Abstract
Chest and lung diseases are among the most serious chronic diseases in the world, and they occur as a result of factors such as smoking, air pollution, or bacterial infection, which would expose the respiratory system and chest to serious disorders. Chest diseases lead to a natural weakness in the respiratory system, which requires the patient to take care and attention to alleviate this problem. Countries are interested in encouraging medical research and monitoring the spread of communicable diseases. Therefore, they advised researchers to perform studies to curb the diseases’ spread and urged researchers to devise methods for swiftly and readily detecting and distinguishing lung diseases. In this paper, we propose a hybrid architecture of contrast-limited adaptive histogram equalization (CLAHE) and deep convolutional network for the classification of lung diseases. We used X-ray images to create a convolutional neural network (CNN) for early identification and categorization of lung diseases. Initially, the proposed method implemented the support vector machine to classify the images with and without using CLAHE equalizer. The obtained results were compared with the CNN networks. Later, two different experiments were implemented with hybrid architecture of deep CNN networks and CLAHE as a preprocessing for image enhancement. The experimental results indicate that the suggested hybrid architecture outperforms traditional methods by roughly 20% in terms of accuracy.
1. Introduction
The last decade witnessed the emergence of many epidemics and serious diseases, and their spread negatively affected people on all levels. Even in the middle of a crisis, all countries must have the ability and resources to reliably collect and interpret health data. The recent COVID-19 pandemic has demonstrated the value of data and science in restoring more resilient health systems and accelerating equitably toward our common global goals. Doctors and researchers started looking for quick and effective ways to diagnose lung diseases [1,2,3,4], and one of the options studied was the use of artificial intelligence in the detection and diagnosis of the disease [5,6]. The researchers used machine learning and neural networks to create networks that can determine whether a patient has a specific lung disease, or is healthy. The diagnosis is made by using X-rays of patients to train the neural network, then testing it and using the results for a diagnosis of the patient’s condition. These networks have extraordinarily high rates of accuracy and sensitivity in identifying lung illness, with a very low error rate, according to the data [7,8,9,10]. The carbon emissions that have recently dominated the world, particularly in third-world countries, have resulted in the worsening of lung diseases, spurring researchers and doctors to look for ways to treat them. However, doctors first had to diagnose the different types of lung diseases and distinguish them from one another, prompting researchers to look for quick ways to diagnose diseases with analysis of information [11,12,13]. Deep learning and its networks are one approach to diagnosing lung illnesses. On a vast number of X-rays, these networks are trained and tested using modern, huge, and scalable data, i.e., open-source CNN. Then, using high-precision diagnostics, they assist healthcare providers and users of these networks in establishing the proper diagnosis [14,15,16,17]. The goals of this study are to (1) employ artificial intelligence to automatically define the patient’s condition and diagnose the exact lung disease; (2) reduce the strain on healthcare infrastructure, such as laboratories and radiology departments, as well as healthcare practitioners’ efforts; (3) conserve the resources through precise detection of lung disorders, with concernabout aggravating the issue of the lack of a correct diagnosis, which might have disastrous effects; (4) research the appropriate literature dealing with the diagnosis of lung disorders, as well as explore all of the current procedures that have been thoroughly applied in this regard; (5) if the condition is COVID-19, lung opacity, normal or viral pneumonia, an accurate diagnosis of lung disorders is required; and (6) create a user interface system that makes it simpler for radiologists and doctors to utilize the proposed system. The remainder of this study is structured as follows: Section 2 examines the literature pertinent to this study. In Section 3, the suggested system is given, and the data gathering and preparation methods are also detailed. Section 4 describes the findings of this study. Finally, further work is suggested in Section 5.
2. Literature Review
For accurate diagnosis, doctors use plain X-rays, CT scans, MRIs, ultrasound, and optical imaging in the clinical setting [18,19], where researchers and clinicians from remote locations can easily participate and search for data and analytics, enhancing their ability to study, diagnose, monitor, and treat disorders [20,21].
The inability of medical practitioners to accurately diagnose, the large number of patients, and the lack of specialized doctors have all increased dramatically in recent years; hence, there is a need foreffectiveness of researchers using artificial intelligence to assist doctors and play a pivotal role in the accurate diagnosis of diseases. Appropriate solutions, developed specifically for the healthcare sector, can be used to provide medical practitioners with essential support as they manage increasing amounts of data, information, and image sizes and types [15,22,23,24]. As a result, in this study, the image processor was used to attain high accuracy through the use of networks. Image processing in intelligent systems is a multi-year research discipline that many people have worked on since the advent of artificial intelligence. In its infancy, image processing requires a large number of manual inputs to provide computer instructions to access the output. These machines were trained to detect images [25]. The biological neural network, from which the artificial neural network was formed, gave rise to the concept of networks. One of the most important deep neural networks is the convolutional neural network, often known as CNN or ConvNet, which is derived from deep learning as a subfield of machine learning [26]. The convolutional neural network was inspired by research into the biological visual cortex, which contains small sections of sensitive cells from the visual field. The researchers demonstrated that some particular neurons in the brain only respond in the presence of edges of a specific orientation, such as vertical or horizontal edges [27,28]. The concept of specialized components inside a system that perform certain jobs is also evident in hardware and CNNs [29].
This study follows the procedures that the researchers examined and implemented in [30,31]. The importance of recognizing the lung disease was investigated, diagnosed, and read using X-ray at the lowest possible cost, utilizing a tiny convolutional network and specific classifications, which were then trained and scaled with a size of 224 × 224 for each channel. The results were 89.2%, including a positive mistake for the existence of additional lung ailments that was misclassified as a network trainer. This research was used to improve results by considering the number of trained network classifications, the size of the dataset, and the size of the X-ray image used in the classification. The researchers in [32] offered CheXNet, which is a 121-layer dense convolutional network. A total of 420 images of lung X-rays were measured for diagnosing 14 diseases and making a comparison between specialist doctors and the model CheXNet to detect all fourteen diseases. CheXNet exceeded specialist doctors’ diagnoses for all 14 diseases where the model was utilized. Ten well-known pretrained infection CNNs were used by [33,34]. The work by [35] used VGG-16 for diagnosing lung diseases, such as COVID-19, pneumonia, and normal cases. The use of two types of convolutional neural networks, CNN and RNN, to diagnose lung patients via text categorization was discussed by [36]. Ref. [37] studied several networks, including the neural network CNN, NLP, and SVM. The best results were attained in the medical field when the CNN was used. As a result, the CNN’s strength and superiority over various networks and models in the medical sector were considered. The work by [38,39,40] diagnosed lung diseases by using several neural networks, such as DNN, Faster R-CNN, and VGG-16. The results for accuracy were the following: DenseNet 96.45%, ResNet50 94.90%, Inception V3 95.79%, AlexNet 96.86%, and Faster R-CNN (proposed) 99.29%. The researchers conceptualized the proposed model Faster R-CNN after conducting the experiments. The best model and highest values had a classification accuracy of 97.36%, a sensitivity of 97.65%, and an accuracy of 99.29%, according to the data.The work offered by [41,42] refined images by inserting them into a CAD scheme. The strategy first works to remove image distortions, then introduces simple coloring, and last, images are entered from three inputs to the original image and the other two images, where they discovered that this method was practicable. The CNN networks proved their effectiveness in reading the X-rays of the lung diseases in terms of accuracy, sensitivity, and classification; however, the result is negative if the image is not clear. The value of this research is that it is important to filter X-rays in order to obtain better outcomes, which is exactly what we present.
3. Methodology
3.1. Preprocessing
Histogram equalization is a straightforward image improvement technique in image processing of contrast adjustment using the image’s histogram. In this scenario, the conventional technique is to remap the image’s grayscales so that the resulting histogram approximates the uniform distribution. This approach assumes that the image quality is uniform across all locations and that a single unique grayscale mapping delivers equivalent improvement for all portions of the image. This assumption, however, is invalid when grayscale distributions vary from area to region. In this scenario, an adaptive histogram equalization algorithm can outperform the usual approach substantially. A more advanced approach of interest that is often used for image enhancement is contrast-limited adaptive histogram equalization (CLAHE), which was proposed by [43]. On medical images, contrast-limitedadaptive histogram equalization yielded good results. This approach is based on partitioning the image into many non-overlapping areas of about similar size (64 tiles in 8 columns and 8 rows is a common choice). This partition produces three distinct sets of areas. The class of corner regions is one group that consists of only four areas (CR). The second category is the border region class (BR). This class includes all regions on the image’s border, excluding the corner regions. The final group is known as the class of inner regions (IR); see Figure 1.
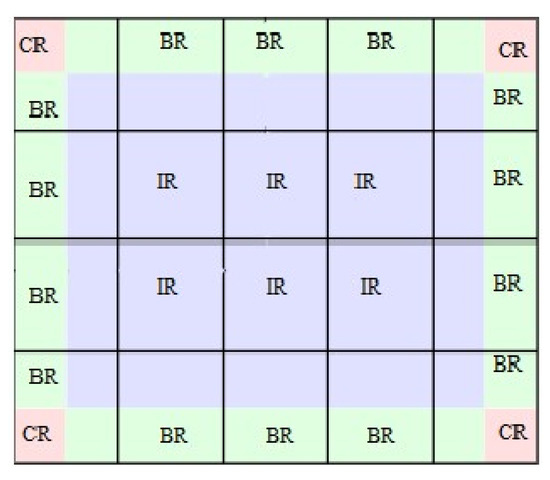
Figure 1.
Structure of regions of an image.
CLAHE processing technology was used in this research to significantly raise the contrast of the image and emphasize the edges of the entire image. However, this processing in part of the homogeneous image raises the noise, so the algorithm (CLAHE) was used with it, where a linear graph equation is often used to keep up with the pixel flow. CLAHE is a variation of adaptive histogram equalization (AHE) that takes care of overamplification of the contrast. CLAHE works on tiny areas of an image called tiles rather than the complete image. When applying CLAHE, there are two parameters to remember: clipLimit—this parameter sets the threshold for contrast-limiting. It is a real scalar in the range [0 1] that determines a contrast-enhancement limit. Higher numbers result in more contrast; tileGridSize—this sets the number of tiles in the row and column. It is a two elements vector of positive integers specifying the number of tiles by row and column, [M N]. Both M and N must be at least 2. The total number of tiles is equal to M * N.
The core idea behind CLAHE is to conduct histogram equalization on non-overlapping image sub-areas, then use interpolation to fix anomalies between boundaries. To remove the false borders, the surrounding tiles are blended using bilinear interpolation. Bilinear interpolation is a fundamental resampling technique used in computer vision and image processing. It is also known as bilinear filtering or bilinear texture mapping. Bilinear interpolation is a method for interpolating two-variable functions (e.g., x and y) using repeated linear interpolation; see Figure 2.
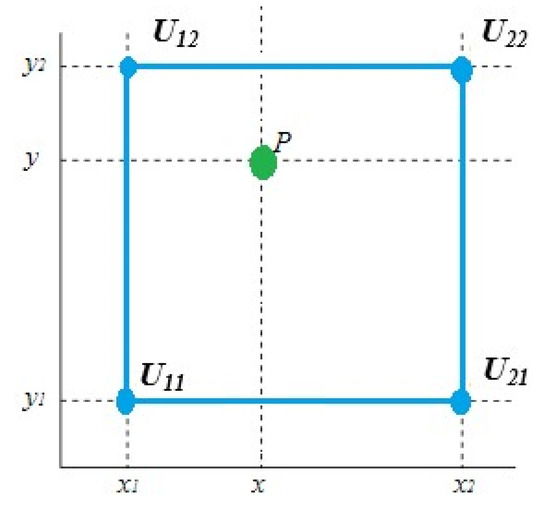
Figure 2.
The four blue dots represent the data points, while the green dot represents the interpolation point.
Assume we wish to determine the value of the unknown function f at the position . The value of f is assumed to be known at the four sites , , , and .
The bilinear interpolation is performed in the x-direction followed by the y-direction. This yields
Then, the desired estimation for p is calculated by
where
It is worth noting that if we interpolate first in the y-direction and then along the x-direction, we will obtain the same result. In CLAHE, the histogram is cut at some threshold and then equalization is applied.
Notice that CLAHE had a significant effect on the original image of the lung, as shown in Figure 3. The plotted histograms of the original image and the enhanced image are shown in Figure 4. They reveal that the values of the original image are spread out between the minimum of 0 and maximum of 255, while in the case of the contrast-enhanced image, most of the pixels are concentrated in the center of the histogram, thus proving the effectiveness of CLAHE in adjusting the contrast of the image.
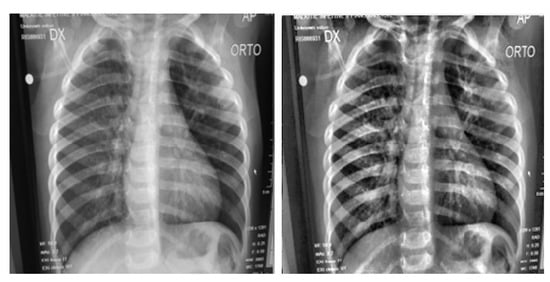
Figure 3.
Original image (left) and contrast-enhanced image (right).
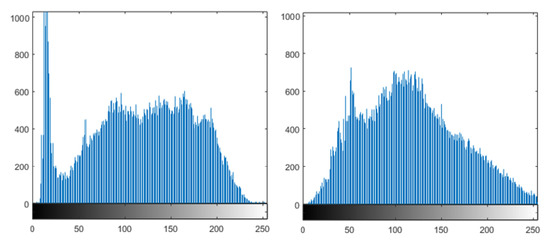
Figure 4.
Original image histogram (left) and contrast-enhanced image histogram (right).
The CLAHE algorithm’s block diagram is shown in Figure 5. It is made up of a tile generation block, an input image buffer block, a pipeline for histogram equalization, and a bilinear interpolation block. To keep up with the incoming data, two histogram equalization pipelines are necessary. They work in a ping-pong fashion. Each pipeline comprises an equal number of histogram equalization modules in the horizontal direction. The histogram equalization modules calculate histogram equalization for each tile in simultaneously. The original input image data are required for the last stage of the histogram equalization module, scaling and mapping. This information is saved in an input image buffer block. To decrease boundary artifacts, the mapped values produced from histogram equalization are scaled and employed in the bilinear interpolation algorithm.
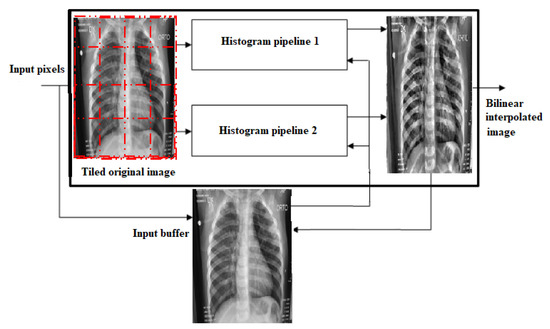
Figure 5.
The CLAHE model on lung X-ray image.
3.2. Proposed System
This study suggests a classification model that is based on convolution neural spectral networks, which is abbreviated as CNSNs. The scheme of the proposed system is shown in Figure 6, in which the images are entered and the processing is performed, then they are called and entered into the network to classify among the diseased cases. In addition, pooling was performed and several operations were used to obtain the result, which is the diagnosis of the disease for this X-ray.
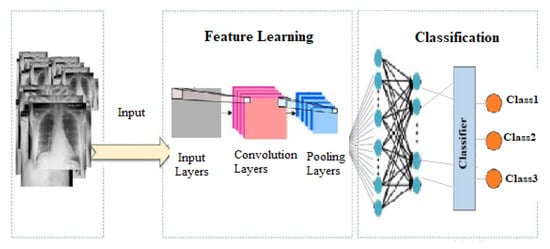
Figure 6.
CNSN Flowchart on lung sample X-ray.
The CNSN model captures spectral properties from an input image at numerous scales, extracts the image features using particular CNSN models, and combines CNSN outputs into a single feature vector that is used as an input for a dense layer. A CNSN takes as input a tensor with a shape. The neighborhood pixels surrounding a central pixel i are recovered at scales n for the input hyperspectral image (i.e., following PCA processing). The spectral dimensions of the input CLAHE data are reduced to using a statistical PCA method. For example, in a setup, CNSN extracts 8 neighboring pixels for the ith pixel, and if , the output feature vector W is . Then, at each scale, specialized CNSN models are used to extract high-level characteristics that are more abstract than W. The size of the CNSN output feature vector W is determined by the CNSN architecture, notably the spatial filter and pooling processes. Finally, the feature vectors are combined into a single feature vector W, which may then be utilized as an input for classification by a fully connected layer. A CNSN, similar to other neural networks, has an input layer, an output layer, and several hidden layers in between. These layers execute processes that change the data with the purpose of learning features particular to the data. Convolution layers, pooling layers, and fully connected layers are three of the most popular layers.
- Convolution layer: This is the first layer in a CNSN. It blends the input of a matrix of the dimensions and then sends the output to the next layer. Convolution layers are made up of neurons that link to subregions of the input images or the preceding layer’s outputs. Each layer convolves the input by sliding the filters vertically and horizontally along the input. While scanning through an image, the layer learns the features localized by these areas. The image is abstracted to a feature map, also known as an activation map, after the tensor is passed through a convolutional layer.
- Pooling layer: A pooling layer’s main function is to minimize the number of parameters in the input tensor; hence it aids in reducing overfitting, extracts representative features from the input tensor, and reduces computation, which improves efficiency. A pooling layer reduces the amount of parameters that the network must learn by conducting nonlinear sampling on the output. It divides the input into rectangular pooling regions, then computes the maximum of each region. By merging the outputs of neuron clusters at one layer into a single neuron in the following layer, pooling layers minimize the dimensionality of data. It is worth knowing that pooling reduces the image’s height and width while keeping the number of channels (depth) constant.
- Fully connected layer: The convolutional neural network’s last layer is the fully connected layer (also known as the hidden layer). It is a feedforward neural network. The output of the final pooling or convolutional layer is flattened and sent into the fully connected layer. Every neuron in one layer is connected to every neuron in the next layer via fully connected layers. It functions similarly to a standard multilayer perceptron neural network (MLP).
These procedures are performed hundreds or thousands of times with each layer to recognize different features.
3.3. Dataset
A team of researchers in collaboration with medical doctors compiled a dataset containing images of lung X-rays for COVID-19-positive cases along with normal and viral pneumonia images [22,44]. The dataset was categorized into four classes: (i) 3615 X-ray images of COVID-19 data, which were collected from different publicly accessible datasets, online sources, and published papers; (ii) 10,192 X-ray images of normal data; (iii) 1345 viral pneumonia data; and (iv) 6012 lung opacity X-ray images collected from the Radiological Society of North America. The final dataset employed in this research has 21,164 images. All the images are in Portable Network Graphics (PNG) file format and resolution is pixels. Sample shots of the four classes are shown in Figure 7.
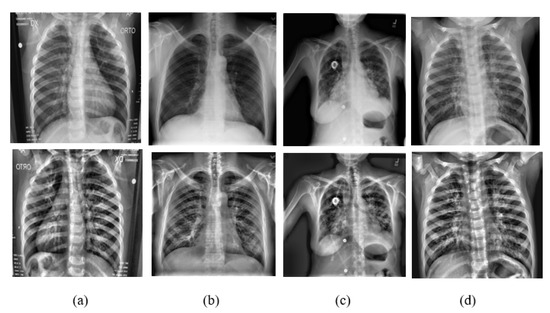
Figure 7.
Shown here are some sampling images of the classes (a) COVID-19 case, (b) normal case, (c) viral pneumonia, and (d) lung opacity. The original images are in the first row and the corresponding CLAHE images are in the second row.
3.4. Algorithm Evaluation
The method used by the researchers split the dataset into train and test sets in MATLAB. The train–test split is used to assess the performance of machine learning algorithms that are appropriate for prediction-based algorithms. This approach is a quick and simple procedure that allows us to compare our proposed machine learning model to machine results. The dataset employed in this research has 21,164 images. A total of 80% of them were used for training, while the remaining 20% were used for test. We evaluated the performance of the proposed model via accuracy, precision, recall, and F1 score metrics. These are the equations used.
where
- : The value of the actual class and the value of the predicted class is yes.
- : The value of the actual class and value of the predicted class is no.
- : The actual class is no and the predicted class is yes.
- : The actual class is yes but predicted class is no.
The observations that are accurately anticipated are known as true positives and true negatives. We aim to keep false positives and negatives to a minimum.
3.5. Implemented Techniques
To boost the contrast of the image and focus on the edges, we used CLAHE for the preprocessing techniques. We used it to check if there was any improvement in the outcomes and whether or not this treatment was effective in improving the experiment’s accuracy and sensitivity. For training, we used the support vector machine, the VGG19, and CNN networks.
4. Results and Discussion
To assess the impact of our suggested technique on performance, two types of tests were carried out. To train the machine to recognize the correct prediction, we used machine learning and deep learning. Deep learning is a step forward in the advancement of machine learning. It makes use of a programmable neural network, which allows machines to make accurate predictions without the need for human intervention. Typically, the results obtained from SVM were, on average, 67.2%. Therefore, it is necessary to resort to more effective learning methods for accurate classification. The deep learning algorithm proved to be a more proficient system compared to the SVM algorithm in classifying the predicted classes. However, deep learning networks involve a large number of input data compared to SVM. The more data that are fed into the network, the more it will better generalize and correctly make predictions with fewer mistakes. The presence of high-speed computers and high storage capacity to train large neural networks encourage researchers to use deep learning. This section presents the experiments of SVM networks and the CNN network. The results of the experiments proved that the accuracy of the proposed technique outperformed the accuracy of the baseline experiments and the other networks used by the researchers. In addition, this section presents and discusses the results and the most important recommendations that the researchers deem appropriate.
4.1. Experiment 1 (Baseline)
The first experiment served as a baseline for comparing the prediction task using support vector machine networks, which are supervised learning models that analyze data for prediction. Referring to Table 1, we found that the accuracy was reasonable 68%, but it was less than the ambitious value we desired. In order to obtain better results, we used four types of well-known features in the world of image processing, which are HOG, MSER, SURF, and BRISK features. The selected features are visualized in Figure 8. Here, we present them briefly:

Table 1.
Summary of the baseline accuracy: SURF features and SVM.
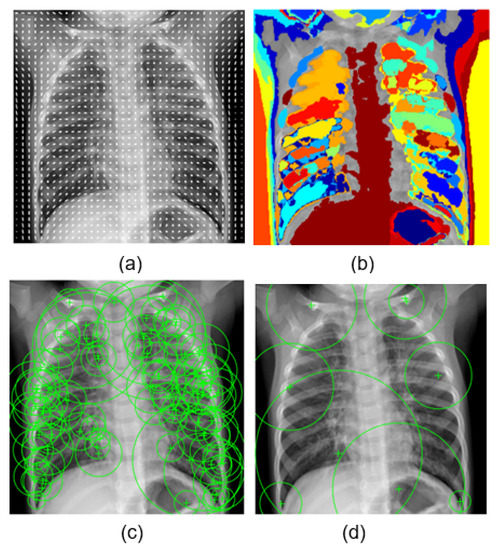
Figure 8.
Sampling of (a) HOG, (b) MSER, (c) SURF, and (d) BRISK features on a lung opacity X-ray image.
- HOG: Histogram of gradient feature, a grid of equally spaced region plots, is used to show HOG features [45]. The grid dimensions are determined by the image size as well as the cell size. The distribution of gradient orientations within an HOG cell is displayed on each region plot. The region used was a block of cell. The learning was performed by using the strongest 17,693 features from each of the image classes.
- MSER: Maximally stable extremal region is a feature object detector [46]. The MSER object examines changes in region area between various intensity thresholds. Threshold values are used to maintain the circular regions, where circular regions have low eccentricity. The circular region range was [30 14000], and the value of the threshold was 2. Decreasing the value of the threshold will return more regions. The learning was carried out using the 28,550 strongest features.
- SURF: Speeded up robust feature is a patented local feature detector and descriptor [47]. SURF finds landmarks and describes them using a vector that is somewhat resistant to distortion, rotation, and scaling. For this experiment, we used these parameters: number of octaves = 5, number of intervals = 4, and threshold = 0.0004. The most powerful 22,895 features from each of the image classes were used to perform the learning.
- BRISK: Binary robust invariant scalable keypoint is a scale- and rotation-invariant feature point detection and description tool [48]. We used the following parameters for this experiment: number of corners = 0.1, number of octaves = 4, and intensity difference = 0.2. The 6953 most effective features from each class were used to carry out the learning.
As illustrated in Table 2, two types of experiments were conducted, the first using the previously mentioned features with CLAHE, and the second without using CLAHE. When using the features singly, SURF performed competitively with HOG and MSER in all classes. The accuracy values ranged between 68.7% and 69.8%. By using BRISK, we obtained a modest accuracy value of 63.1%. On the other hand, the results were sometimes discouraging when using the combined features with CLAHE, which ranged between 35% and 67.2%. Furthermore, we note that the use of CLAHE in the preprocessing and SVM in learning was not beneficial in increasing the accuracy; on the contrary, the use of CLAHE had a negative effect. However, the baseline experiments gave satisfactory results of 86% accuracy in the case of predicting viral pneumonia diseases. Figure 9 shows the comparison of the different correlations of the four lung diseases using CLAHE and SURF features using SVM. Typically, the results obtained from SVM were average. Therefore, it is necessary to resort to more effective learning methods for accurate classification, and this is what we review and address in the next subsections.

Table 2.
Results of accuracy classification and algorithm evaluation for SVM.
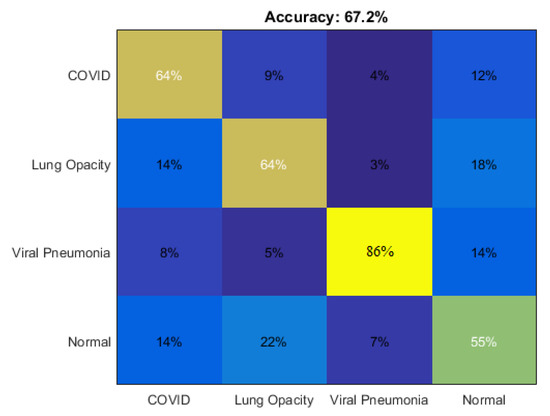
Figure 9.
Comparing the accuracy of different correlation of the four lung diseases using CLAHE and SURF features with SVM learning.
4.2. Experiment 2
The number of lung images acquired by X-ray was over twenty-one thousand images divided into four classes; thus, we used VGG-19 which is a pretrained CNN to access data of enormous size [49]. The VGG-19 network consists of 19 layers (16 convolution layers, 3 fully connected layer, 5 maxpool layers, and 1 softmax layer). We used the VGG-19 network by using images sized , indicating that the matrix was of shape . We employed kernels that were in size and had a stride size of 1 pixel, allowing to span the entire image. To keep the image’s spatial resolution, spatial padding was also applied. The accuracy for training was 95.9%, and it was 84.6% for testing. The results are clarified in Table 3. Figure 10 shows the results of using the VGG-19 network, and the graph shows the accuracy in training and the percentage of error in the test results.

Table 3.
Results of accuracy classification and algorithm evaluation for VGG-19.
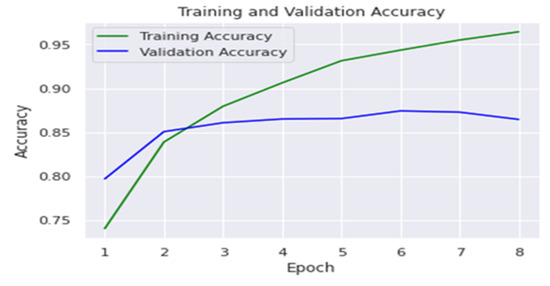
Figure 10.
Training and validation accuracy for VGG-19.
4.3. Experiment 3
The dataset employed in this research, which comprises 21,164 images, consists of 3615 COVID-19 images, 6012 lung opacity images, 1345 viral pneumonia images, and 10,192 normal images. On 80% of them, the model was trained, while the remaining 20% were used for testing. To tune the hyperparameters, the first hyperparameter to tune is the number of neurons in each hidden layer. Then, the rest of the hyperparameters are tuned, which include optimizer, learning rate, batch size, epochs, and layers. For the hyperparameter-tuning demonstration, we set the number of neurons in every layer to be the same. The number of neurons ranged from 5 to 100. One of the hyperparameters is the learning rate of the optimizer. The learning rate controls the step size for a model to reach the minimum loss function. A lower learning rate increases the likelihood of discovering a minimum loss function. A higher learning rate makes the model learn quicker. There are seven optimizers to choose from. We chose the linear optimizer Adam with a learning rate of 0.001. Another hyperparameter is the batch size. Batch size is the amount of training sub-samples for the input, which prevents the model from receiving all of the training data at once. The learning process moves more quickly with the smaller batch size and more slowly in larger batches. The batch size was set to 1000. The next hyperparameter is epochs, which is the number of times a complete dataset is run through the neural network model, that is, one epoch denotes one forward and backward pass of the training dataset through the neural network. Training of layers was performed at 2000 images per step. Training on all classes was run for 500 steps, or 5 epochs, since training of the final layers was converged for all classes. Testing was performed after every step using the test images, and the best-performing model was kept for analysis. In this experiment, the training was ended after 30 epochs of the model because there were no further improvements in accuracy. For the layer hyperparameter, we used five layers. The first layer was convolution (16). The second layer, convolution (32), max pooling, and batch normalization. The third layer, convolution (64), max pooling, and batch normalization, The fourth layer was convolution (128) and max pooling. Here, we applied dropout (0.2) in the fourth layer to improve the test results. The fifth layer was convolution (256), max pooling, and dropout (0.2). Finally, we applied flatten, dense (256), dropout (0.15), and dense again. The accuracy may be affected by various layers. A result with fewer layers may cause underfitting, whereas one with too many layers may cause overfitting.
The accuracy of training was 97.5%, and it was 91.3% for testing. Figure 11 shows the accuracy of using the CNN network in training and validation sets. The results of the proposed method on each class are clarified in Table 4. In addition, a comparison of the proposed method with other methods reported in the literature is presented in Table 5. Table 6 presents the summary of the obtained results.
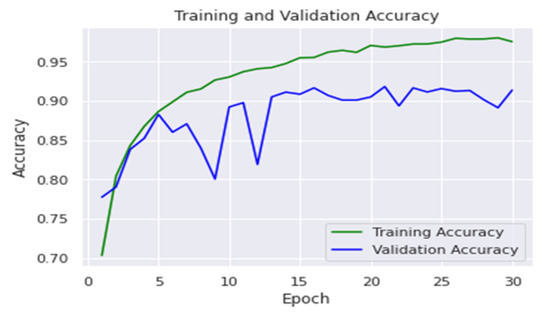
Figure 11.
Training and validation accuracy for CNN.

Table 4.
Results of accuracy classification and algorithm evaluation for CNN.

Table 5.
Comparison of proposed method with other methods reported in the literature.

Table 6.
Summary of results accuracy classification.
Figure 12 shows the accuracy of using the CNN network in the dataset, taking different training and validation partition combinations. The results we obtained were as follows: 92%, 91%, 85%, and 82% when using partitions 90–10, 80–20, 70–30, and 60–40, respectively. However, we followed the state-of-the-art approach called the 80–20 rule. The results of the proposed method for each class are clarified in Table 4. In addition, a comparison of the proposed method with other methods reported in the literature is presented in Table 5. Table 6 presents the summary of the obtained results.
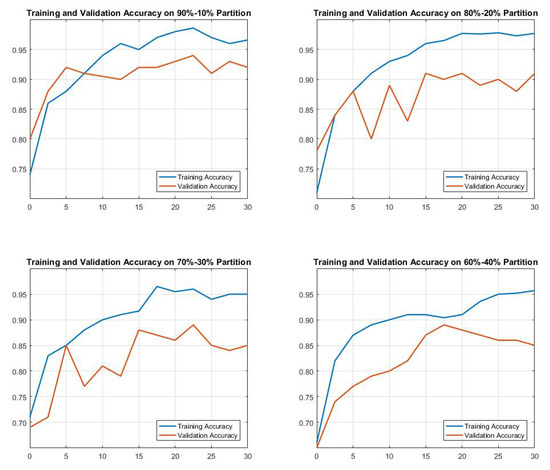
Figure 12.
Training and validation accuracy for CNN by taking different combination of data partitions.
4.4. Discussion
We used various convolutional neural networks in this study, with the goal of presenting the best results and applying them to the classification of lung disorders. The first experiment served as a baseline for comparing the prediction task using support vector machine networks. The accuracy gained was a respectable 68%, but it falls short of the goal we established for ourselves. The second experiment was a pretrained neural network, VGG19, and the results were not acceptable. The accuracy improved to 84% after applying CLAHE. Finally, after a deep study of research in this field, a convolutional neural network was designed from scratch, where the result after using the processor was 91% accuracy. The accuracy results on the training set are illustrated in Table 6. Following the experiments, it is worth noting that the CNN network’s accuracy for a large dataset is lower than for a small dataset. This is because the number of X-rays for diseases is not equal. The suggested model was tested on four classes, three of which were lung diseases and one of which was healthy. The large number of X-rays made categorization challenging, reducing the quality of the results. It was difficult to classify the images since they were not clear and consistent, and because they were open-source. However, the proposed model can be used in hospitals, medical clinics, and radiology clinics, and will assist specialists in identifying lung diseases.
5. Conclusions and Future Work
Chest and lung diseases are among the world’s most dangerous chronic diseases, and they are caused by a variety of factors that expose the respiratory and chest systems to disorders such as smoking, polluted air, or bacterial infection, as well as genetic factors, and these factors affect the respiratory system and lungs, resulting in chest diseases such as asthma. The chest crisis, bronchitis, and other chronic diseases that naturally weaken the effectiveness of the respiratory system necessitate the patient’s care and attention to alleviate this problem, knowing that it cannot be cured, but that it can be treated to avoid damage. It was the most common chest disease in previous centuries. Accordingly, the first step was to look for recent, up-to-date, and reasonable-quality data, so the researchers used the Kaggle website as an open and reliable source, as well as the nerve cells. The convolutional method, as recommended by the researchers, is the best solution to this problem, as it uses a pretrained network, which is VGG19, and the results were acceptable. After using the CLAHE graph equalizer, the accuracy was 84%, and because we were looking for better results, we designed a hybrid model of convolutional neural network (CNN) and CLAHE system, and the result increased to 91%, which was remarkable. For the future, we suggest the following:
- First, we recommend that an application be created to function on mobile phones after it has been designed on a computer.
- Second, we advocate using the program to diagnosis other disorders involving the kidneys, such as kidney cancer, because there are few doctors who specialize in identifying kidney ailments, and doctors’ error rates are high, compared to the machine, which has a 91% accuracy rate.
- Third, experimenting with a variety of networks may lead to more accurate disease diagnosis.
- Fourth, this technique not only assists doctors, caretakers, radiologists, and patients with this disease, but it also provides valuable information to researchers who are diagnosing other diseases in the medical domain.
- Fifth, there is still room for improvement in terms of accuracy by using different preprocessing techniques.
Author Contributions
F.H.: Conceptualization, supervision, methodology, formal analysis, resources, data curation, writing—original draft preparation. A.M.: Conceptualization, supervision, writing—review and editing, project administration, funding acquisition. S.A.: Conceptualization, writing—review and editing, supervision. S.M.E.-S.: Conceptualization, writing—review and editing, supervision. B.A.: Conceptualization, writing—review and editing. L.A.: Conceptualization, writing—review and editing. A.H.G.: Conceptualization, writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- AlZu’bi, S.; Jararweh, Y.; Al-Zoubi, H.; Elbes, M.; Kanan, T.; Gupta, B. Multi-orientation geometric medical volumes segmentation using 3d multiresolution analysis. Multimed. Tools Appl. 2018, 78, 24223–24248. [Google Scholar] [CrossRef]
- AlZu’bi, S.; Al-Qatawneh, S.; Alsmirat, M. Transferable HMM Trained Matrices for Accelerating Statistical Segmentation Time. In Proceedings of the 2018 Fifth International Conference on Social Networks Analysis, Management and Security (SNAMS), Valencia, Spain, 15–18 October 2018; pp. 172–176. [Google Scholar]
- AlZu’bi, S.; Mughaid, A.; Hawashin, B.; Elbes, M.; Kanan, T.; Alrawashdeh, T.; Aqel, D. Reconstructing Big Data Acquired from Radioisotope Distribution in Medical Scanner Detectors. In Proceedings of the 2019 IEEE Jordan International Joint Conference on Electrical Engineering and Information Technology (JEEIT), Amman, Jordan, 9–11 April 2019; pp. 325–329. [Google Scholar]
- AlZu’bi, S.; Aqel, D.; Mughaid, A.; Jararweh, Y. A multi-levels geo-location based crawling method for social media platforms. In Proceedings of the 2019 Sixth International Conference on Social Networks Analysis, Management and Security (SNAMS), Granada, Spain, 22–25 October 2019; pp. 494–498. [Google Scholar]
- Zhao, S.; Lin, Q.; Ran, J.; Musa, S.S.; Yang, G.; Wang, W.; Lou, Y.; Gao, D.; Yang, L.; He, D.; et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. Int. J. Infect. Dis. 2020, 92, 214–217. [Google Scholar] [CrossRef] [PubMed]
- Elbes, M.; Kanan, T.; Alia, M.; Ziad, M. COVD-19 Detection Platform from X-ray Images using Deep Learning. Int. J. Adv. Soft Compu. Appl. 2022, 14, 1. [Google Scholar] [CrossRef]
- Walsh, S.L.; Humphries, S.M.; Wells, A.U.; Brown, K.K. Imaging research in fibrotic lung disease; applying deep learning to unsolved problems. Lancet Respir. Med. 2020, 8, 1144–1153. [Google Scholar] [CrossRef]
- Mughaid, A.; Obeidat, I.; Hawashin, B.; AlZu’bi, S.; Aqel, D. A smart geo-location job recommender system based on social media posts. In Proceedings of the 2019 Sixth International Conference on Social Networks Analysis, Management and Security (SNAMS), Granada, Spain, 22–25 October 2019; pp. 505–510. [Google Scholar]
- Bharati, S.; Podder, P.; Mondal, M.R.H. Hybrid deep learning for detecting lung diseases from X-ray images. Inform. Med. Unlocked 2020, 20, 100391. [Google Scholar] [CrossRef]
- Kieu, S.T.H.; Bade, A.; Hijazi, M.H.A.; Kolivand, H. A survey of deep learning for lung disease detection on medical images: State-of-the-art, taxonomy, issues and future directions. J. Imaging 2020, 6, 131. [Google Scholar] [CrossRef]
- AlZu’bi, S.; Shehab, M.; Al-Ayyoub, M.; Jararweh, Y.; Gupta, B. Parallel implementation for 3d medical volume fuzzy segmentation. Pattern Recognit. Lett. 2020, 130, 312–318. [Google Scholar] [CrossRef]
- Al-Mnayyis, A.; Alasal, S.A.; Alsmirat, M.; Baker, Q.B.; Alzu’bi, S. Lumbar disk 3D modeling from limited number of MRI axial slices. Int. J. Electr. Comput. Eng. 2020, 10, 4101. [Google Scholar] [CrossRef]
- AlZu’bi, S.; Aqel, D.; Mughaid, A. Recent intelligent approaches for managing and optimizing smart blood donation process. In Proceedings of the 2021 International Conference on Information Technology (ICIT), Amman, Jordan, 14–15 July 2021; pp. 679–684. [Google Scholar]
- Sethi, R.; Mehrotra, M.; Sethi, D. Deep learning based diagnosis recommendation for COVID-19 using chest X-rays images. In Proceedings of the 2020 Second International Conference on Inventive Research in Computing Applications (ICIRCA), Coimbatore, India, 15–17 July 2020; pp. 1–4. [Google Scholar]
- Ibrahim, D.M.; Elshennawy, N.M.; Sarhan, A.M. Deep-chest: Multi-classification deep learning model for diagnosing COVID-19, pneumonia, and lung cancer chest diseases. Comput. Biol. Med. 2021, 132, 104348. [Google Scholar] [CrossRef]
- Al-Zu’bi, S.; Hawashin, B.; Mughaid, A.; Baker, T. Efficient 3D medical image segmentation algorithm over a secured multimedia network. Multimed. Tools Appl. 2021, 80, 16887–16905. [Google Scholar] [CrossRef]
- AlZu’bi, S.; Makki, Q.H.; Ghani, Y.A.; Ali, H. Intelligent Distribution for COVID-19 Vaccine Based on Economical Impacts. In Proceedings of the 2021 International Conference on Information Technology (ICIT), Amman, Jordan, 14–15 July 2021; pp. 968–973. [Google Scholar]
- Liu, Q.; Li, N.; Jia, H.; Qi, Q.; Abualigah, L. Modified remora optimization algorithm for global optimization and multilevel thresholding image segmentation. Mathematics 2022, 10, 1014. [Google Scholar] [CrossRef]
- Yousri, D.; Abd Elaziz, M.; Abualigah, L.; Oliva, D.; Al-Qaness, M.A.; Ewees, A.A. COVID-19 X-ray images classification based on enhanced fractional-order cuckoo search optimizer using heavy-tailed distributions. Appl. Soft Comput. 2021, 101, 107052. [Google Scholar] [CrossRef] [PubMed]
- Daradkeh, M.; Abualigah, L.; Atalla, S.; Mansoor, W. Scientometric Analysis and Classification of Research Using Convolutional Neural Networks: A Case Study in Data Science and Analytics. Electronics 2022, 11, 2066. [Google Scholar] [CrossRef]
- AlShourbaji, I.; Kachare, P.; Zogaan, W.; Muhammad, L.; Abualigah, L. Learning Features Using an optimized Artificial Neural Network for Breast Cancer Diagnosis. SN Comput. Sci. 2022, 3, 229. [Google Scholar] [CrossRef]
- Rahman, T.; Khandakar, A.; Qiblawey, Y.; Tahir, A.; Kiranyaz, S.; Kashem, S.B.A.; Islam, M.T.; Al Maadeed, S.; Zughaier, S.M.; Khan, M.S.; et al. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Comput. Biol. Med. 2021, 132, 104319. [Google Scholar] [CrossRef]
- AlZu’bi, S.; AlQatawneh, S.; ElBes, M.; Alsmirat, M. Transferable HMM probability matrices in multi-orientation geometric medical volumes segmentation. Concurr. Comput. Pract. Exp. 2020, 32, e5214. [Google Scholar] [CrossRef]
- AlZu’bi, S.; Aqel, D.; Lafi, M. An intelligent system for blood donation process optimization-smart techniques for minimizing blood wastages. Clust. Comput. 2022, 25, 3617–3627. [Google Scholar] [CrossRef]
- Hussein, F.; Piccardi, M. V-JAUNE: A framework for joint action recognition and video summarization. ACM Trans. Multimed. Comput. Commun. Appl. (TOMM) 2017, 13, 1–19. [Google Scholar] [CrossRef]
- Hubel, D.H.; Wiesel, T.N. Receptive fields, binocular interaction and functional architecture in the cat’s visual cortex. J. Physiol. 1962, 160, 106. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Xu, L.; Ren, J.S.; Liu, C.; Jia, J. Deep convolutional neural network for image deconvolution. Adv. Neural Inf. Process. Syst. 2014, 27, 1790–1798. [Google Scholar]
- Narin, A.; Kaya, C.; Pamuk, Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. Pattern Anal. Appl. 2021, 24, 1207–1220. [Google Scholar] [CrossRef]
- Hall, L.O.; Paul, R.; Goldgof, D.B.; Goldgof, G.M. Finding COVID-19 from chest X-rays using deep learning on a small dataset. arXiv 2020, arXiv:2004.02060. [Google Scholar]
- Sanagavarapu, S.; Sridhar, S.; Gopal, T. COVID-19 identification in CLAHE enhanced CT scans with class imbalance using ensembled resnets. In Proceedings of the 2021 IEEE International IOT, Electronics and Mechatronics Conference (IEMTRONICS), Toronto, ON, Canada, 21–24 April 2021; pp. 1–7. [Google Scholar]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. Chexnet: Radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Ardakani, A.A.; Kanafi, A.R.; Acharya, U.R.; Khadem, N.; Mohammadi, A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020, 121, 103795. [Google Scholar] [CrossRef]
- Dutta, P.; Roy, T.; Anjum, N. COVID-19 detection using transfer learning with convolutional neural network. In Proceedings of the 2021 2nd International Conference on Robotics, Electrical and Signal Processing Techniques (ICREST), Dhaka, Bangladesh, 5–7 January 2021; pp. 429–432. [Google Scholar]
- Sitaula, C.; Hossain, M.B. Attention-based VGG-16 model for COVID-19 chest X-ray image classification. Appl. Intell. 2021, 51, 2850–2863. [Google Scholar] [CrossRef]
- Banerjee, A.; Kulcsar, K.; Misra, V.; Frieman, M.; Mossman, K. Bats and coronaviruses. Viruses 2019, 11, 41. [Google Scholar] [CrossRef]
- Baker, B.; Gupta, O.; Naik, N.; Raskar, R. Designing neural network architectures using reinforcement learning. arXiv 2016, arXiv:1611.02167. [Google Scholar]
- Shibly, K.H.; Dey, S.K.; Islam, M.T.U.; Rahman, M.M. COVID faster R–CNN: A novel framework to Diagnose Novel Coronavirus Disease (COVID-19) in X-ray images. Inform. Med. Unlocked 2020, 20, 100405. [Google Scholar] [CrossRef]
- Bougourzi, F.; Contino, R.; Distante, C.; Taleb-Ahmed, A. CNR-IEMN: A Deep Learning based approach to recognise COVID-19 from CT-scan. In Proceedings of the ICASSP 2021–2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Toronto, ON, Canada, 6–11 June 2021; pp. 8568–8572. [Google Scholar]
- Seum, A.; Raj, A.H.; Sakib, S.; Hossain, T. A comparative study of cnn transfer learning classification algorithms with segmentation for COVID-19 detection from CT scan images. In Proceedings of the 2020 11th International Conference on Electrical and Computer Engineering (ICECE), Dhaka, Bangladesh, 17–19 December 2020; pp. 234–237. [Google Scholar]
- Bikbov, B.; Purcell, C.A.; Levey, A.S.; Smith, M.; Abdoli, A.; Abebe, M.; Adebayo, O.M.; Afarideh, M.; Agarwal, S.K.; Agudelo-Botero, M.; et al. Global, regional, and national burden of chronic kidney disease, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2020, 395, 709–733. [Google Scholar] [CrossRef]
- James, R.M.; Sunyoto, A. Detection of CT-Scan lungs COVID-19 image using convolutional neural network and CLAHE. In Proceedings of the 2020 3rd International Conference on Information and Communications Technology (ICOIACT), Yogyakarta, Indonesia, 24–25 November 2020; pp. 302–307. [Google Scholar]
- Pisano, E.D.; Zong, S.; Hemminger, B.M.; DeLuca, M.; Johnston, R.E.; Muller, K.; Braeuning, M.P.; Pizer, S.M. Contrast limited adaptive histogram equalization image processing to improve the detection of simulated spiculations in dense mammograms. J. Digit. Imaging 1998, 11, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.E.; Rahman, T.; Khandakar, A.; Mazhar, R.; Kadir, M.A.; Mahbub, Z.B.; Islam, K.R.; Khan, M.S.; Iqbal, A.; Al Emadi, N.; et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access 2020, 8, 132665–132676. [Google Scholar] [CrossRef]
- Dalal, N.; Triggs, B. Histograms of oriented gradients for human detection. In Proceedings of the 2005 IEEE computer society conference on computer vision and pattern recognition (CVPR’05), San Diego, CA, USA, 20–25 June 2005; Volume 1, pp. 886–893. [Google Scholar]
- Nistér, D.; Stewénius, H. Linear time maximally stable extremal regions. In Proceedings of the European Conference on Computer Vision, Marseille, France, 12–18 October 2008; pp. 183–196. [Google Scholar]
- Bay, H.; Tuytelaars, T.; Gool, L.V. Surf: Speeded up robust features. In Proceedings of the European Conference on Computer Vision, Graz, Austria, 7–13 May 2006; pp. 404–417. [Google Scholar]
- Leutenegger, S.; Chli, M.; Siegwart, R.Y. BRISK: Binary robust invariant scalable keypoints. In Proceedings of the 2011 International Conference on Computer Vision, Washington, DC, USA, 6–13 November 2011; pp. 2548–2555. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Xue, S.; Abhayaratne, C. COVID-19 diagnostic using 3d deep transfer learning for classification of volumetric computerised tomography chest scans. In Proceedings of the ICASSP 2021–2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Toronto, ON, Canada, 6–11 June 2021; pp. 8573–8577. [Google Scholar]
- Brunese, L.; Martinelli, F.; Mercaldo, F.; Santone, A. Machine learning for coronavirus COVID-19 detection from chest X-rays. Procedia Comput. Sci. 2020, 176, 2212–2221. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).