Abstract
High-throughput microarrays contain a huge number of genes. Determining the relationships between all these genes is a time-consuming computation. In this paper, the authors provide a parallel algorithm for finding the Pearson’s correlation coefficient between genes measured in the Affymetrix microarrays. The main idea in the proposed algorithm, ForkJoinPcc, mimics the well-known parallel programming model: the fork–join model. The parallel MATLAB APIs have been employed and evaluated on shared or distributed multiprocessing systems. Two performance metrics—the processing and communication times—have been used to assess the performance of the ForkJoinPcc. The experimental results reveal that the ForkJoinPcc algorithm achieves a substantial speedup on the cluster platform of 62× compared with a 3.8× speedup on the multicore platform.
1. Introduction
High-throughput microarrays contain a huge number of genes. Determining the relationships between these genes can be conducted using two approaches. In the first one, the dependencies can be calculated between only a set of informative genes [1]. The Pearson’s correlation coefficient, Pcc, and mutual information can be used as measures of such gene dependencies to construct a gene co-network [2]. In the second approach, parallel computing can be used to find the dependencies between all the huge numbers of genes measured in our high-throughput microarray experiments. Parallel computing is essential, as the high-throughput microarrays contain more than 30,000 genes, and the determination of dependencies between the genes is infeasible using existing computing infrastructures. Therefore, in this research, the authors are proposing a solution for determining the relationships between genes measured in high-throughput microarrays. The correlation coefficient measures the strength of a linear relationship between two variables. Therefore, it is used as a similarity measure between two gene expression profiles. A strong association between genes should have Pcc values from ±0.7 to ±1. The Pcc between two expression profiles X and Y is defined as in Equation (1):
The proposed algorithm here, ForkJoinPcc, calculates the correlation matrix between genes expressed in Affymetrix microarrays. The main idea in the ForkJoinPcc algorithm mimics a well-known parallel programming model: the fork–join model [3]. The fork–join programming model, shown in Figure 1, begins as a single process: the master thread, which is executed serially. Then, the master thread creates a team of parallel threads, and each thread performs some linear algebra on its subset of data. When the team threads complete their tasks in the parallel region, they synchronize and terminate, leaving only the master thread. The fork–join model has been demonstrated to have powerful performance in parallelization on the multicore platform [4,5].
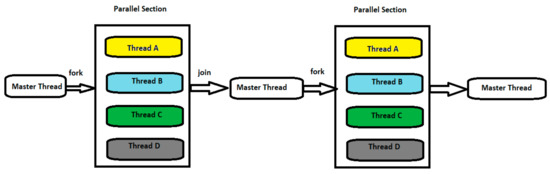
Figure 1.
Master and worker threads in a fork–join programming model.
2. Literature Review
During the last decade, many studies have been focused on utilizing the parallel computing paradigms in solving heavy computational problems in bioinformatics [6]. As the data are growing at a faster rate than the corresponding growth of the performance of the available hardware, new solutions should be provided to process such data [7]. Problems of this form are sometimes referred to as embarrassingly parallel. If multiple processors or computers, which are often available in organizations where statistical computations are carried out, could be easily harnessed to perform these computations in parallel, then computations that would otherwise take hours or days could be performed in minutes or hours. One of the most important goals of research in bioinformatics is the inference of the gene co-expression networks and gene-to-gene interactions [8]. Gene co-expression networks are demonstrations of the relationships between a set of interacting genes [9]. To identify the interactions between a set of genes, the associated gene expressions should be measured. Affymetrix microarrays [10] and RNA sequencing [11] are the dominant approaches for measuring the expression profiles of genes under certain conditions. The output of these approaches is the gene expression matrix, which is the input for the construction of gene co-expression networks.
In [12], the compute unified device architecture (CUDA) was utilized for inferring the gene regularity networks on GPUs. In [13], the R package MPICorMat was introduced. They employed the MPI/OpenMP approach to construct the similarity matrix between a huge number of genes. The Pcc was used as a measure of similarity between the genes in such networks. However, the construction of such a matrix is a heavy computation that may take days to weeks to finish, as all pairwise similarities must be computed, and the complexity of the computation rises quadratically versus the number of genes. An expansion was performed in [14] to the MPICorMat package, which included extra metrics for the measurement of the similarity matrices. In [15], an efficient parallel package (MPIGeneNet) is presented. They utilized the random matrix theory and the Pcc as a measure of dependencies in gene co-expression networks. In [16], MPI was employed for the construction of the mutual information (MI) between genes interacting in a gene co-expression network. Although they used the MI to represent the measure of similarity between genes, this measure is irrelevant for inferring the interactions in such networks, as stated in [17]. In [18], a framework for the R statistical computing language was presented. It provided a simple, powerful programming interface to a computational cluster. The resulting framework allows statisticians to obtain significant speedup for some computations at little additional development cost. However, it does not provide a solution flexible enough to express all classes of parallel algorithms. In [19], GPUs and the CUDA language were used to calculate the pairwise distances (Manhattan distances and Pcc) between genes in estimating the hierarchical clusters of genes measured by a set of microarrays. However, this study was limited to only the creation of pairwise distances, and it did not perform any type of linkage algorithm to create a set of clusters. The work performed in [19] presents a prototype framework (SPRINT) that allows the addition of parallelized functions to R to enable the easy exploitation of high-performance computing systems. SPRINT is a wrapper around such parallelized functions. Their use requires very little modification to existing sequential R scripts and no expertise in parallel computing. The main issue regarding the framework proposed in this study is that functions must be reimplemented, which requires significant effort. In [20], two parallel algorithms for computing the correlation and covariance matrix are proposed. The first algorithm uses the quadrant correlation method, and the second uses the Maronna method. The parallel implementation of these methods made them applicable to much larger problems. However, the parallel implementation of the proposed algorithm in this study was limited to the cluster computer infrastructure.
There are other studies on parallel programming that have been conducted in different applications rather than bioinformatics. The authors in [21] introduced a parallel implementation of the Ranking SVM algorithm. They presented OpenCL and OpenMP versions for the multicore, and manycore platforms, and an evaluation of both parallel programming models was performed. More work was performed in [22] to calculate the Pearson’s correlation matrix using a hybrid programming model of CUDA and MPI. The data matrix was divided into pieces, and a master GPU distributed the pieces on the worker GPUs using the MPI technique. In [23], a GPU-based parallel programming model was introduced to calculate the pairwise Pcc in real MRI data to understand the functional associations in the human brain. Additionally, in [24], the authors presented a novel computation model for determining the scalability of the parallel programming iterative algorithms in a distributed multiprocessing system of computer clusters.
To conclude, the scope of most of the previously mentioned publications was introducing or evaluating different APIs in different parallel computing architectures for solving some of the heavy computations in bioinformatics and non-bioinformatics applications. However, the MATLAB APIs introduced in the MATLAB Parallel Computing Toolbox and the MATLAB Distributed Computing Server (MDCS) have not yet been investigated in the current state of the art. The MathWorks Parallel Computing ToolboxTM and MATLAB Distributed Computing ServerTM are two examples of technologies that provide this functionality [25]. In this study, the ForkJoinPcc algorithm has been developed using the Parallel MATLAB Toolbox and two software structures has been utilized including the single program multiple data model (SPMD) and parallel loop (ParFor). They are abstractions for the message passing function defined in the MPI-2 standard [26]. Parallel SPMD programs are usually built using a message passing library such as MPI. For designing and running parallel programs on the Windows platform, Microsoft MPI [27] is a Microsoft implementation of the MPI standard. Another SPMD implementation is available in many programming tools, such as the Java parallel programming model [28]. The parallel loop (ParFor) construct is analogous to OpenMP parallel loops [29] and the Fortress language specification’s parallel for constructs [30]. The main distinction is that OpenMP parallel loops and Fortress loops run on threads within a single process on a single physical computer, whereas ParFor repetitions run on several processes, perhaps on multiple physically independent systems. The parallel constructs available in the MATLAB parallel computing toolbox abstract information as needed and present to users a language independent of resource allocation and underlying implementation, which is why the suggested method was implemented using the MATLAB API in this work. Most crucially, the toolbox includes the same functionality as more difficult tools like CUDA and C++ but in a MATLAB-like programming style [31]. Since MATLAB is a commonly used tool for researchers in our area, and we believe an implementation with the MATLAB API will be valuable for the community.
The contributions of this work include the following:
- A proposed framework for the ForkJoinPcc algorithm which mimics the fork–join model is introduced here.
- The performance of the ForkJoinPcc algorithm is tested intensively in terms of the processing time and the communication overhead between the worker nodes.
- The ForkJoinPcc algorithm is applied to cover gene expression matrices of hepatocellular carcinoma (HCC), comprising a huge number of genes.
- Four MATLAB constructs including SPMD, distributed arrays, MatlabPool, and ParFor are employed in implementing the ForkJoinPcc algorithm.
- The adequacy of these constructs in achieving good performance on shared or distributed multiprocessing systems (multicore and computer cluster) is evaluated.
3. Parallel MATLAB Toolsets
There have been many attempts [32,33,34] made by researchers in MathWorks to produce some MATLAB utilities for parallel programming. In November 2004, MathWorks presented two toolboxes as an explicit parallel programming utility, including the Parallel Computing Toolbox and the MDCS. The MDCS includes a set of worker nodes managed by a master node connected in a shared and distributed multiprocessing system. The master node in the MATLAB Client propagates a computation task to the workers and labs through a set of message-passing functions. These functions are abstractions for the message-passing function defined in the MPI-2 standard [26]. The master node communicates via a point-to-point communication paradigm with the working labs. The message-passing functions include labSend and labReceive for the point-to-point operations between the MATLAB Client and the worker nodes. Other software constructs that exist in the Parallel MATLAB Toolbox include SPMD, distributed arrays, MatlabPool, and ParFor, which will be described in the following subsections.
3.1. Single Program Multiple Data (SPMD) Model
This is a technique utilized to accomplish parallelism on the shared and distributed memory systems. In a distributed memory architecture, there is a set of independent workers (computing nodes). Each worker executes their task on a different subset of the dataset, and the workers can communicate via MPI by transmitting and receiving messages. In a shared memory multiprocessing system, the workers are CPUs that share a memory space. Workers can perform the same task on different batches of the whole dataset, and they can communicate by placing their results in the shared memory section. As shown in Figure 2, the whole dataset is split among N workers which run simultaneously to attain faster execution of the whole program. To employ SPMD in MATLAB, one must initially generate a pool of parallel workers with a total number of workers that does not exceed the actual number of nodes in the shared CPUs and distributed multiprocessing system. Then, within the frame of the SPMD section, each worker node will have a unique identifier that identifies that node for further communication between its neighboring nodes.
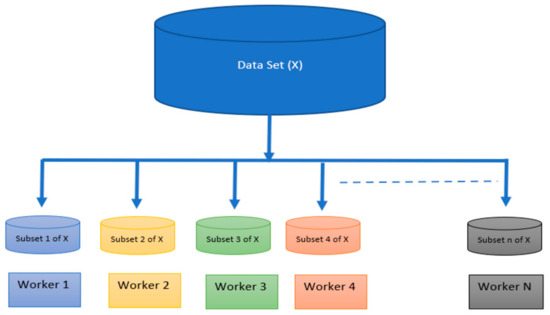
Figure 2.
Splitting a dataset among workers in shared and distributed memory systems.
3.2. Parallel Pool
A parallel pool (MatlabPool) is a set of workers working in parallel on a cluster of computing nodes or a multiprocessor system. Instead of executing a task via a single working node, the pool utilizes several workers to accomplish the same task on different chunks of data simultaneously. The working nodes in the pool can communicate with each of their neighbors throughout the lifetime of the running job. In MATLAB, the pool size and number of workers should be identified in advance. Figure 3 shows a request for creating a parallel pool of four working nodes. Reserved working nodes in a pool are not accessible to other clients, and only a single parallel pool can be created at a time for the client session in MATLAB.
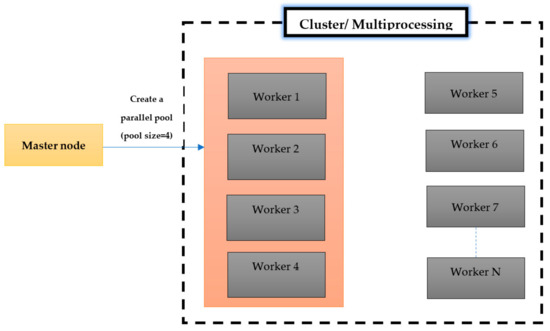
Figure 3.
MatlabPool (pool size = 4).
3.3. Loop Parallelism (ParFor)
This is a kind of parallelism in parallel programming that tries to parallelize the heavy computational tasks in loops. The ability to parallelize the sequential tasks in a loop is possible if the data are stored in a random-access structure. In the parallelized program, multiple threads will be employed to work on multiple indices simultaneously instead of working on one index at a time, as in the sequential program. In this manner, the overall execution for the loop will need less time, and the complexity of its processing is decreased. Parallelism can be performed simply just in the case of independent operations where each iteration in the loop is independent of other iterations. However, numerous algorithms may fail when parallelized due to the dependency between its iterations in a repeated loop. In such cases, some mechanisms like synchronization via message passing and semaphores should be employed.
4. Design and Implementation of ForkJoinPcc
In this section, the complexity of finding the dependencies between huge numbers of genes using Pcc is discussed. The serial algorithm SerialPcc for computing the correlation matrix for a set of genes is shown in Algorithm 1. It consists of three nested loops, which make it a heavy computational process. For example, if there are 20,000 genes measured in 35 microarrays, then there will be 20,000 × 20,000 × 35 iterations to obtain the Pcc between these genes (correlation matrix). It is undesirable to perform this computation serially as it has quadratic complexity.
| Algorithm 1: A serial algorithm for computing the Pcc in gene co-expression networks. |
| Input: Gene expression matrix of N genes expressed in S samples Output: Correlation matrix C (N,N) For I = 1 to N For K = 1 to N For J = 1 to S Cor_sum = Cor_sum + ((GenExp(i, j) − mean(GenExp(i, ;) ∗ (GenExp(k, j) − mean(GenExp(k, ;)))) X_sum = X_sum + ((GenExp(i, j) − mean(GenExp(i, ;)))^2 Y_sum = Y_sum + ((GenExp(k, j) − mean(GenExp(k, ;)))^2 End For End For End For |
The main idea in the ForkJoinPcc algorithm is breaking the nested loops, as shown in Algorithm 1. This was accomplished by dividing the whole computation into modules. Each module has an independent linear algebra. By examining the arithmetic formula of Pcc mentioned in Equation (1), this calculation can be divided into mini-processes as follows:
- Find the mean value of each gene;
- Subtract the mean value from all values in the gene expression matrix;
- Multiply the shifted matrix by its transpose;
- Square the shifted matrix;
- Find the sum of each row in the squared matrix;
- Find the correlation matrix between all genes.
In the ForkJoinPcc algorithm, the fork–join model is mimicked by breaking the mathematical operations into independent mini-operations that can be processed in parallel. Each mini-operation that can be performed in parallel is called a parallel section. The main framework of the ForkJoinPcc algorithm is depicted in Figure 4. The detailed steps for each parallel section are listed in Algorithm 2. It starts by preprocessing the microarrays in serial computation to compute the data array (the gene expression matrix). The data array is 22,277 × 35 in a row-major order of 22,277 genes and 35 microarray experiments. Then, the data array is broadcasted to all cores. In the first parallel section, the mean value of each gene is computed. This parallel section was implemented as follows. A pool of 4 labs (cores) in the multicore system was opened, and 12 cores were used in the cluster computer. A single program multiple data section was initiated, and the mean value of each gene in the gene expression matrix was calculated through a parallel loop inside this section. Each core worked on a subset of genes, and the results were stored in a distributed array. The results were gathered in the master node. In a new parallel section using the SPMD and parallel loop, all values for each gene were shifted by the mean value. In the same manner, the other parallel sections were implemented. However, the implementation of the parallel sections in steps 7 and 13, as listed in Algorithm 2, requires 2 nested loops that yield 22,277 × 22,277 iterations. Therefore, each iteration of the outer 22,277 iterations had a parallel for loop with 22,277 iterations. This way of implementation was undesirable for these parallel sections. Therefore, this obstacle was overcome in two ways: initiating parallel jobs for implementing these sections instead of SPMD and parallel loops. The task of the parallel job for the first parallel section is the multiplication of the shifted matrix by its transpose. The output of this task is a 22,277 × 22,277 matrix. However, such a matrix size cannot be allocated in the multicore platform. Therefore, in the multicore system, six jobs were created for performing this multiplication. Each job worked with 4000 items except the last one, which worked with 2277 items of the shifted matrix. When finished, each worker and lab sent their results to the client lab. In the same manner, the other bottleneck parallel section was implemented.
| Algorithm 2: A parallel algorithm for computing the correlation matrix |
| Input: Gene expression matrix, X, of N genes expressed in S samples Output: Correlation matrix C (N,N) 1 Initialize a pool of worker nodes. 2 Broadcast the gene expression matrix to all workers nodes. 3 In parallel, Compute the mean value of the expression of each gene in all samples, Mean[i] where i =1 to N. 4 Gather the results. 5 In parallel, subtract the mean from each value in X 6 Gather the results. 7 In parallel, multiply X_shifted by its transpose 8 Gather the results. 9 In parallel, multiply each element in Y by itself to get the Squared_Y 10 Gather the results. 11 In parallel, find the sum of each row in the matrix Squared_Y into the variable Sum_Sq_Y 12 Gather the results. 13 In parallel, find the correlation between gene i, and j 14 Gather the results. |
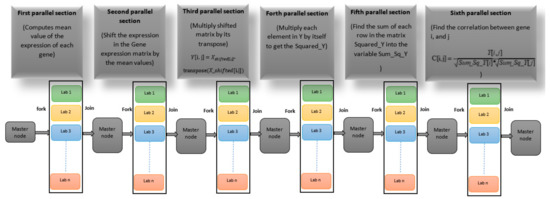
Figure 4.
A fork–join model for computing the correlation matrix in gene co-expression networks.
5. Experimental Set-Up and Dataset
The experiments were conducted on devices at the El Fayoum University High-Performance Computing Center. For the multicore platform, a Core i7 Intel 10750H processors with 2.60 GHz and 16 GB RAM were utilized. The Core i7 processor consists of four hyper-threaded cores. For each hyperthreaded processor core that is physically present, the operating system addresses two virtual processors and shares the workload between them when possible. For the cluster platform, the experiments were conducted on six nodes, with one master and five worker computers. Each node had a Core i7 Intel 10750H processor with 2.60 GHz and 16 GB RAM. The operating system that was utilized was Windows 10 (64-bit operating system with an x64-based processor). For the SDK, the MATLAB Parallel Computing Toolbox and MDCS release 2019a from MathWorks were been employed. The hardware details are shown in Table 1.

Table 1.
The hardware details of the experimental environment.
We conducted our experiments using a real dataset to launch a comparison of the performance of each parallel infrastructure. The gene expression matrix of hepatocellular carcinoma (HCC) was employed to estimate and analyze its gene networks and pathways. Hepatocellular carcinoma is a consequence of the hepatitis C virus (HCV), and it is the most important type of liver cancer. Raw data were downloaded from Gene Expression Omnibus (GEO) (Edgar, Domrachev, and Lash, 2002) and preprocessed via the Affy package [35] offered by Bioconductor [36]. The preprocessed gene expression matrix contained 22,277 genes and 35 normal or tumor samples.
6. Results and Discussion
To assess the adequacy of the ForkJoinPcc algorithm on the multicore system and the cluster computer, the computation time for each parallel section in the proposed algorithm was computed. In addition, the broadcast and gather times for different chunks of data for each parallel section were measured. The broadcast and gather time represents the communication overhead that can be faced when using a parallel infrastructure. On the other hand, the computation time is the useful part that should be decreased with the help of parallel computing. The detailed measured broadcast and gather and computation times for the first parallel section in the ForkJoinPcc algorithm for a different number of working cores are depicted in Table 2. For more illustration of the broadcast and gather time in all phases of the parallel sections of the ForkJoinPcc algorithm, the measured times for the second, third, fourth, fifth, and sixth parallel sections are listed in Table 3.

Table 2.
Broadcast and gather and computation times for the first parallel section.

Table 3.
Broadcast and gather and computation times for the second, third, fourth, fifth, and sixth parallel sections in the ForkJoinPcc algorithm.
In Table 2, the broadcast and gather time was calculated as the total elapsed time for the labSend and labReceive constructs between the master lab and worker lab to finish the first parallel section. It can be observed that the broadcast and gather time increased as the number of cores and labs increased. On the other hand, the computation time for the multicore system was reduced from 830 s to 210 s as the number of cores increased to 4 cores. A much lower computation time of 68.88 s was retrieved for the computer cluster. Such large growth in time for the multi-core execution was due to the third-party MATLAB parallel constructs. As mentioned before, there were three MATLAB parallel constructs employed in the implementation of the first parallel section, including SPMD, ParFor, and distributed arrays. The MATLAB distributed array is an API that was originally implemented on the top of the message-passing infrastructure, and each lab stores a portion of that array. In addition, the execution of the ParFor construct is perofrmed on labs that have been created on a Matpool. The labs in the pool do not share memory, which is a significant feature in the multicore platform.
As depicted in Table 3, the retrieved broadcast and gather time had a slight variation for the different parallel sections and was almost constant. The broadcast and gather time for the cluster platform was approximately double the corresponding time consumed for the multicore platform. It reached approximately 17.6 s for the whole dataset of 22,227 genes on the cluster platform. This reveals that the communication overhead in the cluster platform was high, but this time could be neglected with respect to the computation time, which was much higher than the broadcast and gather time on the cluster platform.
Additionally, the third and sixth parallel sections were the most time-consuming parallel sections in the ForkJoinPcc algorithm, as illustrated in Table 3. The processing time for the third parallel section for the whole dataset reached 16,518 and 883 s on the multicore and cluster platforms, respectively. A time of 19,446 s was needed to run the sixth parallel section on the multicore platform compared with the 900 s needed on the cluster platform. The implementation for these parallel sections was based on using parallel jobs and distributed arrays. The underlying strategy for the parallel jobs in MATLAB was based on physically separated nodes working on a cluster platform, which is inefficient for the shared-memory architecture of the multicore platform.
7. Comparing the Performance
In this section, the performance of the ForkJoinPcc algorithm is compared to Seri-alPcc. The speedup for the ForkJoinPcc algorithm was calculated on the multicore and cluster platforms. For an illustration of the performance of the ForkJoinPcc algorithm, the speedup for different data sizes, a different number of genes (N), and a different number of samples (M), these are listed in Table 4. As illustrated in Table 4, it can be noticed that the speedup of ForkJoinPcc on the cluster platform (17–62×) was much greater than the corresponding one on the multicore platform (3.4–3.9×) for different data sizes. This is because of the higher efficiency and compatibility of the third-party toolset (MATLAB Parallel Computing tools) to the underlying message-passing infrastructure of the cluster platform. The main clue of the offered MATLAB Parallel Computing tools is to extend MATLAB’s capabilities into the parallel computing industry and introduce MATLAB tools that can help in parallelizing the code to be executed in a cluster computing environment [25,37,38]. Figure 5 depicts the key components of the software stack that support both the MATLAB and Parallel Computing Toolbox functionalities, while the significant feature of the multicore platform, shared memory architecture, has not been efficiently utilized using the MATLAB parallel toolsets.

Table 4.
Speedup of the ForkJoinPcc algorithm on different platforms.
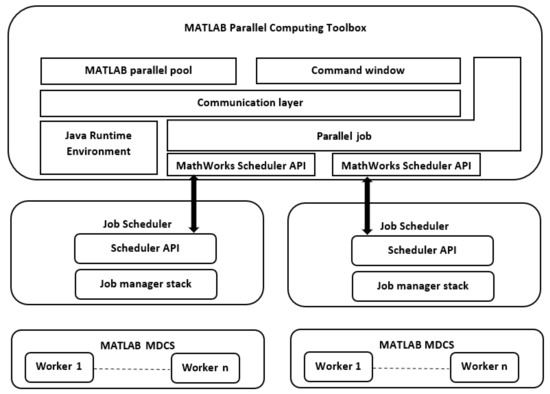
Figure 5.
Parallel computing software stack in MATLAB.
As depicted in Table 4, the performance of the ForkJoinPcc algorithm was compared to our previous contribution in [37] for implementing a parallel implementation for the same application on a cloud platform. Two big data approaches including MapReduce and Spark were introduced for computation of the Pcc similarity matrix in the GCN. For 22,277 genes, the processing of the Pcc matrix consumed 1789.98 and 17,443.98 s on the cloud platform using the Spark and MapReduce techniques, respectively. The speedup can be listed in descending order as follows: 80×, 62×, 8.22×, and 3.8× for Spark and ForkJoinPcc on the cluster platform and MapReduce and ForkJoinPcc on the multicore platform, respectively. when comparing the speedup on the cloud and the multicore and cluster environments, the parallel implementation using Spark on the cloud platform yielded fast processing and an 80.1× speedup of the Pcc matrix for the whole number of genes. The data processing performed in the main memory of the worker nodes in the spark system and the avoidance of unnecessary I/O operations with the disks helped in accomplishing its higher performance with respect to the other techniques.
8. Conclusions
The interactions between the co-expressed genes in a GCN can be identified with the help of computing the correlation coefficients between all genes. The construction of such a matrix is a heavy computation, and the present study proposes a parallel algorithm, ForkJoinPcc, for its implementation on shared and distributed multiprocessing platforms to find the dependencies between all the huge numbers of genes measured in high-throughput microarrays. The main idea in the proposed algorithm mimics a well-known parallel programming model: the fork–join model. The implementation was performed using the parallel MATLAB APIs introduced in the MATLAB Parallel Computing Toolbox and the MATLAB Distributed Computing Server. Four constructs from MATLAB, including SPMD, distributed arrays, MatlabPool, and ParFor, were utilized. Our evaluation for the parallel MATLAB tools based on the high values of the speedup attained on the cluster platform implies that these tools are more compatible with the underlying message-passing infrastructure of the cluster platform than the multicore platform. However, the pros of the multicore platform, such as a shared memory architecture, have not been effectively employed using the MATLAB parallel toolsets.
In this study, we created a different parallel algorithm and implementation for the same application: the correlation matrix in gene co-expression networks. We employed the fork–join model in the introduced algorithm, which had a similar idea to MapReduce and Spark. The fork–join approach utilizes the divide-and-conquer algorithms that recursively fork processes running in parallel, waits for them to finish, and then merges their results [38]. However, we obtained negative results compared with the yielded results we found in our previous work using Spark.
Author Contributions
Conceptualization, A.A.A., H.N.A., G.A., N.H.S., R.A.A.A.A.S., O.S.A., V.F.G. and N.A.S.; methodology, A.A.A., H.N.A., G.A., N.H.S., R.A.A.A.A.S., O.S.A., V.F.G. and N.A.S.; software, N.A.S.; validation, A.A.A., H.N.A., G.A., N.H.S., R.A.A.A.A.S., O.S.A., V.F.G. and N.A.S.; formal analysis, A.A.A., H.N.A., G.A., N.H.S., R.A.A.A.A.S., O.S.A., V.F.G. and N.A.S.; investigation, A.A.A., H.N.A., G.A., N.H.S., R.A.A.A.A.S., O.S.A., V.F.G. and N.A.S.; resources, N.A.S.; data curation, N.A.S.; writing—original draft preparation, A.A.A., H.N.A., G.A., N.H.S., R.A.A.A.A.S., O.S.A., V.F.G. and N.A.S.; writing—review and editing, A.A.A., H.N.A., G.A., N.H.S., R.A.A.A.A.S., O.S.A., V.F.G. and N.A.S.; visualization, N.A.S.; supervision, N.H.S. and R.A.A.A.A.S.; project administration, A.A.A.; funding acquisition, A.A.A. All authors have read and agreed to the published version of the manuscript.
Funding
Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R308), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors express their gratitude to Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R308), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Abdel Samee, N.M.; Solouma, N.H.; Kadah, Y.M. Detection of Biomarkers for Hepatocellular Carcinoma Using a Hybrid Univariate Gene Selection Methods. Theor. Biol. Med. Model. 2012, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Samee, N.M.A.; Solouma, N.H.; Kadah, Y.M. Gene Network Construction and Pathways Analysis for High Throughput Microarrays. In Proceedings of the National Radio Science Conference, NRSC, Cairo, Egypt, 10–12 April 2012; pp. 649–658. [Google Scholar]
- De Wael, M.; Marr, S.; Van Cutsem, T. Fork/Join Parallelism in the Wild: Documenting Patterns and Anti-Patterns in Java Programs Using the Fork/Join Framework. In Proceedings of the PPPJ ’14 International Conference on Principles and Practices of Programming on the Java Platform: Virtual Machines, Languages, and Tools, Krakow, Poland, 23–26 September 2014; Association for Computing Machinery: New York, NY, USA, 2014; Volume 13, pp. 39–50. [Google Scholar]
- Francis, N.; Mathew, J. Implementation of Parallel Clustering Algorithms Using Join and Fork Model. In Proceedings of the 2016 Online International Conference on Green Engineering and Technologies, IC-GET 2016, Online, 19 November 2016; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2017. [Google Scholar]
- Jacob, A.C.; Eichenberger, A.E.; Sung, H.; Antao, S.F.; Bercea, G.T.; Bertolli, C.; Bataev, A.; Jin, T.; Chen, T.; Sura, Z.; et al. Efficient Fork-Join on GPUs through Warp Specialization. In Proceedings of the 24th IEEE International Conference on High Performance Computing, HiPC 2017, Jaipur, India, 18–21 December 2017; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2018; Volume 2017, pp. 358–367. [Google Scholar]
- Liang, M.; Zhang, F.; Jin, G.; Zhu, J. FastGCN: A GPU Accelerated Tool for Fast Gene Co-Expression Networks. PLoS ONE 2015, 10, e0116776. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Schmidt, B.; Liu, W.; Müller-Wittig, W. Parallel Mutual Information Estimation for Inferring Gene Regulatory Networks on GPUs. BMC Res. Notes 2011, 4, 189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Horvath, S. A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4. [Google Scholar] [CrossRef]
- Cai, Y.; Ma, F.; Qu, L.H.; Liu, B.; Xiong, H.; Ma, Y.; Li, S.; Hao, H. Weighted Gene Co-Expression Network Analysis of Key Biomarkers Associated with Bronchopulmonary Dysplasia. Front. Genet. 2020, 11, 539292. [Google Scholar] [CrossRef]
- DeRisi, J.; Penland, L.; Brown, P.O.; Bittner, M.L.; Meltzer, P.S.; Ray, M.; Chen, Y.; Su, Y.A.; Trent, J.M. Use of a CDNA Microarray to Analyse Gene Expression Patterns in Human Cancer. Nat. Genet. 1996, 14, 457–460. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- García-Calvo, R.; Guisado, J.L.; Diaz-del-Rio, F.; Córdoba, A.; Jiménez-Morales, F. Graphics Processing Unit–Enhanced Genetic Algorithms for Solving the Temporal Dynamics of Gene Regulatory Networks. Evol. Bioinform. 2018, 14. [Google Scholar] [CrossRef]
- González-Domínguez, J.; Martín, M.J. Fast Parallel Construction of Correlation Similarity Matrices for Gene Co-Expression Networks on Multicore Clusters. Procedia Comput. Sci. 2017, 108, 485–494. [Google Scholar] [CrossRef]
- Casal, U.; González-Domínguez, J.; Martín, M.J. Analysis of the Construction of Similarity Matrices on Multi-Core and Many-Core Platforms Using Different Similarity Metrics. In Proceedings of the Lecture Notes in Computer Science; Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics; Springer: Berlin/Heidelberg, Germany, 2019; Volume 11536 LNCS, pp. 168–181. [Google Scholar]
- Gonzalez-Dominguez, J.; Martin, M.J. MPIGeneNet: Parallel Calculation of Gene Co-Expression Networks on Multicore Clusters. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 15, 1732–1737. [Google Scholar] [CrossRef]
- Zola, J.; Aluru, M.; Sarje, A.; Aluru, S. Parallel Information-Theory-Based Construction of Genome-Wide Gene Regulatory Networks. IEEE Trans. Parallel Distrib. Syst. 2010, 21, 1721–1733. [Google Scholar] [CrossRef]
- Song, L.; Langfelder, P.; Horvath, S. Comparison of Co-Expression Measures: Mutual Information, Correlation, and Model Based Indices. BMC Bioinform. 2012, 13, 328. [Google Scholar] [CrossRef] [PubMed]
- Rossini, A.J.; Tierney, L.; Li, N. Simple Parallel Statistical Computing in R. J. Comput. Graph. Stat. 2007, 16, 399–420. [Google Scholar] [CrossRef][Green Version]
- Chang, D.J.; Desoky, A.H.; Ouyang, M.; Rouchka, E.C. Compute Pairwise Manhattan Distance and Pearson Correlation Coefficient of Data Points with GPU. In Proceedings of the 10th ACIS Conference on Software Engineering, Artificial Intelligence, Networking and Parallel/Distributed Computing, SNPD 2009, in Conjunction with IWEA 2009 and WEACR 2009, Daegu, Korea, 27–29 May 2009; pp. 501–506. [Google Scholar]
- Chilson, J.; Ng, R.; Wagner, A.; Zamar, R. Parallel Computation of High-Dimensional Robust Correlation and Covariance Matrices. Algorithmica 2006, 45, 403–431. [Google Scholar] [CrossRef][Green Version]
- Zhu, H.; Li, P.; Zhang, P.; Luo, Z. A High Performance Parallel Ranking SVM with OpenCL on Multicore and Many-Core Platforms. Int. J. Grid High Perform. Comput. 2019, 11, 12. [Google Scholar] [CrossRef]
- Kijsipongse, E.; U-Ruekolan, S.; Ngamphiw, C.; Tongsima, S. Efficient Large Pearson Correlation Matrix Computing Using Hybrid MPI/CUDA. In Proceedings of the 2011 8th International Joint Conference on Computer Science and Software Engineering, JCSSE 2011, Nakhon Pathom, Thailand, 11–13 May 2011; pp. 237–241. [Google Scholar]
- Eslami, T.; Saeed, F. Fast-GPU-PCC: A GPU-Based Technique to Compute Pairwise Pearson’s Correlation Coefficients for Time Series Data—FMRI Study. High-Throughput 2018, 7, 11. [Google Scholar] [CrossRef]
- Sokolinsky, L.B. BSF: A Parallel Computation Model for Scalability Estimation of Iterative Numerical Algorithms on Cluster Computing Systems. J. Parallel Distrib. Comput. 2021, 149, 193–206. [Google Scholar] [CrossRef]
- Sharma, G.; Martin, J. MATLAB®: A Language for Parallel Computing. Int. J. Parallel Program. 2009, 37, 3–36. [Google Scholar] [CrossRef]
- Kepner, J. Parallel Programming with MatlabMPI. arXiv 2001, arXiv:astro-ph/0107406. [Google Scholar] [CrossRef]
- Microsoft MPI—Message Passing Interface. Microsoft Docs. Available online: https://docs.microsoft.com/en-us/message-passing-interface/microsoft-mpi (accessed on 20 February 2022).
- Hummel, S.F.; Ngo, T.; Srinivasan, H. SPMD Programming in Java. Concurr. Pract. Exp. 1997, 9, 621–631. [Google Scholar] [CrossRef]
- Chandra, R.; Dagum, L.; Kohr, D.; Maydan, D.; McDonald, J.; Menon, R. Parallel Programming in OpenMP; Morgan Kaufmann Publishers: San Francisco, CA, USA, 2001. [Google Scholar]
- Allen, E.; Chase, D.; Hallett, J.; Luchangco, V.; Maessen, J.-W.; Ryu, S.; Steele, G.; Tobin-Hochstadt, S. The Fortress Language Specification; Sun Microsystems: Santa Clara, CA, USA, 2007. [Google Scholar]
- Stripinis, L.; Žilinskas, J.; Casado, L.G.; Paulavičius, R. On MATLAB Experience in Accelerating DIRECT-GLce Algorithm for Constrained Global Optimization through Dynamic Data Structures and Parallelization. Appl. Math. Comput. 2021, 390, 125596. [Google Scholar] [CrossRef]
- Travinin Bliss, N.; Kepner, J. PMATLAB Parallel MATLAB Library. Int. J. High Perform. Comput. Appl. 2007, 21, 336–359. [Google Scholar] [CrossRef]
- Kepner, J.; Ahalt, S. MatlabMPI. J. Parallel Distrib. Comput. 2004, 64, 997–1005. [Google Scholar] [CrossRef]
- Hudak, D.E.; Ludban, N.; Gadepally, V.; Krishnamurthy, A. Developing a Computational Science IDE for HPC Systems. In Proceedings of the ICSE 2007 Workshops: Third International Workshop on Software Engineering for High Performance Computing Applications, SE-HPC’07, Minneapolis, MN, USA, 26 May 2007; pp. 5–9. [Google Scholar]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Affy-Analysis of Affymetrix GeneChip Data at the Probe Level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open Software Development for Computational Biology and Bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed]
- Samee, N.A.; Osman, N.H.; Seoud, R.A.A.A.A. Comparing MapReduce and Spark in Computing the PCC Matrix in Gene Co-Expression Networks. Int. J. Adv. Comput. Sci. Appl. 2021, 12, 2021. [Google Scholar] [CrossRef]
- Rosales, E.; Rosà, A.; Binder, W. FJProf: Profiling Fork/Join Applications on the Java Virtual Machine. In Proceedings of the VALUETOOLS’20: 13th EAI International Conference on Performance Evaluation Methodologies and Tools, Tsukuba, Japan, 18–20 May 2020; ACM International Conference Proceeding Series. Association for Computing Machinery: New York, NY, USA, 2020; pp. 128–135. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).