | ||
|---|---|---|---|---|
| mIOU | Corr | MAE (mm) | MAPE (%) | |
| S1 | X | 0.833 | 1.844 | 9.3 |
| S2 | 0.936 | 0.907 | 1.397 | 7.0 |
| S3 | 0.936 | 0.911 | 1.317 | 6.6 |
| S4 (Proposed) | 0.935 | 0.915 | 1.275 | 6.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.-J.; Baek, J.-H.; Kim, Y.-K.; Lee, J.H.; Lee, M.; Park, W.; Lee, S.H.; Koh, Y.J. BTENet: Back-Fat Thickness Estimation Network for Automated Grading of the Korean Commercial Pig. Electronics 2022, 11, 1296. https://doi.org/10.3390/electronics11091296
Lee H-J, Baek J-H, Kim Y-K, Lee JH, Lee M, Park W, Lee SH, Koh YJ. BTENet: Back-Fat Thickness Estimation Network for Automated Grading of the Korean Commercial Pig. Electronics. 2022; 11(9):1296. https://doi.org/10.3390/electronics11091296
Chicago/Turabian StyleLee, Hyo-Jun, Jong-Hyeon Baek, Young-Kuk Kim, Jun Heon Lee, Myungjae Lee, Wooju Park, Seung Hwan Lee, and Yeong Jun Koh. 2022. "BTENet: Back-Fat Thickness Estimation Network for Automated Grading of the Korean Commercial Pig" Electronics 11, no. 9: 1296. https://doi.org/10.3390/electronics11091296
APA StyleLee, H.-J., Baek, J.-H., Kim, Y.-K., Lee, J. H., Lee, M., Park, W., Lee, S. H., & Koh, Y. J. (2022). BTENet: Back-Fat Thickness Estimation Network for Automated Grading of the Korean Commercial Pig. Electronics, 11(9), 1296. https://doi.org/10.3390/electronics11091296








