Segmentation of Spectral Plant Images Using Generative Adversary Network Techniques
Abstract
:1. Introduction
- In this paper, hyperspectral images—which consist of spatial information and spectral information—are considered to obtain more detailed information about the leaf images. Using this spectral information, we have detected whether the plant image is a fake image or a real image with a higher accuracy.
- In this proposed work, the segmentation of the hyperspectral image is performed using the p2p GAN model. The main advantage of p2p GAN segmentation is that along with segmentation, a verification of the hyperspectral image can be obtained; in our research, the overall accuracy obtained by p2p GAN was 99.1%.
2. Literature Review
3. Materials and Methods
3.1. Data Set
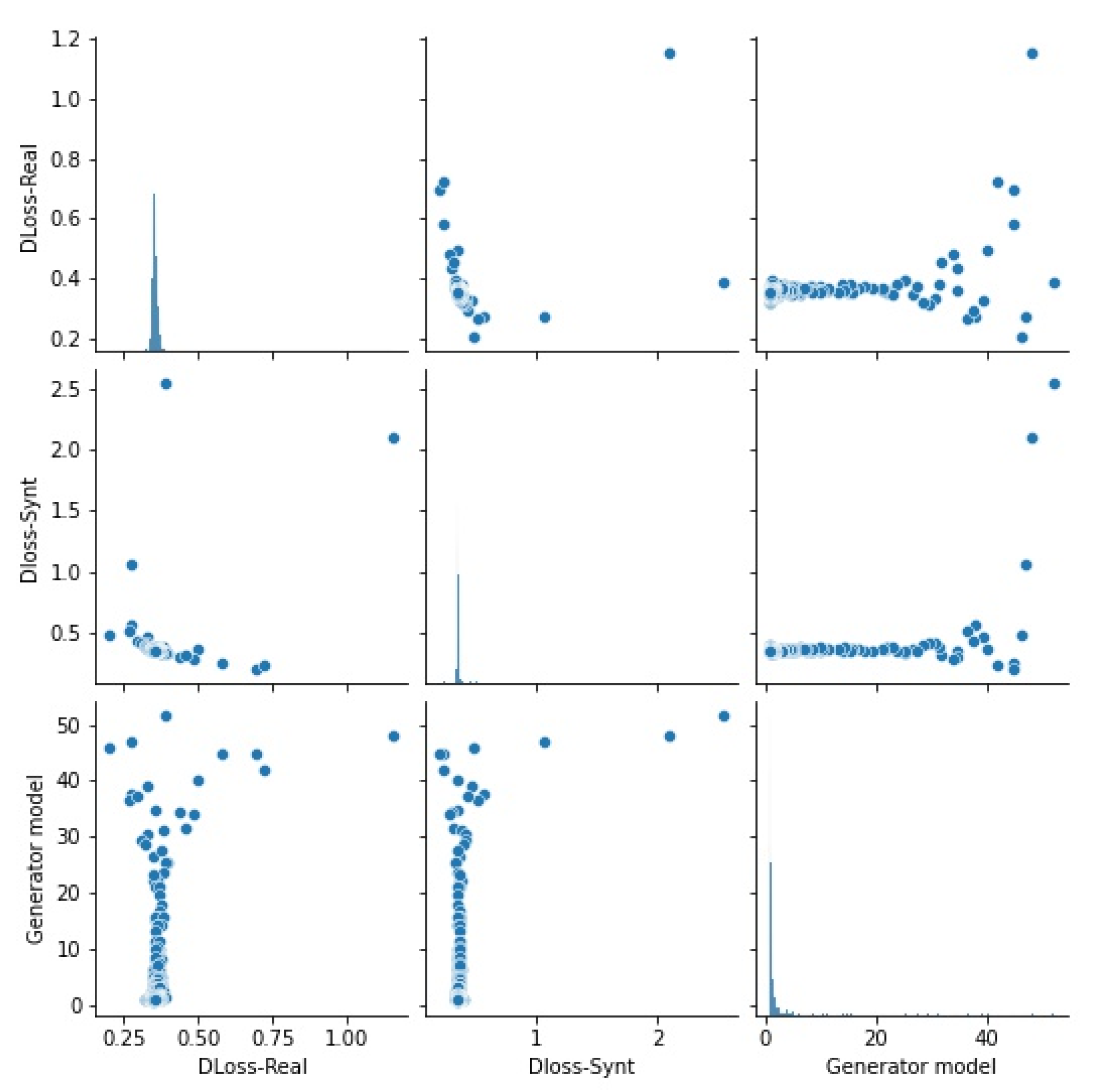
3.2. To Translate Pictures from Pixel to Pixel, a Conditional Generative Adversarial Network Is Utilized
3.3. When Compared to the Beginning
4. Results and Discussion
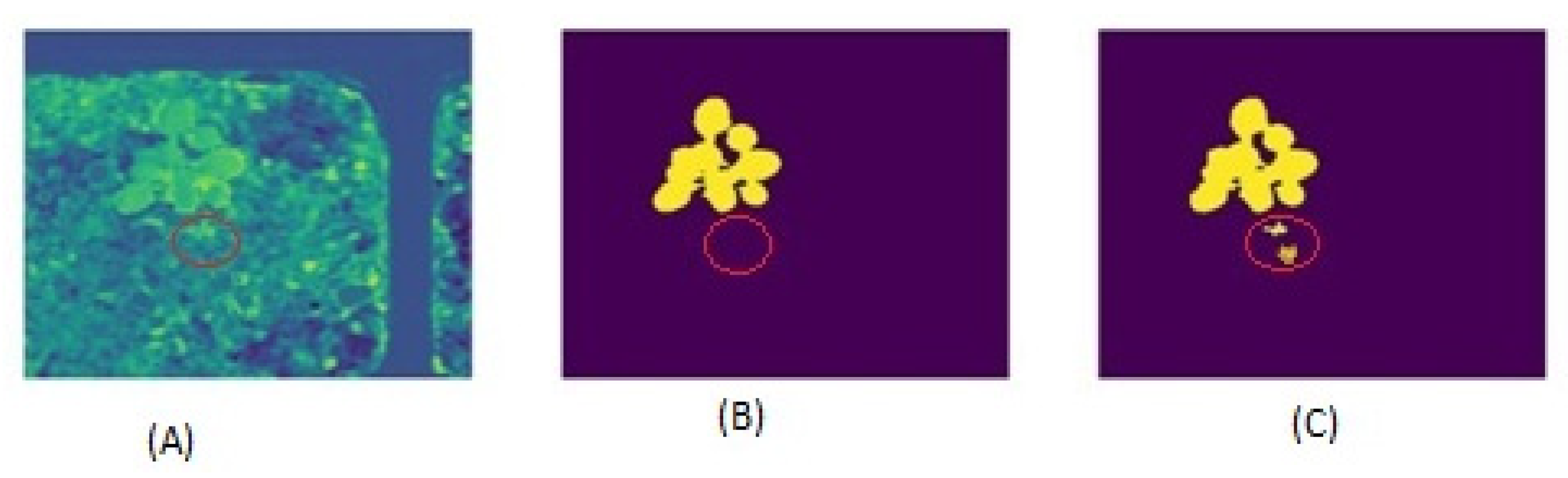
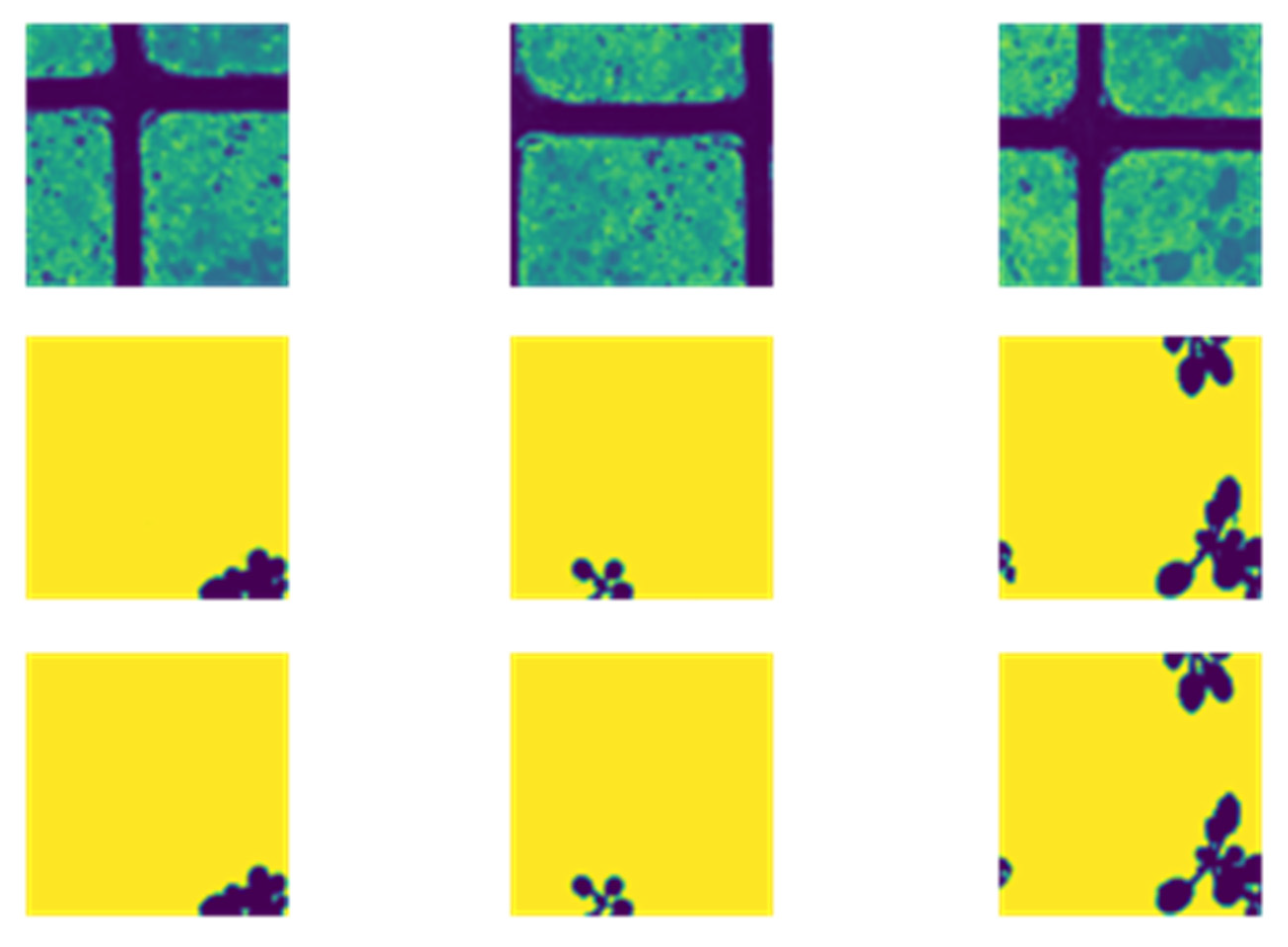
4.1. A Plant Picture with a Dirt Backdrop to Show the Limitations of Threshold-Based Segmentation Analysis with Threshold
4.2. Image Segmentation from Pixels to Pixels Using Provisional Generative Adversarial Networks
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Coic, L.; Sacré, P.-Y.; Dispas, A.; De Bleye, C.; Fillet, M.; Ruckebusch, C.; Hubert, P.; Ziemons, E. Pixel-based Raman hyperspectral identification of complex pharmaceutical formulations. Anal. Chim. Acta 2021, 1155, 338361. [Google Scholar] [CrossRef] [PubMed]
- Shan, J.; Zhao, J.; Zhang, Y.; Liu, L.; Wu, F.; Wang, X. Simple and rapid detection of microplastics in seawater using hyperspectral imaging technology. Anal. Chim. Acta 2019, 1050, 161–168. [Google Scholar] [CrossRef]
- Adao, T.; Hruška, J.; Pádua, L.; Bessa, J.; Peres, E.; Morais, R.; Sousa, J.J. Hyperspectral imaging: A review on UAV-based sensors, data processing and applications for agriculture and forestry. Remote Sens. 2017, 9, 1110. [Google Scholar] [CrossRef] [Green Version]
- Arendse, E.; Fawole, O.A.; Magwaza, L.S.; Opara, U.L. Non-destructive prediction of internal and external quality attributes of fruit with thick rind: A review. J. Food Eng. 2018, 217, 11–23. [Google Scholar] [CrossRef]
- Botelho, B.G.; Oliveira, L.S.; Franca, A.S. Fluorescence spectroscopy as tool for the geographical discrimination of coffees produced in different regions of Minas Gerais State in Brazil. Food Control. 2017, 77, 25–31. [Google Scholar] [CrossRef]
- Mishra, P.; Schmuck, M.; Roth, S.; Nicol, A.; Nordon, A. Homogenising and segmenting hyperspectral images of plants and testing chemicals in a high-throughput plant phenotyping setup. In Proceedings of the 2019 10th Workshop on Hyperspectral Imaging and Signal Processing: Evolution in Remote Sensing (WHISPERS), Amsterdam, The Netherlands, 24–26 September 2019; IEEE: New York, NY, USA; pp. 1–5. [Google Scholar]
- Asaari, M.S.M.; Mishra, P.; Mertens, S.; Dhondt, S.; Inzé, D.; Wuyts, N.; Scheunders, P. Close-range hyperspectral image analysis for the early detection of stress responses in individual plants in a high-throughput phenotyping platform. ISPRS J. Photogramm. Remote Sens. 2018, 138, 121–138. [Google Scholar] [CrossRef]
- Fahlgren, N.; Gehan, M.A.; Baxter, I. Lights, camera, action: High-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015, 24, 93–99. [Google Scholar] [CrossRef] [Green Version]
- Mishra, P.; Polder, G.; Gowen, A.; Rutledge, D.N.; Roger, J.-M. Utilising variable sorting for normalisation to correct illumination effects in close-range spectral images of potato plants. Biosyst. Eng. 2020, 197, 318–323. [Google Scholar] [CrossRef]
- Mishra, P.; Karami, A.; Nordon, A.; Rutledge, D.N.; Roger, J.-M. Automatic de-noising of close-range hyperspectral images with a wavelength-specific shearlet-based image noise reduction method. Sens. Actuators B Chem. 2019, 281, 1034–1044. [Google Scholar] [CrossRef]
- Kandpal, L.M.; Tewari, J.; Gopinathan, N.; Boulas, P.; Cho, B.K. In-process control assay of pharmaceutical microtablets using hyperspectral imaging coupled with multivariate analysis. Anal. Chem. 2016, 88, 11055–11061. [Google Scholar] [CrossRef]
- Ferreira, K.B.; Oliveira, A.G.G.; Gonçalves, A.S.; Gomes, J.A. Evaluation of hyperspectral imaging visible/near infrared spectroscopy as a forensic tool for automotive paint distinction. Forensic Chem. 2017, 5, 46–52. [Google Scholar] [CrossRef]
- Chen, G.; Qian, S.E. Denoising of hyperspectral imagery using principal component analysis and wavelet shrinkage. IEEE Trans. Geosci. Remote Sens. 2010, 49, 973–980. [Google Scholar] [CrossRef]
- Wahabzada, M.; Mahlein, A.K.; Bauckhage, C.; Steiner, U.; Oerke, E.C.; Kersting, K. Plant phenotyping using probabilistic topic models: Uncovering the hyperspectral language of plants. Sci. Rep. 2016, 6, 22482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heylen, R.; Burazerovic, D.; Scheunders, P. Fully constrained least squares spectral unmixing by simplex projection. IEEE Trans. Geosci. Remote Sens. 2011, 49, 4112–4122. [Google Scholar] [CrossRef]
- De Beer, T.; Bodson, C.; Dejaegher, B.; Walczak, B.; Vercruysse, P.; Burggraeve, A.; Lemos, A.; Delattre, L.; Heyden, Y.V.; Remon, J.; et al. Raman spectroscopy as a process analytical technology (PAT) tool for the in-line monitoring and understanding of a powder blending process. J. Pharm. Biomed. Anal. 2008, 48, 772–779. [Google Scholar] [CrossRef] [PubMed]
- Sacré, P.-Y.; Lebrun, P.; Chavez, P.-F.; De Bleye, C.; Netchacovitch, L.; Rozet, E.; Klinkenberg, R.; Streel, B.; Hubert, P.; Ziemons, E. A new criterion to assess distributional homogeneity in hyperspectral images of solid pharmaceutical dosage forms. Anal. Chim. Acta 2014, 818, 7–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cailletaud, J.; Bleye, C.; Dumont, E.; Sacré, P.Y.; Gut, Y.; Bultel, L.; Ginot, Y.M.; Hubert, P.; Ziemons, E. Towards a spray-coating method for the detection of low-dose compounds in pharmaceutical tablets using surface-enhanced Raman chemical imaging (SER-CI). Talanta 2018, 188, 584–592. [Google Scholar] [CrossRef]
- El-Hagrasy, A.S.; Delgado-Lopez, M.; Drennen, J.K., III. A process analytical technology approach to near-infrared process control of pharmaceutical powder blending: Part II: Qualitative near-infrared models for prediction of blend homogeneity. J. Pharm. Sci. 2006, 95, 407–421. [Google Scholar] [CrossRef]
- Alexandrino, G.L.; Poppi, R. NIR imaging spectroscopy for quantification of constituents in polymers thin films loaded with paracetamol. Anal. Chim. Acta 2013, 765, 37–44. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; ElMasry, G.; Sun, D.-W.; Allen, P. Prediction of some quality attributes of lamb meat using near-infrared hyperspectral imaging and multivariate analysis. Anal. Chim. Acta 2012, 714, 57–67. [Google Scholar] [CrossRef]
- Duponchel, L. Exploring hyperspectral imaging data sets with topological data analysis. Anal. Chim. Acta 2018, 1000, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Li, J.; Wu, C.; Plaza, A. Regional clustering-based spatial preprocessing for hyperspectral unmixing. Remote Sens. Environ. 2018, 204, 333–346. [Google Scholar] [CrossRef]
- Fauteux-Lefebvre, C.; Lavoie, F.; Gosselin, R. A Hierarchical Multivariate Curve Resolution Methodology to Identify and Map Compounds in Spectral Images. Anal. Chem. 2018, 90, 13118–13125. [Google Scholar] [CrossRef] [PubMed]
- Bøtker, J.; Wu, J.X.; Rantanen, J. Hyperspectral imaging as a part of pharmaceutical product design. In Data Handling in Science and Technology; Elsevier: Amsterdam, The Netherlands, 2020; Volume 32, pp. 567–581. [Google Scholar]
- Biancolillo, A.; Boqué, R.; Cocchi, M.; Marini, F. Data fusion strategies in food analysis. In Data Handling in Science and Technology; Elsevier: Amsterdam, The Netherlands, 2020; Volume 31, pp. 271–310. [Google Scholar]
- Mitsutake, H.; Castro, S.R.; de Paula, E.; Poppi, R.J.; Rutledge, D.N.; Breitkreitz, M.C. Comparison of different chemometric methods to extract chemical and physical information from Raman images of homogeneous and heterogeneous semi-solid pharmaceutical formulations. Int. J. Pharm. 2018, 552, 119–129. [Google Scholar] [CrossRef]
- de Juan, A. Multivariate curve resolution for hyperspectral image analysis. In Data Handling in Science and Technology; Elsevier: Amsterdam, The Netherlands, 2020; Volume 32, pp. 115–150. [Google Scholar]
- Xu, K.; Zhao, Y.; Zhang, L.; Gao, C.; Huang, H. Spectral–Spatial Residual Graph Attention Network for Hyperspectral Image Classification. IEEE Geosci. Remote Sens. Lett 2021, 19, 5509305. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, S.; Kansal, S.; Alkinani, M.H.; Elaraby, A.; Garg, S.; Natarajan, S.; Sharma, V. Segmentation of Spectral Plant Images Using Generative Adversary Network Techniques. Electronics 2022, 11, 2611. https://doi.org/10.3390/electronics11162611
Kumar S, Kansal S, Alkinani MH, Elaraby A, Garg S, Natarajan S, Sharma V. Segmentation of Spectral Plant Images Using Generative Adversary Network Techniques. Electronics. 2022; 11(16):2611. https://doi.org/10.3390/electronics11162611
Chicago/Turabian StyleKumar, Sanjay, Sahil Kansal, Monagi H. Alkinani, Ahmed Elaraby, Saksham Garg, Shanthi Natarajan, and Vishnu Sharma. 2022. "Segmentation of Spectral Plant Images Using Generative Adversary Network Techniques" Electronics 11, no. 16: 2611. https://doi.org/10.3390/electronics11162611
APA StyleKumar, S., Kansal, S., Alkinani, M. H., Elaraby, A., Garg, S., Natarajan, S., & Sharma, V. (2022). Segmentation of Spectral Plant Images Using Generative Adversary Network Techniques. Electronics, 11(16), 2611. https://doi.org/10.3390/electronics11162611






