Ensemble Self-Paced Learning Based on Adaptive Mixture Weighting
Abstract
1. Introduction
2. Background
2.1. Self-Paced Learning
| Algorithm 1: Algorithm of SPL |
Input: The training dataset . Output: Model parameter .  |
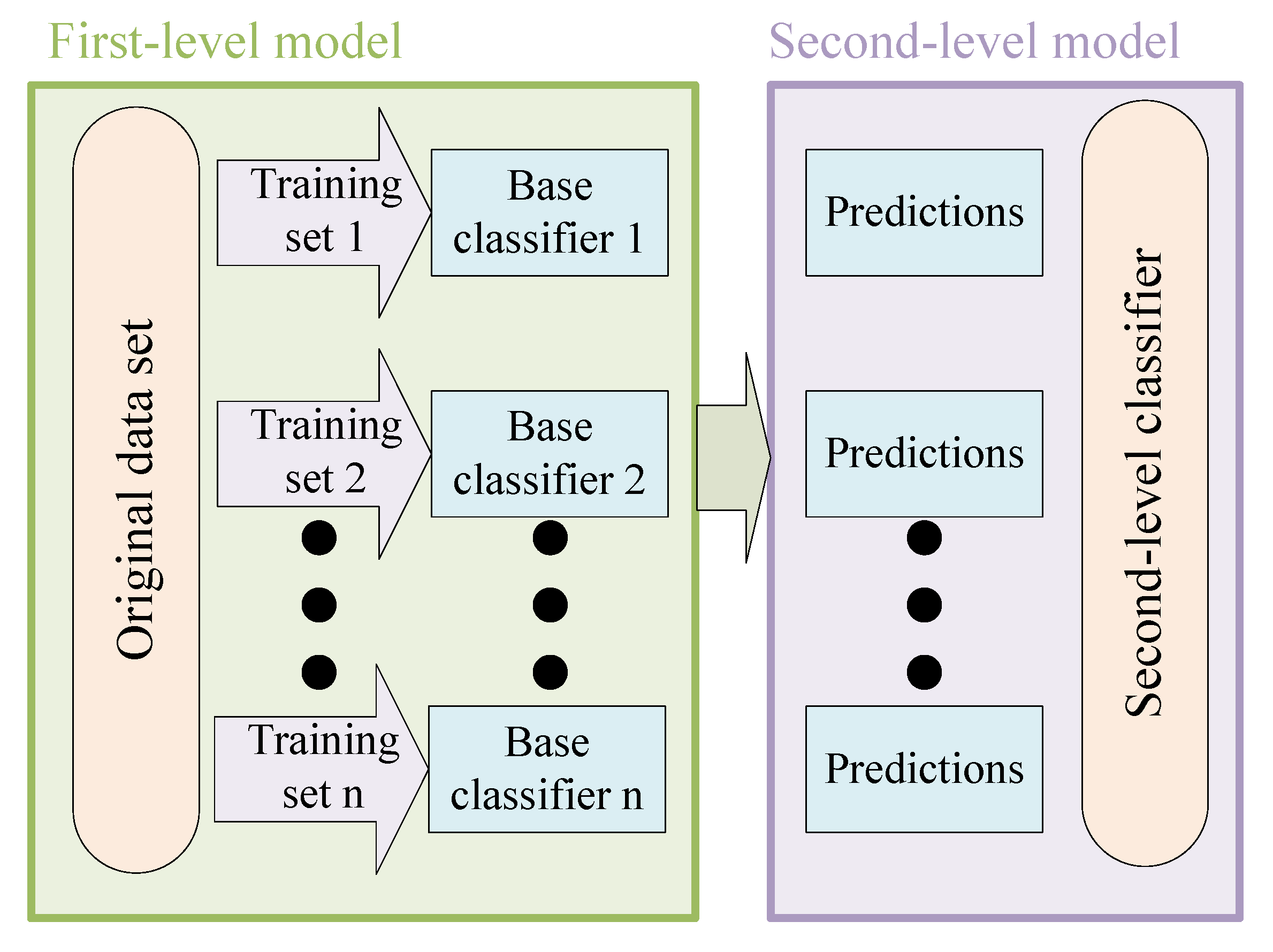
2.2. Ensemble Stacking Learning
3. Methodology
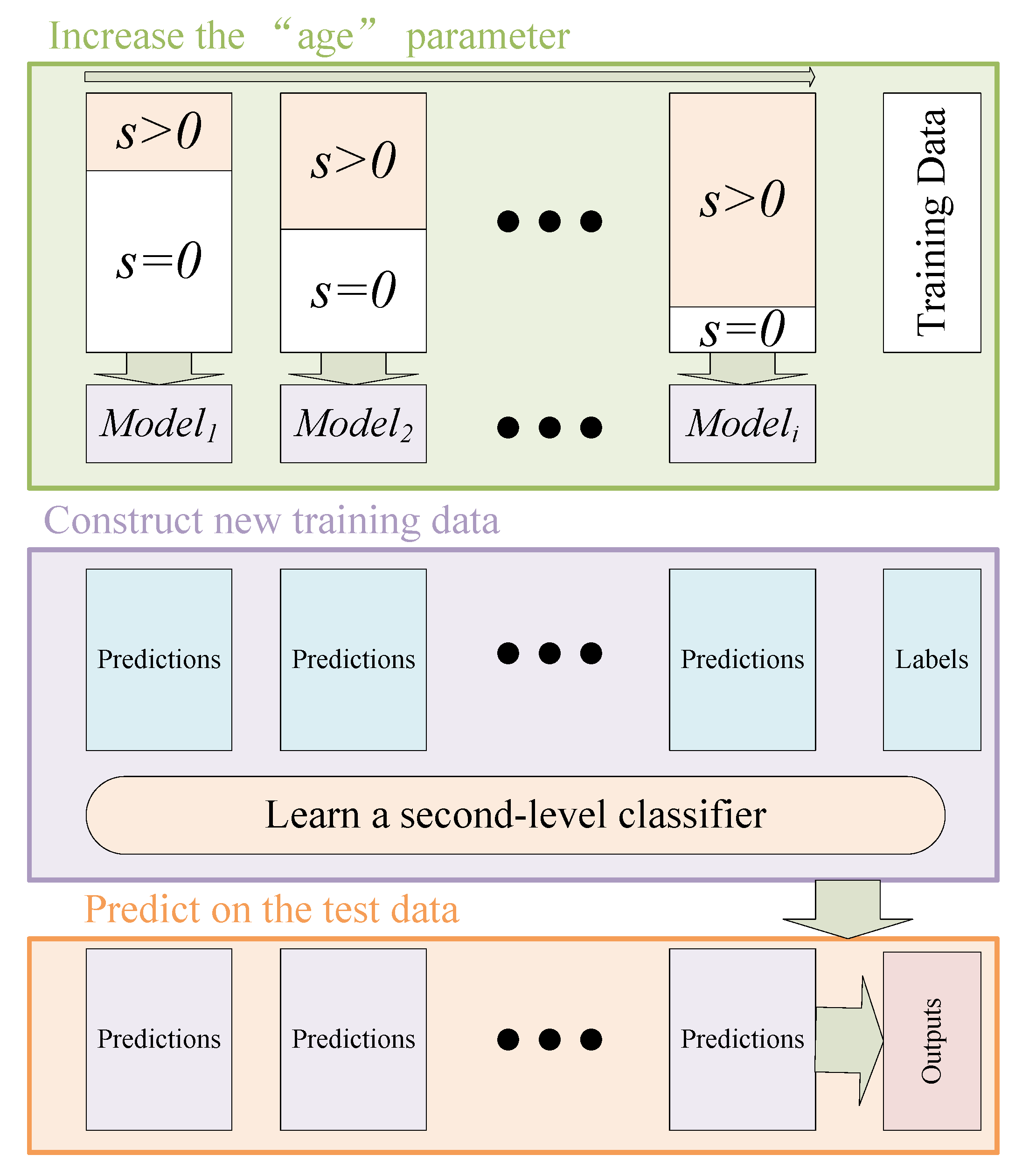
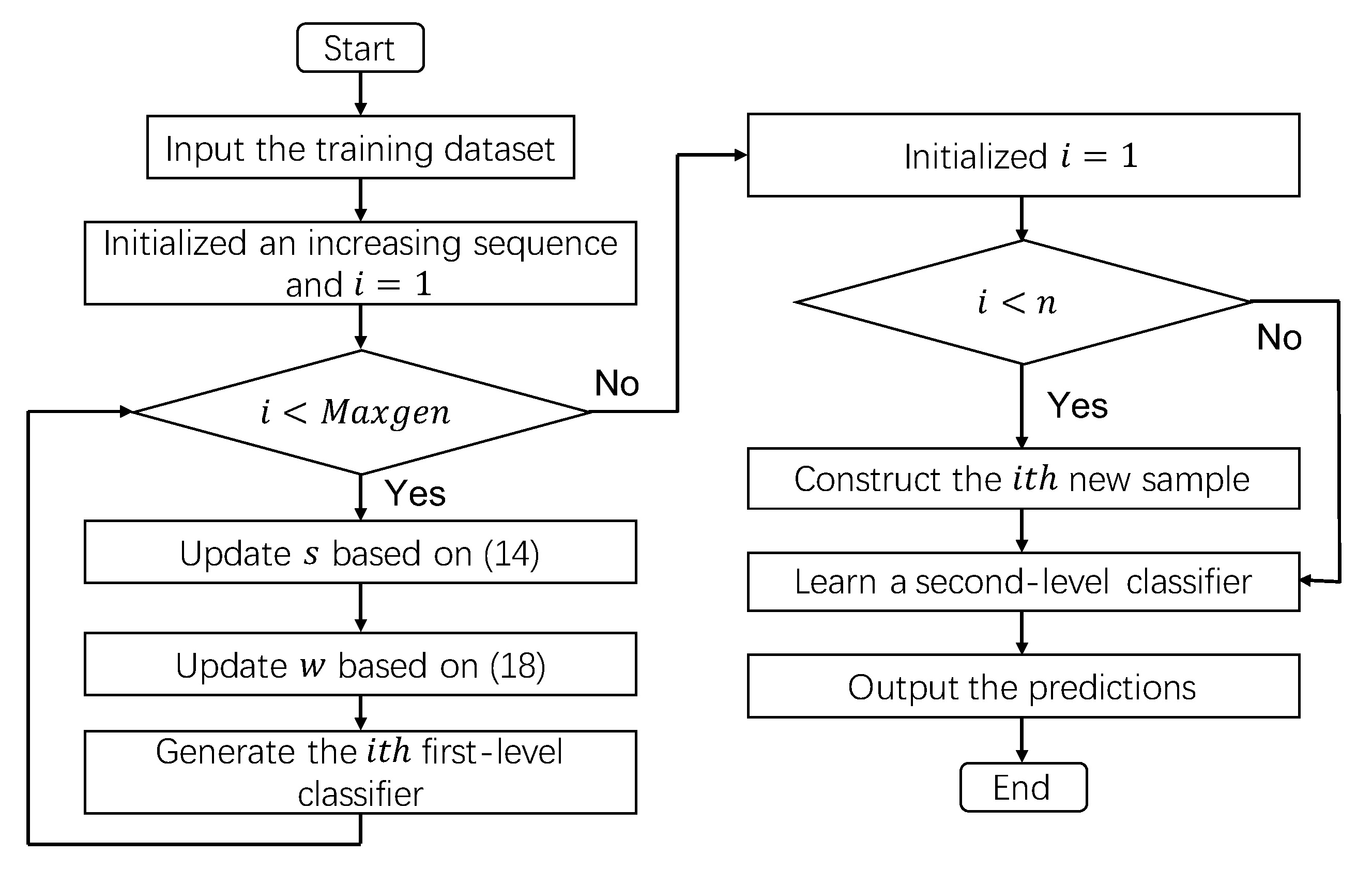
3.1. Ensemble Self-Paced Learning
| Algorithm 2: Algorithm of Ensemble SPL |
Input: The training dataset . Output: An ensemble clssifier H.  |
3.2. Adaptive Mixture Weighting
3.3. Optimization and Analysis
3.3.1. Majorization Step
3.3.2. Minimization Step
3.3.3. Parameter Analysis
3.4. Analysis
4. Experimental Studies
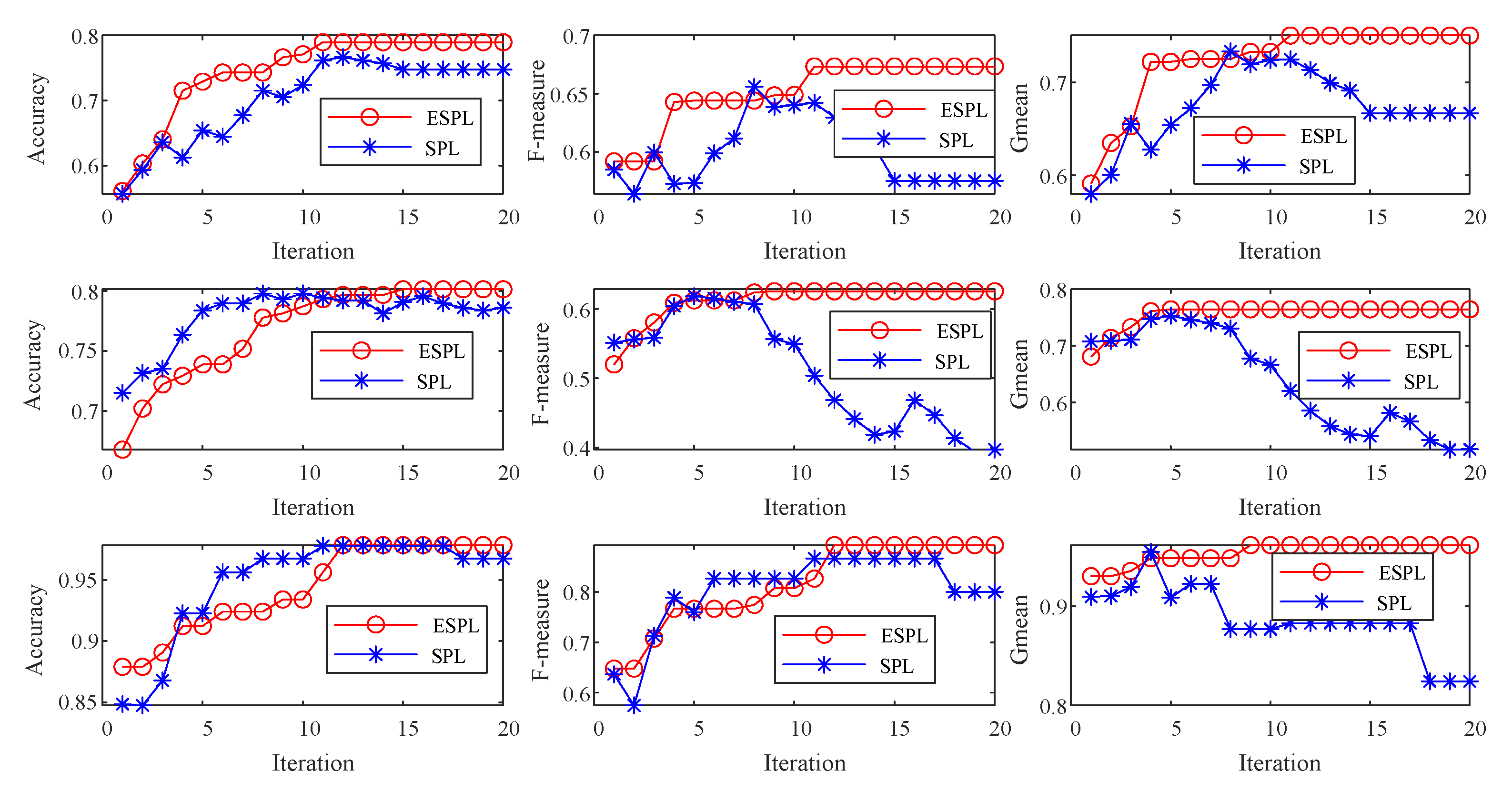
4.1. Test of the Proposed Ensemble Strategy
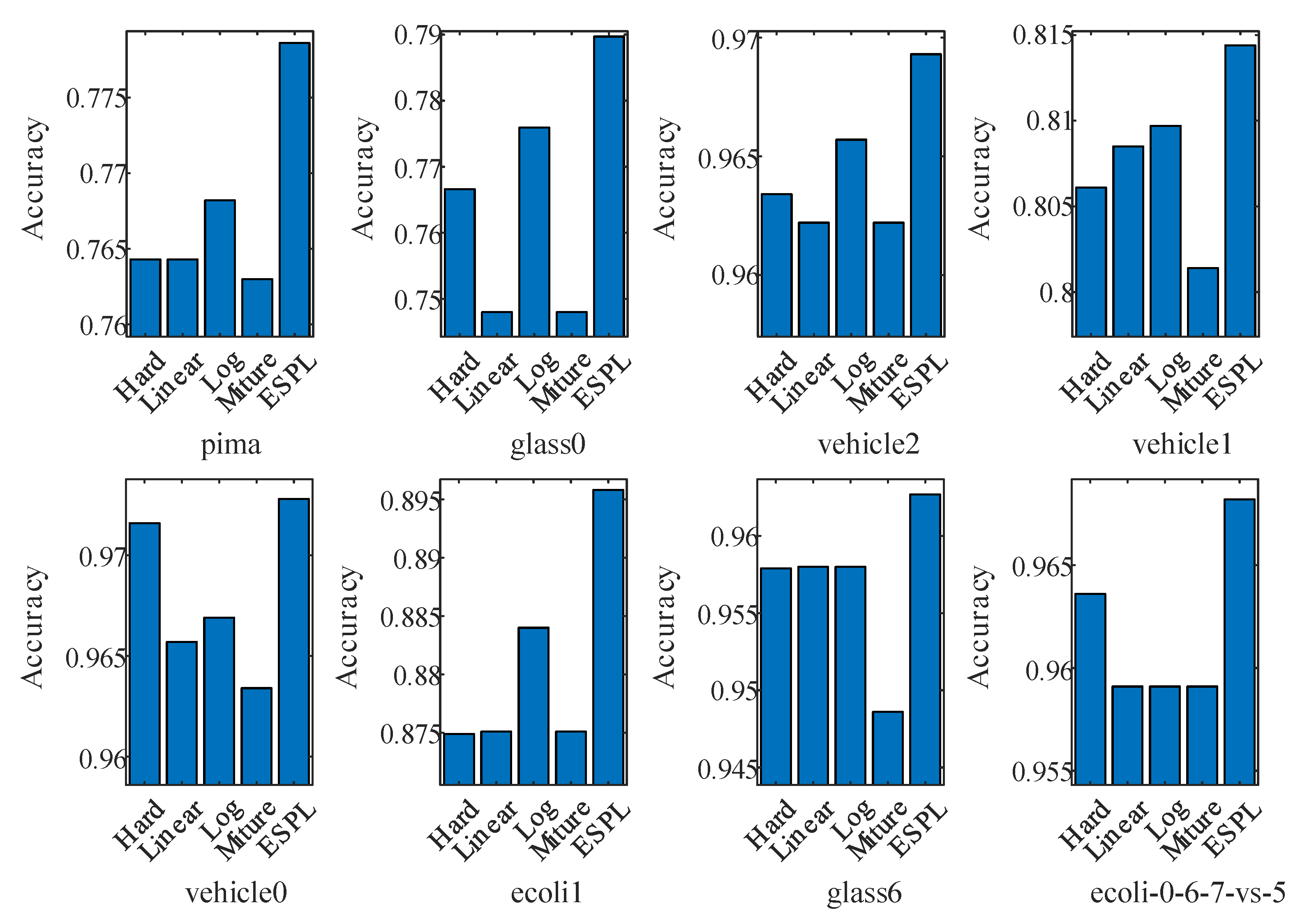
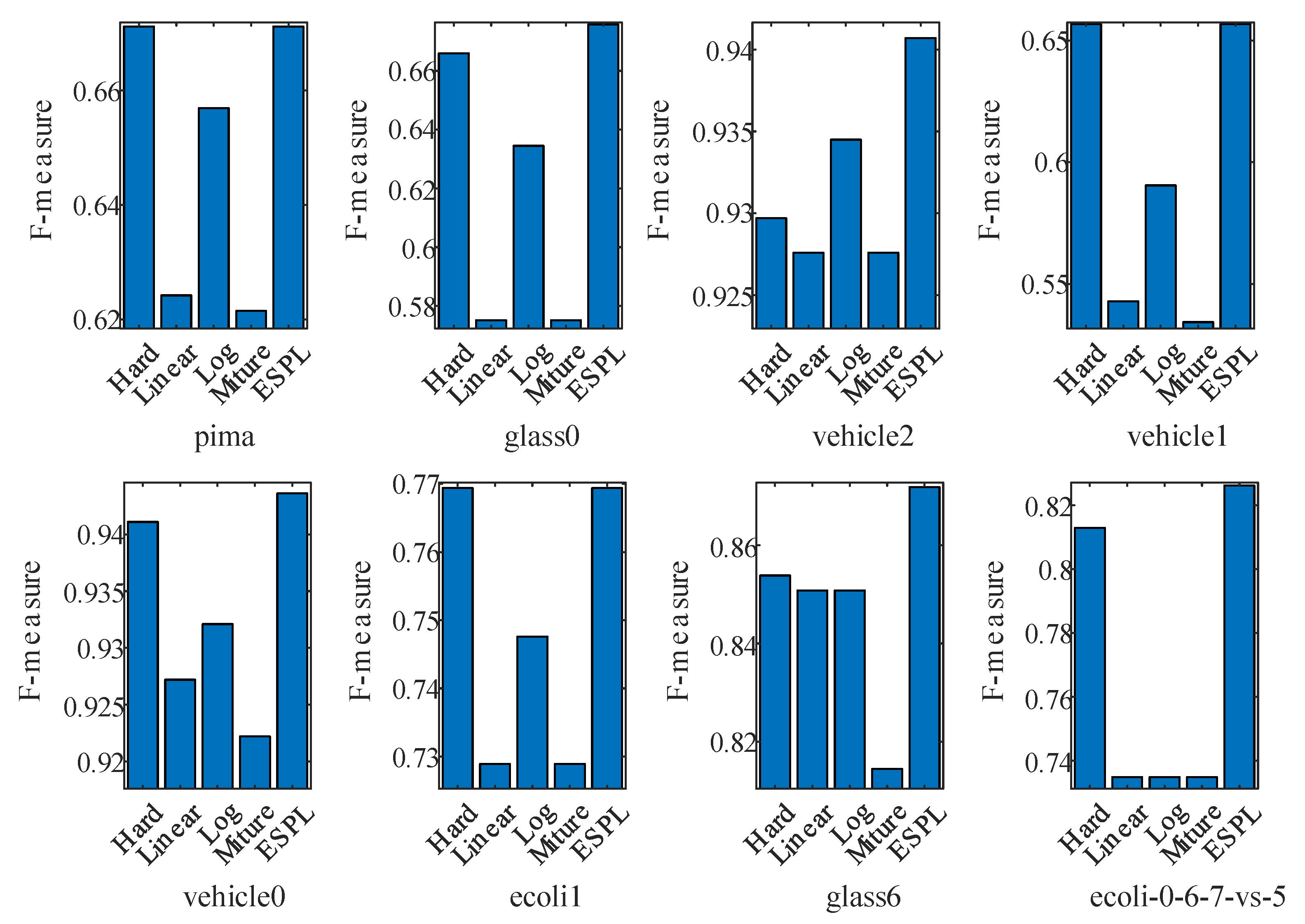
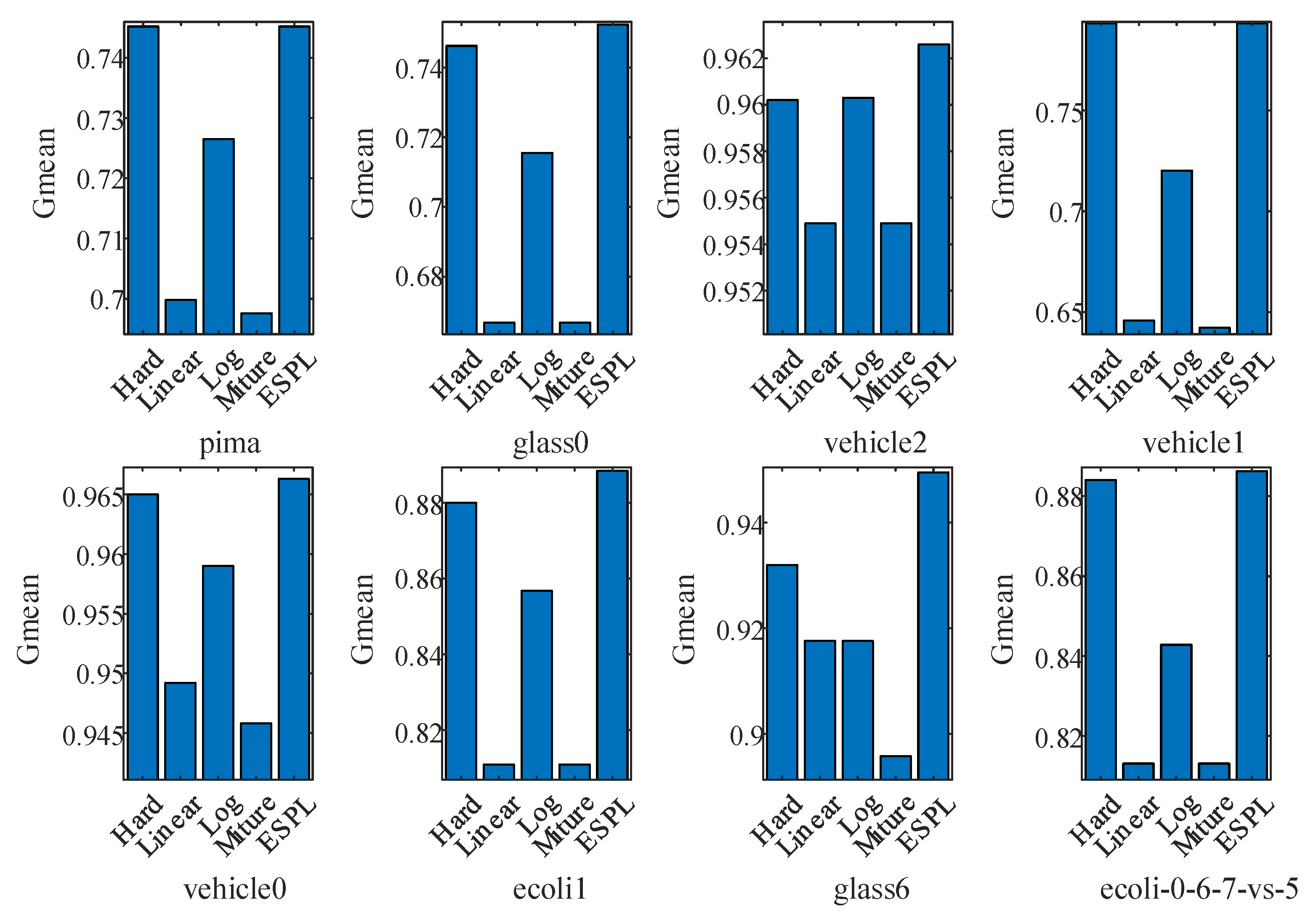
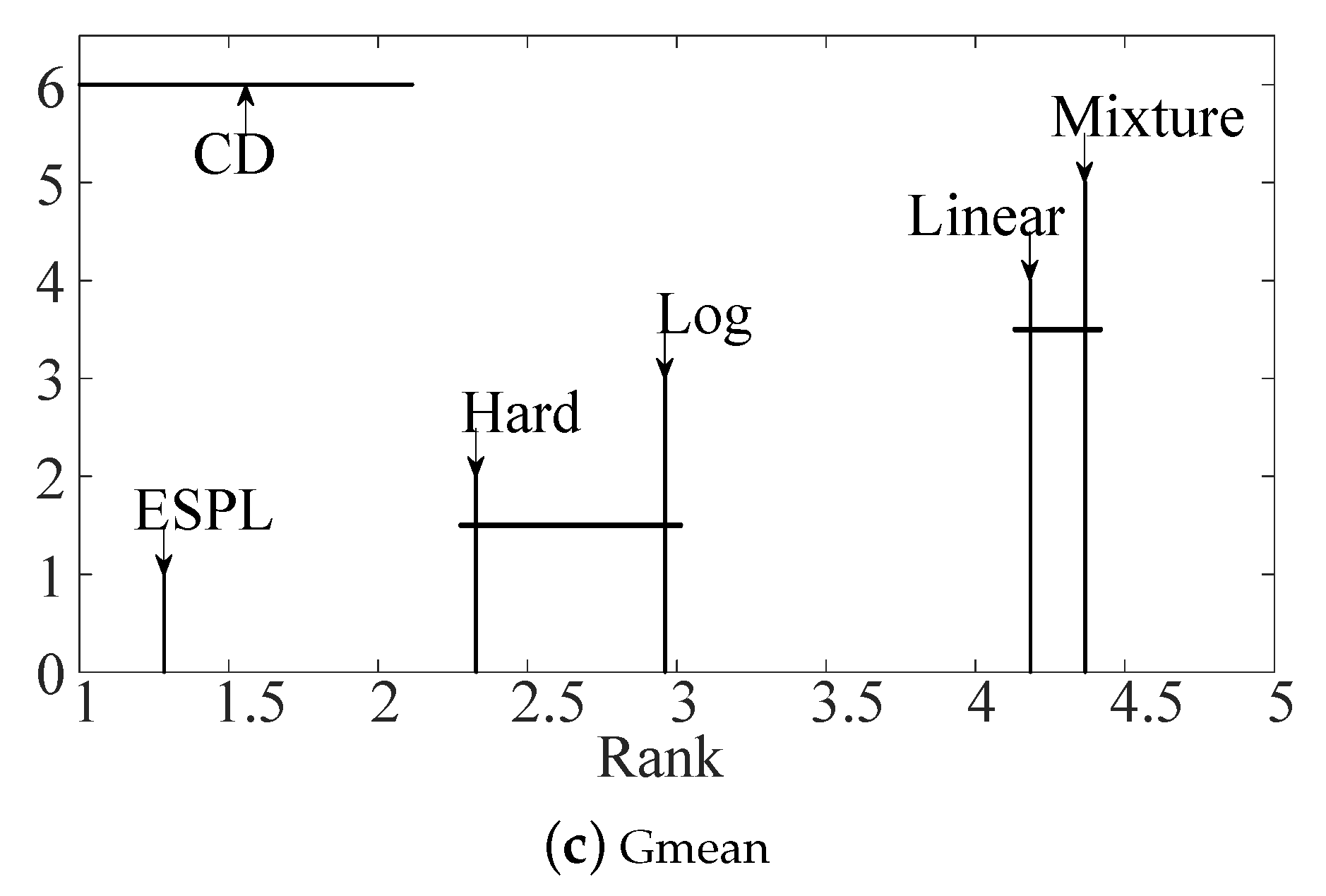
4.2. Test of the Proposed Adaptive Mixture Weighting Method
4.3. Experimental Results
5. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kumar, M.P.; Packer, B.; Koller, D. Self-paced learning for latent variable models. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 6–9 December 2010; pp. 1189–1197. [Google Scholar]
- Bengio, Y.; Louradour, J.; Collobert, R.; Weston, J. Curriculum learning. In Proceedings of the 26th Annual International Conference on Machine Learning, Montreal, QC, Canada, 14–18 June 2009; pp. 41–48. [Google Scholar]
- Bae, J.; Kim, T.; Lee, W.; Shim, I. Curriculum learning for vehicle lateral stability estimations. IEEE Access 2021, 9, 89249–89262. [Google Scholar] [CrossRef]
- Wang, X.; Chen, Y.; Zhu, W. A survey on curriculum learning. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 44, 4555–4576. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Meng, D.; Zhao, Q.; Shan, S.; Hauptmann, A.G. Self-paced curriculum learning. In Proceedings of the 29th AAAI Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015; pp. 2694–2700. [Google Scholar]
- Wang, W.; Feng, R.; Chen, J.; Lu, Y.; Chen, T.; Yu, H.; Chen, D.Z.; Wu, J. Nodule-plus R-CNN and deep self-paced active learning for 3D instance segmentation of pulmonary nodules. IEEE Access 2019, 7, 128796–128805. [Google Scholar] [CrossRef]
- Jiang, L.; Meng, D.; Mitamura, T.; Hauptmann, A.G. Easy samples first: Self-paced reranking for zero-example multimedia search. In Proceedings of the 22nd ACM International Conference on Multimedia, Orlando, FL, USA, 3–7 November 2014; pp. 547–556. [Google Scholar]
- Li, H.; Gong, M.; Meng, D.; Miao, Q. Multi-objective self-paced learning. In Proceedings of the 30th AAAI Conference on Artificial Intelligence, Phoenix, AZ, USA, 12–17 February 2016; pp. 1802–1808. [Google Scholar]
- Zhao, Q.; Meng, D.; Jiang, L.; Xie, Q.; Xu, Z.; Hauptmann, A.G. Self-paced learning for matrix factorization. In Proceedings of the 29th AAAI Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015; pp. 3196–3202. [Google Scholar]
- Li, H.; Gong, M.; Wang, C.; Miao, Q. Pareto self-paced learning based on differential evolution. IEEE Trans. Cybern. 2021, 51, 4187–4200. [Google Scholar] [CrossRef]
- Supancic, J.S.; Ramanan, D. Self-paced learning for long-term tracking. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Portland, OR, USA, 23–28 June 2013; pp. 2379–2386. [Google Scholar]
- Ge, D.; Song, J.; Qi, Y.; Wang, C.; Miao, Q. Self-paced dense connectivity learning for visual tracking. IEEE Access 2019, 7, 37181–37191. [Google Scholar] [CrossRef]
- Peng, J.; Wang, P.; Desrosiers, C.; Pedersoli, M. Self-paced contrastive learning for semi-supervised medical image segmentation with meta-labels. In Proceedings of the Advances in Neural Information Processing Systems, Virtual, 7–10 December 2021; Volume 34. [Google Scholar]
- Li, H.; Li, J.; Zhao, Y.; Gong, M.; Zhang, Y.; Liu, T. Cost-sensitive self-paced learning with adaptive regularization for classification of image time series. IEEE J. Sel. Top. Appl. Earth Observ. Remote Sens. 2021, 14, 11713–11727. [Google Scholar] [CrossRef]
- Pan, X.; Wei, D.; Zhao, Y.; Ma, M.; Wang, H. Self-paced learning with diversity for medical image segmentation by using the query-by-committee and dynamic clustering techniques. IEEE Access 2020, 9, 9834–9844. [Google Scholar] [CrossRef]
- Zhang, D.; Meng, D.; Li, C.; Jiang, L.; Zhao, Q.; Han, J. A self-paced multiple-instance learning framework for co-saliency detection. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 594–602. [Google Scholar]
- Meng, D.; Zhao, Q.; Jiang, L. A theoretical understanding of self-paced learning. Inform. Sci. 2017, 414, 319–328. [Google Scholar] [CrossRef]
- Klink, P.; Abdulsamad, H.; Belousov, B.; D’Eramo, C.; Peters, J.; Pajarinen, J. A probabilistic interpretation of self-paced learning with applications to reinforcement learning. J. Mach. Learn. Res. 2021, 22, 1–52. [Google Scholar]
- Jiang, L.; Meng, D.; Yu, S.-I.; Lan, Z.; Shan, S.; Hauptmann, A. Self-paced learning with diversity. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; Volume 27, pp. 2078–2086. [Google Scholar]
- Lin, L.; Wang, K.; Meng, D.; Zuo, W.; Zhang, L. Active self-paced learning for cost-effective and progressive face identification. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 7–19. [Google Scholar] [CrossRef]
- Sun, L.; Zhou, Y. FSPMTL: Flexible self-paced multi-task learning. IEEE Access 2020, 8, 132012–132020. [Google Scholar] [CrossRef]
- Li, H.; Gong, M.; Zhang, M.; Wu, Y. Spatially self-paced convolutional networks for change detection in heterogeneous images. IEEE J. Sel. Topics Appl. Earth Observ. Remote Sens. 2021, 14, 4966–4979. [Google Scholar] [CrossRef]
- Gong, M.; Li, H.; Meng, D.; Miao, Q.; Liu, J. Decomposition-based evolutionary multi-objective optimization to self-paced learning. IEEE Trans. Evol. Comput. 2019, 23, 288–302. [Google Scholar] [CrossRef]
- Huang, Z.; Ren, Y.; Liu, W.; Pu, X. Self-paced multi-view clustering via a novel soft weighted regularizer. IEEE Access 2019, 7, 168629–168636. [Google Scholar] [CrossRef]
- Gu, B.; Zhai, Z.; Li, X.; Huang, H. Finding age path of self-paced learning. In Proceedings of the 2021 IEEE International Conference on Data Mining, Auckland, New Zealand, 7–10 December 2021; pp. 151–160. [Google Scholar]
- Dong, X.; Yu, Z.; Cao, W.; Shi, Y.; Ma, Q. A survey on ensemble learning. Front. Comput. Sci. 2020, 14, 241–258. [Google Scholar] [CrossRef]
- Galar, M.; Fernandez, A.; Barrenechea, E.; Bustince, H.; Herrera, F. A review on ensembles for the class imbalance problem: Bagging-, boosting-, and hybrid-based approaches. IEEE Trans. Syst. Man Cybern. C Appl. Rev. 2011, 42, 463–484. [Google Scholar] [CrossRef]
- Khoshgoftaar, T.M.; Van Hulse, J.; Napolitano, A. Comparing boosting and bagging techniques with noisy and imbalanced data. IEEE Trans. Syst. Man Cybern. A Syst. Humans 2010, 41, 552–568. [Google Scholar] [CrossRef]
- Martínez-Muñoz, G.; Suárez, A. Out-of-bag estimation of the optimal sample size in bagging. Pattern Recogn. 2010, 43, 143–152. [Google Scholar] [CrossRef]
- Mordelet, F.; Vert, J.-P. A bagging SVM to learn from positive and unlabeled examples. Pattern Recogn. Lett. 2014, 37, 201–209. [Google Scholar] [CrossRef]
- Prokhorenkova, L.; Gusev, G.; Vorobev, A.; Dorogush, A.V.; Gulin, A. CatBoost: Unbiased boosting with categorical features. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 2–18 December 2018; Volume 31, pp. 6638–6648. [Google Scholar]
- Shen, C.; Li, H. On the dual formulation of boosting algorithms. IEEE Trans. Pattern Anal. Mach. Intell. 2010, 32, 2216–2231. [Google Scholar] [CrossRef]
- Akyol, K. Stacking ensemble based deep neural networks modeling for effective epileptic seizure detection. Expert Syst. Appl. 2020, 148, 113239. [Google Scholar] [CrossRef]
- Jiang, W.; Chen, Z.; Xiang, Y.; Shao, D.; Ma, L.; Zhang, J. SSEM: A novel self-adaptive stacking ensemble model for classification. IEEE Access 2019, 7, 120337–120349. [Google Scholar] [CrossRef]
- Alcalá-Fdez, J.; Fernández, A.; Luengo, J.; Derrac, J.; García, S.; Sánchez, L.; Herrera, F. KEEL data-mining software tool: Data set repository, integration of algorithms and experimental analysis framework. J. Mult. Valued Log. Soft Comput. 2011, 17, 255–287. [Google Scholar]
- Ramírez-Gallego, S.; García, S.; Benítez, J.M.; Herrera, F. Multivariate discretization based on evolutionary cut points selection for classification. IEEE Trans. Cybern. 2016, 46, 595–608. [Google Scholar] [CrossRef]
- Rosales-Pérez, A.; García, S.; Gonzalez, J.A.; Coello, C.A.C.; Herrera, F. An evolutionary multiobjective model and instance selection for support vector machines with Pareto-based ensembles. IEEE Trans. Evol. Comput. 2017, 21, 863–877. [Google Scholar] [CrossRef]
- Demšar, J. Statistical comparisons of classifiers over multiple data sets. J. Mach. Learn. Res. 2006, 7, 1–30. [Google Scholar]
- Polat, K.; Güneş, S. A novel hybrid intelligent method based on C4. 5 decision tree classifier and one-against-all approach for multi-class classification problems. Expert Syst. Appl. 2009, 36, 1587–1592. [Google Scholar] [CrossRef]
- Ioffe, S. Probabilistic linear discriminant analysis. In Proceedings of the European Conference on Computer Vision, Graz, Austria, 7–13 May 2006; pp. 531–542. [Google Scholar]
- Rätsch, G.; Onoda, T.; Müller, K.-R. Soft margins for adaboost. Mach. Learn. 2001, 42, 287–320. [Google Scholar] [CrossRef]


| Dataset | Instances | Attributes |
|---|---|---|
| wisconsin | 683 | 10 |
| pima | 768 | 9 |
| glass0 | 214 | 10 |
| haberman | 306 | 4 |
| vehicle2 | 846 | 19 |
| vehicle1 | 846 | 19 |
| vehicle3 | 846 | 19 |
| vehicle0 | 846 | 19 |
| ecoli1 | 336 | 8 |
| new_thyroid1 | 215 | 6 |
| new_thyroid2 | 215 | 6 |
| ecoli2 | 336 | 8 |
| glass6 | 214 | 10 |
| ecoli-0-3-4_vs_5 | 200 | 8 |
| ecoli-0-2-3-4_vs_5 | 202 | 8 |
| yeast-0-3-5-9_vs_7-8 | 506 | 9 |
| ecoli-0-4-6_vs_5 | 203 | 7 |
| ecoli-0-3-4-6_vs_5 | 205 | 8 |
| ecoli-0-3-4-7_vs_5-6 | 257 | 8 |
| ecoli-0-1_vs_2-3-5 | 244 | 8 |
| yeast_2_vs_4 | 514 | 9 |
| glass-0-4_vs_5 | 92 | 10 |
| ecoli-0-2-6-7_vs_3-5 | 224 | 8 |
| vowel0 | 988 | 14 |
| ecoli-0-6-7_vs_5 | 220 | 7 |
| ecoli-0-1-4-7_vs_2-3-5-6 | 336 | 8 |
| ecoli-0-1_vs_5 | 240 | 7 |
| led7digit-0-2-4-5-6-7-8-9_vs_1 | 443 | 8 |
| ecoli-0-1-4-7_vs_5-6 | 332 | 7 |
| page_blocks_1_3_vs_4 | 472 | 11 |
| shuttle_c2_vs_c4 | 129 | 10 |
| dermatology-6-5-5tst | 358 | 35 |
| Dataset | Decision | LDA | ADA | ESPL |
|---|---|---|---|---|
| Tree | Boost | |||
| wisconsin | 0.9459 | 0.9590 | 0.9678 | 0.9693 |
| pima | 0.7227 | 0.7682 | 0.7643 | 0.7786 |
| glass0 | 0.8175 | 0.7617 | 0.7666 | 0.7897 |
| haberman | 0.6601 | 0.7418 | 0.7383 | 0.7548 |
| vehicle2 | 0.9527 | 0.9681 | 0.9634 | 0.9693 |
| vehicle1 | 0.7459 | 0.8002 | 0.8061 | 0.8144 |
| vehicle3 | 0.7364 | 0.7955 | 0.7908 | 0.8014 |
| vehicle0 | 0.9456 | 0.9515 | 0.9669 | 0.9728 |
| ecoli1 | 0.8930 | 0.8841 | 0.8749 | 0.8958 |
| new_thyroid1 | 0.9442 | 0.9488 | 0.9907 | 0.9907 |
| new_thyroid2 | 0.9628 | 0.9395 | 0.9907 | 0.9953 |
| ecoli2 | 0.9167 | 0.9137 | 0.9018 | 0.9167 |
| glass6 | 0.9301 | 0.9628 | 0.9579 | 0.9627 |
| ecoli-0-3-4_vs_5 | 0.9300 | 0.9400 | 0.9500 | 0.9550 |
| ecoli-0-2-3-4 | 0.9256 | 0.9554 | 0.9604 | 0.9654 |
| _vs_5 | ||||
| yeast-0-3-5-9 | 0.8893 | 0.9170 | 0.7546 | 0.9170 |
| _vs_7-8 | ||||
| ecoli-0-4-6_vs_5 | 0.9460 | 0.9657 | 0.9656 | 0.9656 |
| ecoli-0-3-4-6 | 0.9512 | 0.9512 | 0.9659 | 0.9659 |
| _vs_5 | ||||
| ecoli-0-3-4-7 | 0.9378 | 0.9456 | 0.9689 | 0.9728 |
| _vs_5-6 | ||||
| ecoli-0-1_vs | 0.9425 | 0.9589 | 0.9548 | 0.9589 |
| _2-3-5 | ||||
| yeast_2_vs_4 | 0.9436 | 0.9572 | 0.9417 | 0.9533 |
| glass-0-4_vs_5 | 0.9889 | 0.9561 | 0.9673 | 0.9784 |
| ecoli-0-2-6-7 | 0.9376 | 0.9598 | 0.9687 | 0.9687 |
| _vs_3-5 | ||||
| vowel0 | 0.9858 | 0.9494 | 0.9727 | 0.9727 |
| ecoli-0-6-7_vs_5 | 0.9500 | 0.9591 | 0.9591 | 0.9682 |
| ecoli-0-1-4-7 | 0.9494 | 0.9583 | 0.9643 | 0.9673 |
| _vs_2-3-5-6 | ||||
| ecoli-0-1_vs_5 | 0.9583 | 0.9667 | 0.9500 | 0.9542 |
| led7digit-0-2-4 | 0.9594 | 0.9617 | 0.9594 | 0.9616 |
| -5-6-7-8-9_vs_1 | ||||
| ecoli-0-1-4-7 | 0.9488 | 0.9610 | 0.9669 | 0.9699 |
| _vs_5-6 | ||||
| page_blocks | 0.9915 | 0.9640 | 0.9534 | 0.9746 |
| _1_3_vs_4 | ||||
| shuttle_c2_vs_c4 | 0.9923 | 0.9766 | 1.0000 | 1.0000 |
| dermatology | 0.9944 | 0.9972 | 0.9972 | 0.9972 |
| -6-5-5tst |
| Dataset | Decision | LDA | ADA | ESPL |
|---|---|---|---|---|
| Tree | Boost | |||
| wisconsin | 0.9227 | 0.9399 | 0.9544 | 0.9565 |
| pima | 0.5852 | 0.6277 | 0.6399 | 0.6712 |
| glass0 | 0.7037 | 0.5802 | 0.6291 | 0.6758 |
| haberman | 0.2873 | 0.2303 | 0.4836 | 0.5319 |
| vehicle2 | 0.9090 | 0.9379 | 0.9297 | 0.9407 |
| vehicle1 | 0.4920 | 0.5721 | 0.5905 | 0.6567 |
| vehicle3 | 0.4728 | 0.5475 | 0.5724 | 0.6318 |
| vehicle0 | 0.8866 | 0.8971 | 0.9321 | 0.9436 |
| ecoli1 | 0.7681 | 0.7594 | 0.7430 | 0.7694 |
| new_thyroid1 | 0.8364 | 0.8030 | 0.9713 | 0.9713 |
| new_thyroid2 | 0.8857 | 0.7547 | 0.9713 | 0.9867 |
| ecoli2 | 0.7000 | 0.7139 | 0.6837 | 0.7200 |
| glass6 | 0.7345 | 0.8576 | 0.8508 | 0.8718 |
| ecoli-0-3-4_vs_5 | 0.6254 | 0.7021 | 0.7524 | 0.7835 |
| ecoli-0-2-3-4 | 0.6176 | 0.7754 | 0.7857 | 0.8143 |
| _vs_5 | ||||
| yeast-0-3-5-9 | 0.4298 | 0.3360 | 0.2866 | 0.3612 |
| _vs_7-8 | ||||
| ecoli-0-4-6_vs_5 | 0.6881 | 0.8214 | 0.8063 | 0.8254 |
| ecoli-0-3-4-6 | 0.7381 | 0.7529 | 0.8040 | 0.8040 |
| _vs_5 | ||||
| ecoli-0-3-4-7 | 0.6264 | 0.6864 | 0.8311 | 0.8529 |
| _vs_5-6 | ||||
| ecoli-0-1_vs | 0.6633 | 0.7396 | 0.7064 | 0.7521 |
| _2-3-5 | ||||
| yeast_2_vs_4 | 0.7129 | 0.7518 | 0.6342 | 0.7292 |
| glass-0-4_vs_5 | 0.9333 | 0.7667 | 0.8076 | 0.8933 |
| ecoli-0-2-6-7 | 0.6494 | 0.7600 | 0.7944 | 0.7944 |
| _vs_3-5 | ||||
| vowel0 | 0.9178 | 0.7265 | 0.8383 | 0.8425 |
| ecoli-0-6-7_vs_5 | 0.7543 | 0.7611 | 0.7348 | 0.8262 |
| ecoli-0-1-4-7 | 0.6798 | 0.7188 | 0.7800 | 0.7921 |
| _vs_2-3-5-6 | ||||
| ecoli-0-1_vs_5 | 0.7244 | 0.7944 | 0.7024 | 0.7310 |
| led7digit-0-2-4 | 0.7614 | 0.7889 | 0.7614 | 0.7766 |
| -5-6-7-8-9_vs_1 | ||||
| ecoli-0-1-4-7 | 0.6340 | 0.6810 | 0.7721 | 0.7988 |
| _vs_5-6 | ||||
| page_blocks | 0.9381 | 0.5958 | 0.6683 | 0.8426 |
| _1_3_vs_4 | ||||
| shuttle_c2_vs_c4 | 0.9333 | 0.8000 | 1.0000 | 1.0000 |
| dermatology | 0.9492 | 0.9714 | 0.9778 | 0.9778 |
| -6-5-5tst |
| Dataset | Decision | LDA | ADA | ESPL |
|---|---|---|---|---|
| Tree | Boost | |||
| wisconsin | 0.9416 | 0.9486 | 0.9665 | 0.9686 |
| pima | 0.6741 | 0.7021 | 0.7137 | 0.7452 |
| glass0 | 0.7731 | 0.6681 | 0.7129 | 0.7523 |
| haberman | 0.4539 | 0.3673 | 0.6184 | 0.6718 |
| vehicle2 | 0.9405 | 0.9572 | 0.9580 | 0.9626 |
| vehicle1 | 0.6334 | 0.6816 | 0.7202 | 0.7931 |
| vehicle3 | 0.6220 | 0.6649 | 0.6997 | 0.7662 |
| vehicle0 | 0.9304 | 0.9325 | 0.9590 | 0.9663 |
| ecoli1 | 0.8381 | 0.8428 | 0.8263 | 0.8885 |
| new_thyroid1 | 0.9042 | 0.8226 | 0.9824 | 0.9824 |
| new_thyroid2 | 0.9303 | 0.7834 | 0.9824 | 0.9972 |
| ecoli2 | 0.7882 | 0.8120 | 0.8522 | 0.8834 |
| glass6 | 0.8342 | 0.9022 | 0.9176 | 0.9495 |
| ecoli-0-3-4_vs_5 | 0.7652 | 0.8392 | 0.8557 | 0.9143 |
| ecoli-0-2-3-4 | 0.7550 | 0.8758 | 0.8543 | 0.8811 |
| _vs_5 | ||||
| yeast-0-3-5-9 | 0.6258 | 0.4595 | 0.4743 | 0.6092 |
| _vs_7-8 | ||||
| ecoli-0-4-6_vs_5 | 0.7843 | 0.8816 | 0.8565 | 0.9221 |
| ecoli-0-3-4-6 | 0.8206 | 0.8493 | 0.8560 | 0.9047 |
| _vs_5 | ||||
| ecoli-0-3-4-7 | 0.7331 | 0.7849 | 0.8860 | 0.9069 |
| _vs_5-6 | ||||
| ecoli-0-1_vs | 0.7774 | 0.8252 | 0.8122 | 0.8470 |
| _2-3-5 | ||||
| yeast_2_vs_4 | 0.8332 | 0.8098 | 0.7947 | 0.8801 |
| glass-0-4_vs_5 | 0.9940 | 0.8724 | 0.9410 | 0.9743 |
| ecoli-0-2-6-7 | 0.7675 | 0.8284 | 0.8385 | 0.8947 |
| _vs_3-5 | ||||
| vowel0 | 0.9393 | 0.8477 | 0.9216 | 0.9599 |
| ecoli-0-6-7_vs_5 | 0.6442 | 0.8520 | 0.8131 | 0.8863 |
| ecoli-0-1-4-7 | 0.7958 | 0.7844 | 0.8457 | 0.8803 |
| _vs_2-3-5-6 | ||||
| ecoli-0-1_vs_5 | 0.8362 | 0.8796 | 0.8356 | 0.8785 |
| led7digit-0-2-4 | 0.8704 | 0.8993 | 0.8704 | 0.8917 |
| -5-6-7-8-9_vs_1 | ||||
| ecoli-0-1-4-7 | 0.7547 | 0.7553 | 0.8607 | 0.9021 |
| _vs_5-6 | ||||
| page_blocks | 0.9955 | 0.6859 | 0.9235 | 0.9863 |
| _1_3_vs_4 | ||||
| shuttle_c2_vs_c4 | 0.9414 | 0.9332 | 1.0000 | 1.0000 |
| dermatology | 0.9717 | 0.9732 | 0.9985 | 0.9985 |
| -6-5-5tst |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Wang, Z.; Bai, J.; Yang, X.; Yang, Y.; Zhou, J. Ensemble Self-Paced Learning Based on Adaptive Mixture Weighting. Electronics 2022, 11, 3154. https://doi.org/10.3390/electronics11193154
Liu L, Wang Z, Bai J, Yang X, Yang Y, Zhou J. Ensemble Self-Paced Learning Based on Adaptive Mixture Weighting. Electronics. 2022; 11(19):3154. https://doi.org/10.3390/electronics11193154
Chicago/Turabian StyleLiu, Liwen, Zhong Wang, Jianbin Bai, Xiangfeng Yang, Yunchuan Yang, and Jianbo Zhou. 2022. "Ensemble Self-Paced Learning Based on Adaptive Mixture Weighting" Electronics 11, no. 19: 3154. https://doi.org/10.3390/electronics11193154
APA StyleLiu, L., Wang, Z., Bai, J., Yang, X., Yang, Y., & Zhou, J. (2022). Ensemble Self-Paced Learning Based on Adaptive Mixture Weighting. Electronics, 11(19), 3154. https://doi.org/10.3390/electronics11193154







