Future Image Synthesis for Diabetic Retinopathy Based on the Lesion Occurrence Probability
Abstract
1. Introduction
- We propose a future prediction framework for future fundus image synthesis of patients with DR to observe a progression of the disease by preserving the vessel identity of patients based on an adversarial learning mechanism. The framework, which consists of a fundus generator and a lesion probability predictor, effectively addresses the issue of longitudinal patient data shortages for future image visualization by separating the training sequence.
- We combine the reconstruction loss and an adversarial loss to improve the performance of fundus image synthesis to effectively control the pathological information by using the lesion and vessel mask in fundus generator training. Furthermore, in the lesion probability predictor training step, we introduce a regularization loss function to accurately predict the lesion occurrence probability by calculating the structural similarity between the current vessel and target future vessel at the pixel level.
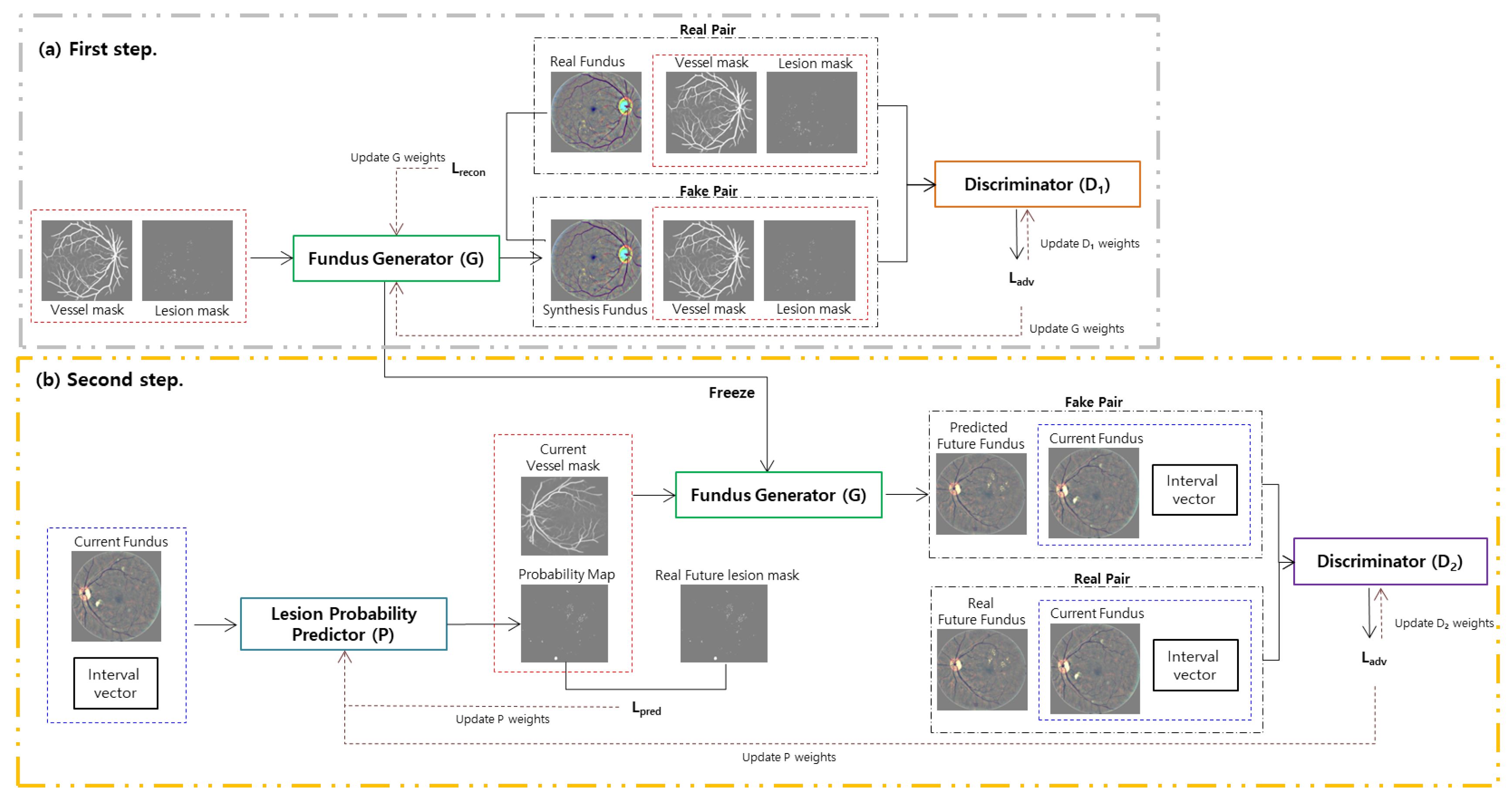
2. Methods
2.1. First Step: Fundus Generator Training
2.2. Second Step: Lesion Probability Predictor Training
3. Experiments and Result
3.1. Datasets
3.2. Pre-Processing
3.3. Implementation
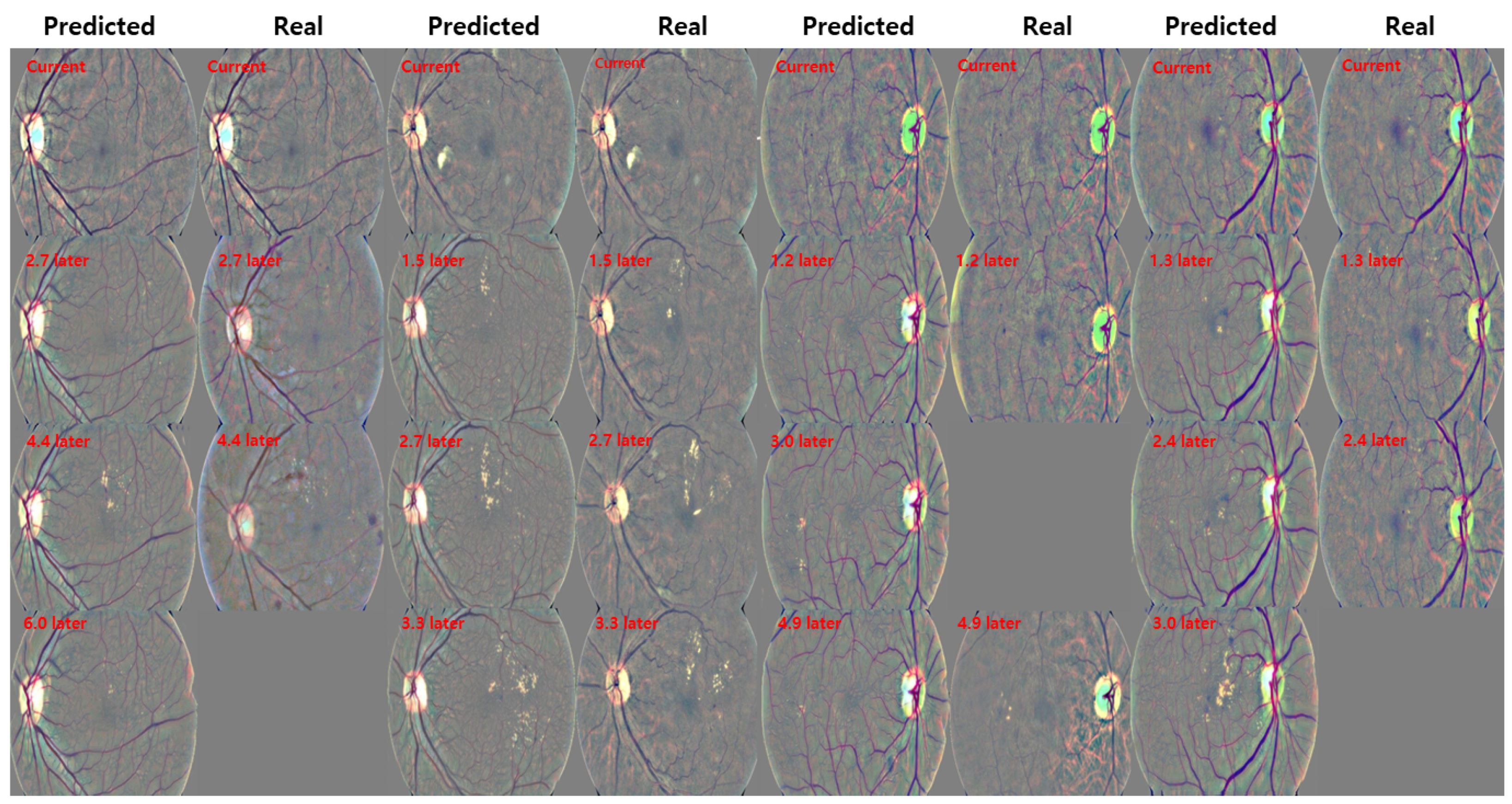
3.4. Qualitative Results
3.5. Quantitative Results
4. Discussion and Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rachmadi, M.F.; Valdés-Hernández, M.D.C.; Makin, S.; Wardlaw, J.M.; Komura, T. Predicting the Evolution of White Matter Hyperintensities in Brain MRI Using Generative Adversarial Networks and Irregularity Map. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019; Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.T., Khan, A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 146–154. [Google Scholar]
- Xia, T.; Chartsias, A.; Tsaftaris, S.A. Consistent Brain Ageing Synthesis. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019; Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.T., Khan, A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 750–758. [Google Scholar]
- Schlegl, T.; Seeböck, P.; Waldstein, S.M.; Langs, G.; Schmidt-Erfurth, U. f-AnoGAN: Fast unsupervised anomaly detection with generative adversarial networks. Med. Image Anal. 2019, 54, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, C.F.; Kamnitsas, K.; Matthew, J.; Fletcher, T.P.; Smith, S.; Koch, L.M.; Kainz, B.; Rueckert, D. SonoNet: Real-Time Detection and Localisation of Fetal Standard Scan Planes in Freehand Ultrasound. IEEE Trans. Med. Imaging 2017, 36, 2204–2215. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.; Loew, M. Breast cancer detection using synthetic mammograms from generative adversarial networks in convolutional neural networks. In Proceedings of the 14th International Workshop on Breast Imaging (IWBI 2018), Atlanta, GA, USA, 8–11 July 2018; Krupinski, E.A., Ed.; International Society for Optics and Photonics, SPIE: Bellingham, WA, USA, 2018; Volume 10718, pp. 228–236. [Google Scholar] [CrossRef]
- Khojasteh, P.; Passos Júnior, L.A.; Carvalho, T.; Rezende, E.; Aliahmad, B.; Papa, J.P.; Kumar, D.K. Exudate detection in fundus images using deeply-learnable features. Comput. Biol. Med. 2019, 104, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, Y.; Wang, Y.; Chen, W.B. Diabetic Retinopathy Stage Classification Using Convolutional Neural Networks. In Proceedings of the 2018 IEEE International Conference on Information Reuse and Integration (IRI), Salt Lake City, UT, USA, 6–9 July 2018; pp. 465–471. [Google Scholar]
- Zhao, Z.; Zhang, K.; Hao, X.; Tian, J.; Heng Chua, M.C.; Chen, L.; Xu, X. BiRA-Net: Bilinear Attention Net for Diabetic Retinopathy Grading. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; pp. 1385–1389. [Google Scholar] [CrossRef]
- Zhou, Y.; He, X.; Huang, L.; Liu, L.; Zhu, F.; Cui, S.; Shao, L. Collaborative Learning of Semi-Supervised Segmentation and Classification for Medical Images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 2019, Long Beach, CA, USA, 15–20 June 2019; pp. 2074–2083. [Google Scholar] [CrossRef]
- Ren, F.; Cao, P.; Zhao, D.; Wan, C. Diabetic macular edema grading in retinal images using vector quantization and semi-supervised learning. Technol. Health Care 2018, 26, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Syed, A.M.; Akram, M.U.; Akram, T.; Muzammal, M.; Khalid, S.; Khan, M.A. Fundus Images-Based Detection and Grading of Macular Edema Using Robust Macula Localization. IEEE Access 2018, 6, 58784–58793. [Google Scholar] [CrossRef]
- Arcadu, F.; Benmansour, F.; Maunz, A.; Willis, J.; Zdenka Haskova, M.P. Deep learning algorithm predicts diabetic retinopathy progression in individual patients. Digit. Med. 2019, 2, 92. [Google Scholar] [CrossRef] [PubMed]
- Zou, K.; Warfield, S.; Bharatha, A.; Tempany, C.; Kaus, M.; Haker, S.; Wells, W.; Jolesz, F.; Kikinis, R. Statistical Validation of Image Segmentation Quality Based on a Spatial Overlap Index. Acad. Radiol. 2004, 11, 178–189. [Google Scholar] [CrossRef]
- Diabetic Retinopathy—Segmentation and Grading Challenge. Available online: https://idrid.grand-challenge.org/ (accessed on 19 March 2021).
- Kaggle Diabetic Retinopathy Detection Competition. Available online: https://www.kaggle.com/c/diabetic-retinopathy-detection (accessed on 19 March 2021).
- Son, J.; Park, S.J.; Jung, K.H. Retinal Vessel Segmentation in Fundoscopic Images with Generative Adversarial Networks. arXiv 2017, arXiv:1706.09318. [Google Scholar]
- DRIVE: Digital Retinal Images for Vessel Extraction. Available online: https://drive.grand-challenge.org/ (accessed on 19 March 2021).
- Mirza, M.; Osindero, S. Conditional Generative Adversarial Nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- Zhao, H.; Li, H.; Maurer-Stroh, S.; Cheng, L. Synthesizing retinal and neuronal images with generative adversarial nets. Med. Image Anal. 2018, 49, 14–26. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Tan, M.; Le, Q. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; Chaudhuri, K., Salakhutdinov, R., Eds.; PMLR: Cambridge, MA, USA, 2019; Volume 97, pp. 6105–6114. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]



| Model | F1 Score |
|---|---|
| ResNet-50 [20] | 0.68 |
| EfficientNet-B0 [21] | 0.72 |
| Se-ResNet-50 [22] | 0.79 |
| Method | Accuracy | Specificity | Sensitivity | F1 Score |
|---|---|---|---|---|
| cGAN [18] | 0.64 | 0.73 | 0.46 | 0.47 |
| Tub-sGAN [19] | 0.78 | 0.84 | 0.69 | 0.62 |
| (n) | 0.80 | 0.85 | 0.70 | 0.69 |
| (r) | 0.83 | 0.87 | 0.74 | 0.74 |
| Data | MILD | SEVERE | PDR | AVG |
|---|---|---|---|---|
| Predicted Future Fundus | 0.80 | 0.71 | 0.70 | 0.74 |
| Real Future Fundus | 0.82 | 0.78 | 0.78 | 0.79 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahn, S.; Pham, Q.T.M.; Shin, J.; Song, S.J. Future Image Synthesis for Diabetic Retinopathy Based on the Lesion Occurrence Probability. Electronics 2021, 10, 726. https://doi.org/10.3390/electronics10060726
Ahn S, Pham QTM, Shin J, Song SJ. Future Image Synthesis for Diabetic Retinopathy Based on the Lesion Occurrence Probability. Electronics. 2021; 10(6):726. https://doi.org/10.3390/electronics10060726
Chicago/Turabian StyleAhn, Sangil, Quang T.M. Pham, Jitae Shin, and Su Jeong Song. 2021. "Future Image Synthesis for Diabetic Retinopathy Based on the Lesion Occurrence Probability" Electronics 10, no. 6: 726. https://doi.org/10.3390/electronics10060726
APA StyleAhn, S., Pham, Q. T. M., Shin, J., & Song, S. J. (2021). Future Image Synthesis for Diabetic Retinopathy Based on the Lesion Occurrence Probability. Electronics, 10(6), 726. https://doi.org/10.3390/electronics10060726






