1. Introduction
The skin is the largest human organ, and it is the outer covering of the body. The skin is the first line of defense in the human body [
1]. It has the role of (i) protecting the internal organs from external environmental influences, (ii) regulating body temperatures, (iii) providing immunity against many diseases, and (iv) providing beauty to the body [
2]. The human body is protected by the skin from harmful ultraviolet rays from the sun, although the essential vitamin D is also produced by this organ when the body is exposed to sunlight. Skin color (body pigmentation) and moisture (from oily to dry skin) vary from person to person according to the hot and cold regions in the world [
3]. Cellular DNA is damaged if the body is exposed to sunlight or ultraviolet rays for a long time, decreasing skin pigmentation and the incidence of malignant skin diseases. Skin cancer (melanoma) is a fatal skin disease with no early diagnosis. In the early stages of the disease, it is not detected due to the similarity of the cancer cells to other skin cells. Abnormal cancerous cells divide rapidly, penetrating the lower skin layers and becoming incurable malignant melanomas [
4].
Disease treatment is challenging when melanoma cells spread throughout the body. Therefore, early diagnosis is required to save lives. According to dermoscopy images, there are many types of skin diseases. The two main types of skin disease can be identified as melanocytic and nonmelanocytic. Melanocytic diseases have two types: melanoma and melanocytic nevi [
5,
6]. In contrast, nonmelanocytic skin diseases contain five types: basal cell carcinoma (BCC), squamous cell carcinoma (SCC), vascular (VASC), benign keratosis lesions (BKL), and dermatofibroma (df). Melanoma (mel) is one of the most dangerous and deadly skin diseases. It is incurable in its late stages and is one of the most common cancers worldwide.
There are many challenges, including different skin colors from one person to another and artifacts, hair, and air bubbles. In addition, there are many similar features between types of skin diseases, especially in the early stages. More than 420 million people are suffering from skin diseases worldwide, representing a limitation in terms of insufficient medical resources. Furthermore, the high treatment cost represents a challenge, especially in developing countries, which causes delays in early diagnosis and leads to the deterioration of life and social development [
7].
According to the American Cancer Society (ACS), there were more than 70,000 cases in the United States in 2017. In 2020, 100,350 cases of melanoma were diagnosed in that country, with 60,190 cases occurring in men and 40,160 occurring in women [
8]. Because of lesion’s early-stage similarity, experts and clinicians find distinguishing melanoma and benign early-stage lesions difficult. Therefore, early diagnosis using computer-aided diagnostic techniques (e.g., artificial intelligence) has become essential. Computer-aided diagnosis is critical to helping physicians and experts save time and effort during early disease diagnosis. In our research, we used artificial intelligence techniques for the early diagnosis of skin lesions. With many layers and complex neurons, specific samples are trained using artificial intelligence techniques to solve particular problems. Deep learning techniques have been used to solve complex diagnostic problems that can hardly be solved using machine learning techniques [
9].
Artificial intelligence techniques have been applied to classify many types of images and medical records, such as dermatoscopy, magnetic resonance imaging, computed tomography (CT) scans, and medical records [
10,
11]. Artificial intelligence techniques have been used to improve the quality of the entered images by using pre-processing methods, identifying lesion areas, and isolating them from healthy skin to focus on the disease area. Then, essential features (e.g., shape, texture, color, and shape) from each image are extracted and distinguished from other images in which these features are stored. Finally, the features are fed into the classification stage to diagnose each lesion in the appropriate class.
The main contributions of this study are as follows:
Extract the important features from each image with LBP, GLCM, and DWT algorithms, combining the extracted features into one vector to obtain representative features for each image; diagnose the images using artificial neural networks ANN and FFNN classifiers.
The capabilities of deep learning networks lie in imparting acquired skills to solve new, relevant problems.
In this research, the early diagnosis of skin lesions and distinguishing benign images from malignant ones are considered.
Machine learning algorithms (ANN and FFNN) achieved better results than CNN models (ResNet-50 and AlexNet).
Machine and deep learning techniques will help medical doctors in the early detection of skin lesions, enhancing the confidence of doctors and reducing the number of biopsies and surgeries.
The remainder of the manuscript is organized as follows: related work is discussed in
Section 2. In
Section 3, the analysis of materials and methodology, containing subsections of image processing methods, is discussed. Later, the results achieved by machine learning algorithms and deep learning are described and compared in
Section 4. Then, the discussion of the research is summarized in
Section 5. Finally, in
Section 6, conclusions regarding this manuscript are provided.
2. Related Work
Many researchers from interested scientific communities have worked on diagnosing skin lesions using artificial intelligence techniques in recent years. Qin et al. (2020) discussed generative adversarial networks (GANs), where a generator and discriminator would be tuned by the network to produce high-resolution images and remove noise. The network evaluation was conducted through a set of images produced by the GANs, and the network achieved high performance [
12]. Tschandl et al. (2019) retrained the ResNet-34 model on the skin lesion dataset through ImageNet and fine-tuned the ResNet-34 Jaccard system through ImageNet, achieving the highest random tuning [
13].
Sreelatha et al. (2019) discussed the gradient and feature adaptive contour (GFAC) method for lesion zone segmentation and early detection of melanoma [
14]. Chatterjee et al. (2019) proposed a recursive feature elimination (RFE) method based on multilayer images. Color, texture, and shape features were extracted by GLCM and fractal-based regional texture analysis (FRTA) methods and feature classification by the support vector machine (SVM) algorithm [
15]. Al-Masni et al. (2020) proposed a technique that combines both lesion segmentation and classification. First, a full-resolution convolutional network (FrCN) was used to segment the melanoma. Then, deep learning models were used to categorize segmented lesions [
16]. Alzubaidi et al. (2021) proposed a new approach for transferring learning to train an extensive ISIC dataset and transfer knowledge to a target dermatology dataset. Furthermore, CNNs, in which recent developments are combined, were used to diagnose skin lesions with high accuracy [
17].
Liu et al. (2021) proposed a multiscale ensemble of convolutional neural networks (MECNN), which consists of three branches. In the first branch, the lesion area is defined by selecting the most points around the lesion. Then, the search area of a region of interest is reduced using the MECNN method. Finally, the outline was divided into two inputs for the other two branches [
18]. Ding et al. (2021) proposed conditional GANs (CGANs) to obtain high-resolution images. The segmentation mask and class labels were combined to establish an efficient mapping of the pathological markers of interest in that study. Translation from an image to a matrix is performed in CGAN methods. Then, such a matrix is assigned to a label as an input for each image [
19]. Surówka et al. (2021) proposed the wavelet packet method for feature extraction through four wavelet decomposition channels. The features are classified by a logistic classifier that can extract high-resolution wavelet properties [
20]. Iqbal et al. (2021) proposed a carefully designed deep convolutional neural network model with multiple layers and various filter sizes [
21].
Sikkandar et al. (2021) proposed a diagnostic model based on lesion segmentation using the GrabCut method and the adaptive neuro-fuzzy classifier [
22]. Ali et al. (2021) introduced the DCNN model, in which noise and artifacts were first removed from the images. In this method, image normalization and deep feature extraction are performed. Then data augmentation during the training phase was applied for the model to acquire a large number of images [
23]. Kim et al. (2021) proposed a hair removal method from the lesion area on the image itself. Thus, coarse hair is removed by the algorithm while preserving the features of the lesion [
24].
Tyagi et al. (2020) proposed an intelligent prognostic model for disease prediction and classification using a combination of CNN with particle swarm optimization (PSO) [
25]. Ahmad et al. (2021) discussed the generative adversarial networks (GANs) method for training a convolutional neural network on a balanced dataset [
26]. Molina-Molina et al. (2020) proposed a 1D fractal signature method for extracting texture features and combining them with features extracted using the Densenet-201 model. Such features would then be used as feature vectors to both classifiers K-nearest neighbors and SVM for diagnosing skin lesions [
27]. Adegun et al. (2020) proposed a framework for the segmentation and classification of skin lesions. This method consists of two stages. In the first stage of the encoder-decoder fully convolutional network, complex and heterogeneous features are learned. In the second stage, the coarse texture is extracted at the encoder stage, and the lesion boundaries are extracted during the decoding stage [
28].
Khan et al. (2021) presented a High-Frequency approach with a Multilayered Feed-Forward Neural Network (HFaFFNN) to integrate all images, enhance images with a log-opening-based activation function. Pre-trained CNNs Darknet-53 and NasNet-mobile have been implemented, and parameters are tuned for high performance. Finally, a parallel max entropy correlation (PMEC) algorithm was used to fuse the extracted features [
29]. Muhammad et al. (2021) presented a two-stage framework for segmentation and classification. For lesions segmentation, a hybrid technique was used through the complementary strengths of two CNNs to produce a region of interest (RoI). To classify lesions by CNNs, 30 layers were trained on the HAM10000 dataset. The most important features were selected using the Regular Falsi method; the system achieved a good performance in diagnosing skin lesions [
30]. Muhammad et al. (2021) presented a two-way CNN information fusion framework for diagnosing melanoma. Image contrast was improved based on fusion, then improved features were extracted by the skewness-controlled moth-flame optimization method. The second frame uses the MobileNetV2 model to extract the features. Finally, the features extracted from the two methods are fused by the new parallel multimax coefficient correlation algorithm. The system achieved superior performance in diagnosing skin cancer [
31]. Muhammad et al. (2021) presented a robust system for diagnosing skin lesions through several stages. Firstly, image enhancement using local color-controlled histogram intensity values (LCcHIV). Secondly, pest segmentation by novel Deep Saliency. The threshold function is applied to obtain binary images. Thirdly, the most important features were extracted through the improved moth flame optimization (IMFO) algorithm. Finally, the system achieved high performance in diagnosing skin lesions by incorporating features and categorizing them using the Kernel Extreme Learning Machine [
32].
Previous studies contain many challenges such as hair, air bubbles, artifacts, and light reflections. Researchers also face challenges such as the similarity of characteristics between types of diseases, which constitute a major challenge in the diagnosis and distinction between diseases. Therefore, the proposed systems in this study addressed all the challenges of the previous studies. Many enhancement techniques improved the images by removing artifacts, air bubbles, skin lines, and reflections by applying two filters together, namely Laplacian and average filters. Furthermore, the Dullrazor technology works with images containing hair and removes hair with high accuracy. As for the challenges of similar features between some diseases, three hybrid algorithms were applied to extract the features from each image and combine the features extracted from the three methods into one vector for each image. Thus, each disease is represented by its representative features. Parameters and weights of two models, ResNet-50 and AlexNet, were also adjusted to extract each disease’s deep and representative characteristics.
3. Materials and Methodology
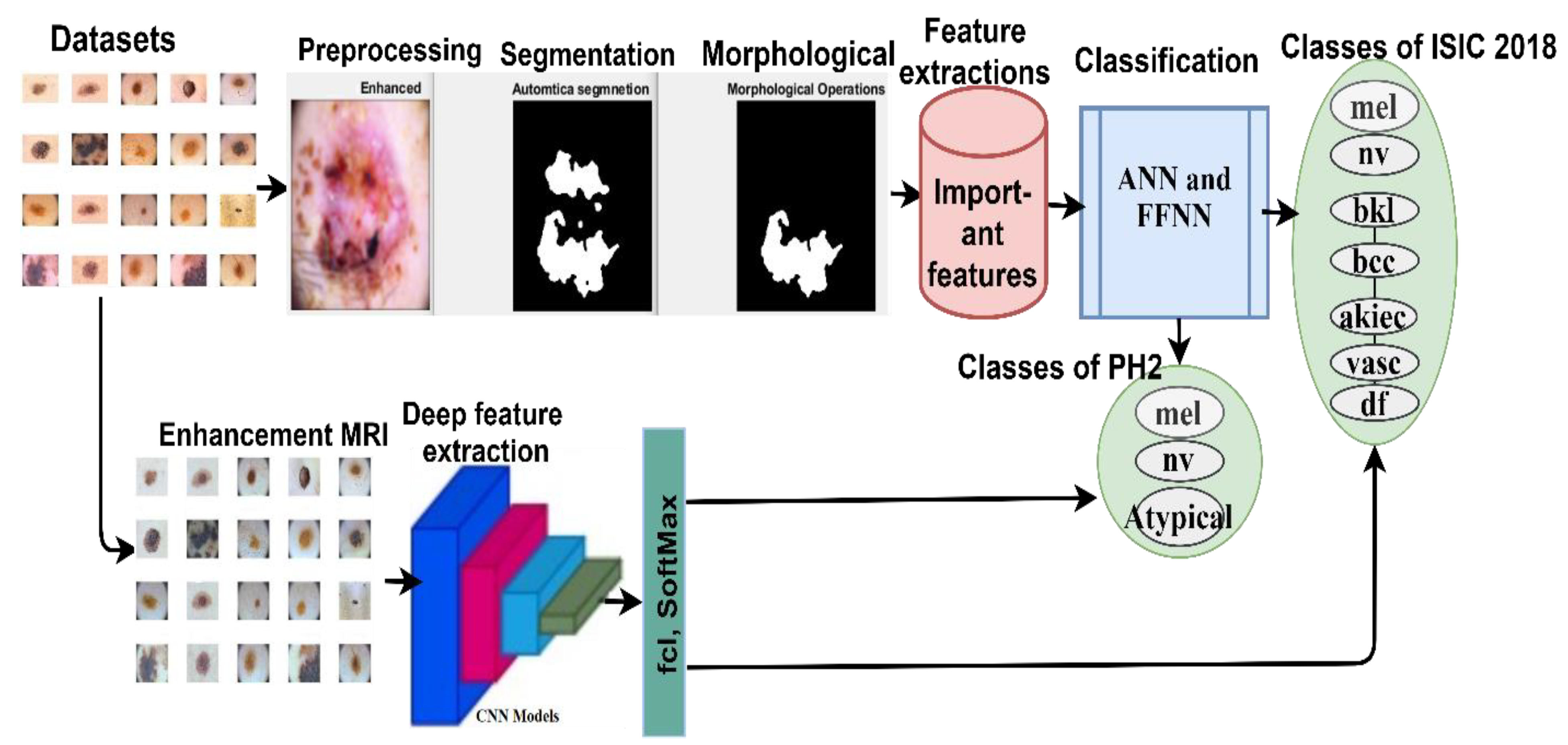
The evaluated proposed systems were applied to two datasets: ISIC 2018 and PH2. Each of these datasets was collected under different conditions and had different characteristics. The use of each dataset has several inherent challenges. The most important of which are (i) isolating the lesion from healthy skin (segmentation), (ii) localizing the features and patterns, (iii) and extracting and classifying the features of each lesion. Therefore, skin lesions may be detected by the systems, and skin cancer may be distinguished from other kinds of lesions. In
Figure 1, the mechanism of action of the proposed method for diagnosing skin diseases is described. Images were enhanced, and the noise was removed using Laplacian and average filters and hair removal with the Dullrazor technique. The lesion segmentation was performed using the adopted region growth algorithm. Feature extraction was conducted in two different ways, in which traditional and deep learning were considered. In traditional methods, features were extracted by combining the extracted features through three algorithms: LBP, GLCM, and DWT. The deep feature maps were also extracted by CNN for the ResNet-50 and AlexNet models. The features extracted using traditional methods were classified using ANN and FFNN. At the same time, the deep feature maps were classified by two pre-trained CNNs, namely ResNet-50 and AlexNet models.
3.1. Dataset
In this study, the two standard datasets, namely, ISIC 2018 and PH2, were used for diagnosing dermatological diseases, which are explained as follows.
3.1.1. International Skin Imaging Collaboration (ISIC 2018) Dataset
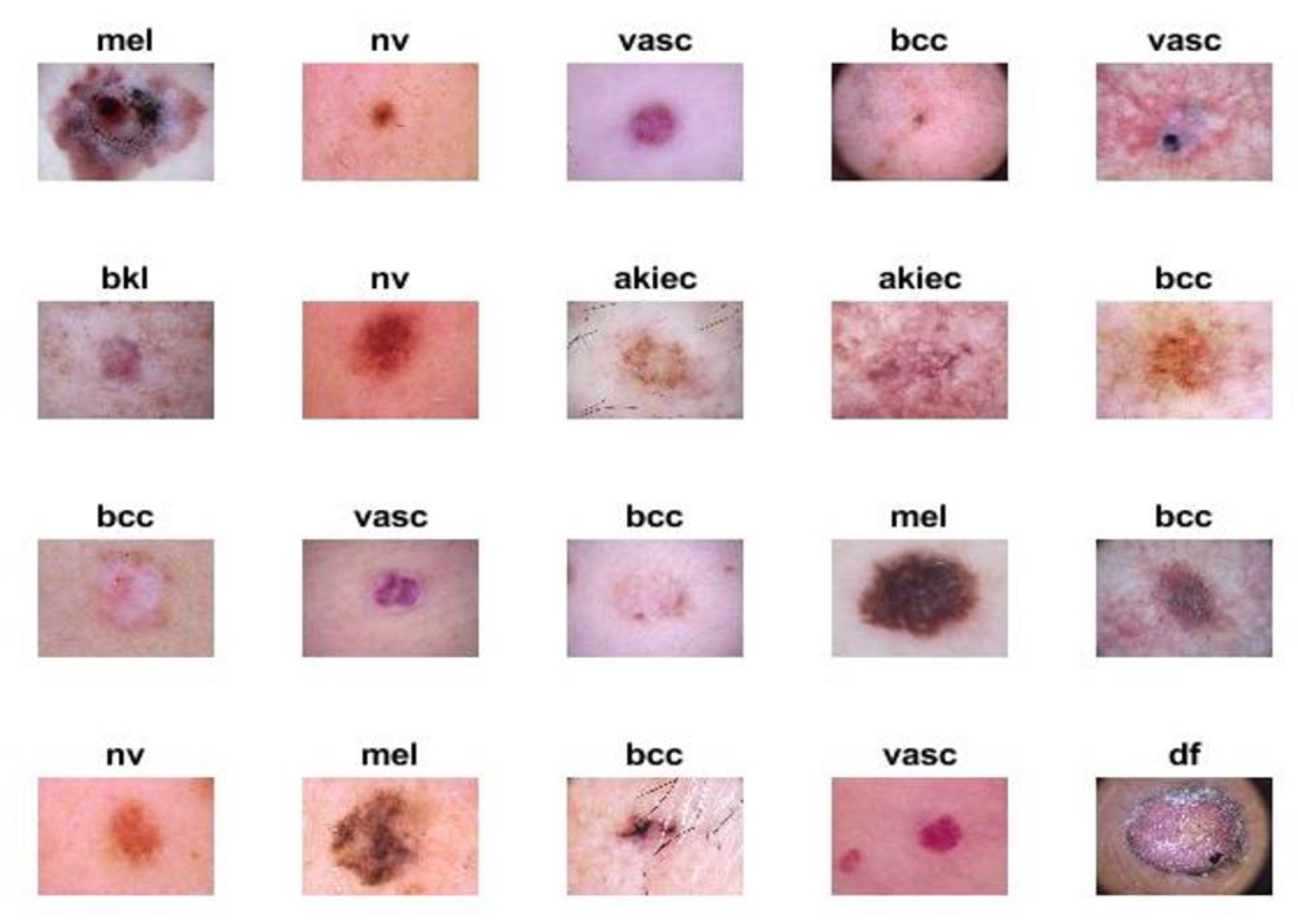
The proposed systems were evaluated using the ISIC 2018 dataset. The criteria for endoscopic devices were assessed to obtain high-resolution images and techniques, such as illumination, size, calibration markers, poses, magnification, terminology as diagnoses, lesion site, and morphology. The ISIC 2018 dataset, also known as HAM-10000, contains seven unbalanced diseases. In this study, the different kinds of diseases evaluated from this dataset and the amount of images considered in each condition were the following: actinic keratoses (AKIEC; n = 200 images), basal cell carcinoma (BCC; n = 200 images), benign keratosis lesions (BKL; n = 200 images), dermatoma (DF; n = 100 images), melanoma (MEL; n = 200 images), melanocytic nevi (NV; n = 200 images), and vascular (VASC; n = 100 images). In
Figure 2, the samples from the ISIC 2018 dataset for seven diseases are described. The data from ISIC 2018 used in this study may be obtained [
33].
3.1.2. PH2 Dataset
The proposed systems were evaluated on the PH2 dataset obtained from the Dermatology Service of Hospital Pedro Hispano (Matosinhos, Portugal). All images were obtained under the same conditions and instrumentation resolution. The dataset consisted of 200 images divided into three diseases: melanocytic nevi (NV; n = 80 images), atypical (n = 80 images), and melanoma (mel; n = 40 images). Where melanocytic nevi are benign tumors and melanoma is a malignant tumor, while atypical have characteristics of benign tumors but may develop into malignant tumors. In
Figure 3, samples from the PH2 dataset are described. The data from the PH2 dataset used in this study may be obtained [
34].
3.2. Pre-Processing
The pre-processing process is the first stage of image processing. In this section, the following information is provided: a description of the filters applied to enhance the images and the hair removal method from the images.
3.2.1. Laplacian and Average Filter Methods
Image enhancement is the first step in image processing. During this process, some noisy features such as hair, air bubbles, skin lines, and reflections due to lighting, etc., in the image are fixed to obtain a more precise image. In this study, Laplacian and average filters were used to remove noise and artifacts, enhance edges, and treat low contrast between lesions and healthy skin. First, an average filter was applied. The image was smoothed by the average filter with a reduction in the differences between adjacent pixels. The filter was applied to image frames of 5 × 5 pixels each time. The process continues until the entire image is covered. Then, the value of each pixel in the image was replaced with an average value based on the adjacent values. In Equation (1), the mechanism of action of the intermediate filter is described.
where
z(
m) is the input,
y(
m − 1) is the previous input, and
M is the number in the average filter.
The Laplacian filter, which is an edge detection filter, was then used. This filter detects the changing areas in the image (e.g., edges of skin lesions). In Equation (2), the general functioning of the Laplacian filter is explained.
where
f is a second-order differential equation and
x,
y represents the coordinates in a 2D matrix.
Finally, the image enhanced by the Laplacian filter is subtracted from the image enhanced by the averaging filter to obtain a more improved image.
3.2.2. Hair Removal Technique
Hair is one of the challenges in diagnosing skin lesions. DullRazor is a pre-processing technology that removes hair from the lesion area. The presence of hair in the lesion area causes confusion for the segmentation methods, as well as the feature extraction algorithm, and the presence of hair causes the feature extraction algorithms to add some features of hair in addition to the features of the lesion; therefore, the resulting features will be inaccurate because they contain the features of both the lesion and the hair. Therefore, the DullRazor technique removes hair before the process of segmentation and extraction of features [
35]. The following three steps were necessary for hair removal:
The location of dark hair is determined by the process of morphological closing of the images of the two datasets that contain hair;
The structure of long or thin hair is checked using bilinear interpolation and substitution of specified pixel values;
The new pixels are then smoothed by a medium filter.
In
Figure 4, a sample of the dataset images containing hair is shown. After applying the Dullrazor tool, the image was processed, and the hair was removed.
3.3. Adopted Region Growth Algorithm (Segmentation)
Dermatoscopy images consisted of an affected portion (skin lesion) and a healthy portion. Therefore, extracting features from the entire image, including healthy skin, leads to incorrect classification results [
36]. Consequently, it is necessary to isolate the lesion region from normal skin. In this study, we used the adopted region growth algorithm. Groups of similar pixels were treated with this algorithm. The following conditions are needed for the successful segmentation process by the algorithm:
First, the segmentation process needs to be completed. Second, similar pixel units must be separated into different groups, and the union of all groups represents the whole image. Third, similar pixel units must be corrected. Fourth, no two pixels should be the same and belong to two different regions. The algorithm works on a bottom-up principle, where it starts from pixels and grows to form regions. Each region contains similar pixels. The basic idea is that the algorithm begins each region with a single-pixel seed. Then, each region grows with similar pixels, and the border regions grow with similar units to represent the boundaries of the lesion and separate it from healthy skin. In
Figure 5 and
Figure 6, samples from the ISIC 2018 and PH2 dataset are described, respectively. In these figures, the process is shown after the optimization, hair removal, segmentation process by isolating the lesion region from normal skin, and morphology method to further enhance the images, and the gaps of which were filled after the segmentation process.
3.4. Feature Extraction
In this study, we combined three feature extraction methods, the LBP, GLCM, and DWT algorithms, to extract the most critical features of skin lesions from the images. The LBP algorithm is one of the simplest and most effective feature extraction algorithms. The central (target) pixel was determined using the algorithm. Then, a frame of 3 × 3 neighboring pixels was selected for each central pixel, known as the parameter R, representing the radius. This parameter was responsible for determining the number of adjacent pixels. Two-dimensional textures were described using the LBP algorithm [
37]. In Equation (3), the decomposition of the center pixel by adjacent pixels is described, and the substitution of the resulting value of the center pixel. After this first step, the method continues for all pixels of the image. A total of 203 features were extracted for each image by the LBP algorithm.
where
gc is the center pixel,
gp is the neighboring pixel,
R is the radius around the central pixel, and
P is the number of neighbors. The binary threshold function
x is defined in Equation (4) as follows:
The internal structure of the lesion area was displayed in gray levels using the GLCM algorithm. Then, the algorithm extracted texture features from an area of interest. Since the lesion area had a smooth and rough texture, the smooth area had pixel values close to each other, while the rough area had different pixels. Then, texture features from the spatial gray levels of a lesion were extracted using the algorithm. Textile metrics were calculated from spatial and statistical information. The location of a pixel from another pixel was determined by the spatial information in terms of a distance d and a direction θ. There were four values of θ: 0°, 45°, 90°, and 135°. Additionally, d = 1 when θ is horizontal or vertical (θ = 0° or θ = 90°), and d = √2 when θ (θ = 45° or θ = 135°). A total of 13 features was extracted for each image: contrast, energy, mean, entropy, correlation, kurtosis, standard deviation, smoothness, homogeneity, RMS, skewness, and variance.
Four features from each image were extracted using the DWT method, in which the input signal was analyzed into two signals with different frequencies using square mirror filters. These two signals were compatible with low-and high-pass filters. Approximation coefficients were produced by low-pass (LL) filters, while detailed coefficients (horizontal, vertical, and diagonal) were produced by high-pass filters (LH, HL, and HH). A total of 220 hybrid features were extracted using all the algorithms (LBP, GLCM and DWT). Such features were combined in one vector for each image. The produced vector was fed to the classification stage to train the classifier. In
Figure 7, the hybrid process for feature extraction is described.
3.5. Classification
In this section, the ISIC 2018 and PH2 datasets were evaluated according to two traditional classification algorithms (e.g., ANN and FFNN) and convolutional neural networks (CNNs) (e.g., ResNet-50 and AlexNet) models for diagnosing skin diseases.
3.5.1. ANN and FFNN
ANN is a type of neural network of soft computing. It is a group of layers consisting of interconnected and internally connected neurons. It has a superior ability to interpret and analyze complex data and to produce clear and explanatory patterns. The error between actual and predicted probabilities is also minimized by ANN [
38]. Information is propagated between neurons and stored as connecting points between them called weights. The objective of the ANN is to update the specified weights w to obtain the minimum square error between the actual output
x and predicted output
y as given by the mean square error (MSE), as described in Equation (5):
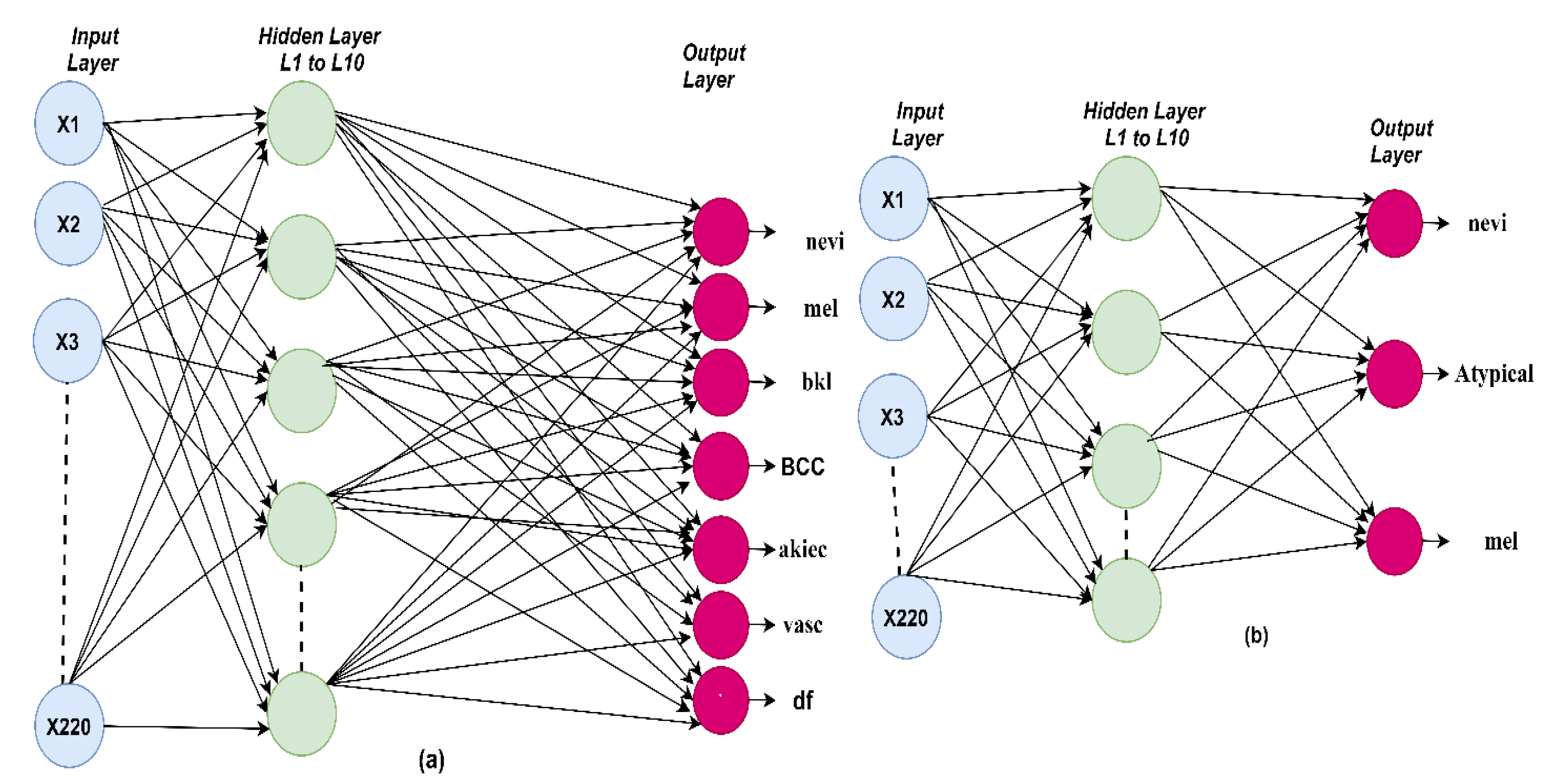
The ANN algorithm was evaluated on the ISIC 2018 and PH2 datasets for diagnosing skin diseases. A model was trained on 220 features through 10 hidden layers between the input and output layers. In
Figure 8a, the architecture of the ANN algorithm is shown for the ISIC 2018 dataset. Seven classes were produced. In
Figure 8b, the architecture of the ANN algorithm is shown for the PH2 dataset, in which three classes were produced.
The FFNN algorithm is similar to the ANN algorithm in solving complex computational problems. The hidden layer neurons are interconnected by w weights. The algorithm works, and information between neurons is fed in the forward direction. The results of each neuron were obtained based on the weight associated with it multiplied by the output of the previous neuron [
39]. The weights were updated in the forward direction from the hidden layer to the output layer. In each iteration, the weights were updated until the minimum squared error was obtained between the expected and actual output. The criteria were used to select the algorithms ANN and FFNN. It is known that these algorithms are among the best machine learning algorithms and are distinguished from the rest of the machine learning algorithms by several criteria such as: (1) they contain many layers such as the input layer to receive the features (220 features in this study) and many layers of hidden (10 hidden layers in this study) and output layers (seven neurons in the case of the ISIC 2018 dataset or three neurons in the case of the PH2 dataset). (2) Interconnected neurons. (3) The weights that connect each neuron with the other neurons. (4) Mean square error compares the actual and predicted output and repeatedly works until the lowest ratio between the predicted and actual output is obtained by changing the weights frequently [
40].
3.5.2. Convolutional Neural Networks (CNNs)
CNNs are deep learning methods used in many areas, including signal processing and image processing, to recognize patterns, classify objects, and detect regions of interest [
41]. In this study, the two datasets, ISIC 2018 and PH2, were evaluated on ResNet-50 and AlexNet for diagnosing skin diseases. Several CNN structures for diagnosing skin lesions have been established, including several layers, training steps, activation functions, and learning rates. The most important layers in a CNN are the convolutional layers, max, average pooling layers, the fully connected layer, and activation functions [
42].
When the image was inputted into the CNN structure, the image was represented as image height × image width. After the image passed through the convolutional layers, the feature map contained the feature depth, represented as image height × image width × image depth. Filter size, step, and zero padding were the most critical parameters of the convolutional layers that affected the performance of the convolutional layers. Convolutional layers wrap with the filter size around the image, learn the weights during the training phase, process the input, and pass it to the next layer.
Zero padding was the process of filling neurons with zeros to maintain the size of the resulting neurons. When zero padding was one, the neurons were padded with a row and a column around the edges. The output in each neuron was input to the next neuron. This output was calculated according to Equation (6) as follows:
where
W represents the volume of the input neuron,
K represents the filter volume in a convolutional layer,
P represents the volume of the input padding, and
S represents the step. Rectified linear unit (ReLU) layers were also used after convolutional layers for image processing. The purpose of ReLU was to pass the positive output, suppress the negative output, and convert it to zero [
43]. Equation (7) showns how a ReLU layer works.
The dimensions were reduced by the pooling layer, as the dimensions of the image were reduced by grouping many neurons and representing them in one neuron according to the maximum or average method, which is called the max-pooling layer or average pooling layer. The maximum value of the groups of neurons was selected using the maximum method, and the average value of the neurons was chosen using the average method. CNNs have millions of parameters, and there was an overfitting problem. Therefore, overfitting was prevented by the dropout layer by stopping 50% of the neurons, while the remaining neurons were turned on in each iteration. However, the training time was doubled by this technique due to repetition by an amount of two. In the fully connected layers, the last layer of convolutional neural networks, each neuron was connected to all neurons. Feature maps were converted to flat representations (unidirectional). Each image was diagnosed by a fully connected layer in its related class. Thus, the network takes a long time during the training and testing phases, and many fully connected layers can be used in the same network.
Softmax is the activation function used in the last stage of the convolutional neural network model. It is nonlinear and is used by multiple classes. In Equation (8), the functioning of the softmax function is described. Respectively, seven and three classes were produced for both the ISIC 2018 and PH2 datasets by the softmax function.
where
y is the output of softmax, and n is the output total number. Two CNN models of transfer learning, the ResNet-50 and AlexNet models, were implemented in this work.
ResNet50 Model
The ResNet-50 model contained 16 blocks with 177 layers divided into 49 convolutional layers, ReLU, batch normalization, one max-pooling layer, one average-pooling layer, one fully connected layer, and the softmax function. Seven and three classes were produced by the softmax function for the ISIC 2018 and PH2 datasets, respectively. ResNet-50 also contains 23.9 million parameters [
44].
Figure 9 describes the basic architecture of ResNet-50 for diagnosing the ISIC 2018 dataset.
Table 1 describes the number of layers, the size of each filter, and the parameters of the ResNet-50 model.
AlexNet Model
The AlexNet model contained 25 layers divided into five convolutional layers, three max-pooling layers, three fully connected layers, a classification-output layer, two leaking layers, and a softmax activation function. Seven and three classes were produced for the ISIC 2018 and PH2 datasets, respectively [
45]. AlexNet contained 62 million parameters, 630 million connections, and 650,000 neurons. In
Figure 10, the basic architecture of AlexNet for diagnosing the ISIC 2018 dataset is described. The number of layers, the size of each filter, and the parameters of the AlexNet model are described in
Table 2.
CNNs have many layers to extract feature maps from the input images. The transfer learning technique was applied with the aim of transferring the experience gained from pre-training to perform new tasks on the ISIC 2018 and PH2 data sets. The knowledge gained when training convolutional neural networks were stored with this technology for more than a million images to obtain more than a thousand classes. Learning is transferred to solve new problems related to the classification of skin diseases. We aim to use CNNs for diagnosing skin diseases by comparing the results with traditional neural networks ANN and FFNN.
6. Conclusions
Skin diseases are spreading nowadays in many countries due to long-term exposure to the sun and weather changes. Many skin diseases must be diagnosed and treated early to avoid severe consequences for the health of affected individuals. Melanoma (skin cancer) is considered one of the most dangerous types of skin disease, and it must be diagnosed before it penetrates the internal tissues of the skin and spreads from one place to another in the body. In this work, we developed diagnostic systems based on artificial intelligence to diagnose the images of two standard datasets, ISIC 2018 and PH2, for the early detection of skin diseases. The images in the two data sets were divided into 80% for training and validation (80% and 20%, respectively) and 20% for testing. In the first step of the proposed early detection using these proposed systems, ANN and FFNN algorithms were implemented to diagnose the features extracted by hybrid methods (e.g., LBP, GLCM, and DWT). The features of the three methods were combined and collected in a features matrix so that each vector (image) contained 220 essential features representing the disease types. In the second step, CNNs models were implemented, ResNet-50 and AlexNet, based on transfer learning. The results obtained with traditional neural networks (ANN and FFNN) were compared with CNN networks (ResNet-50 and AlexNet). It was noted that ANN and FFNN algorithms performed better than two CNN models, ResNet-50 and AlexNet. Despite applying many optimization techniques and extracting the features by hybrid methods between three algorithms, there are some limitations and challenges encountered in the study, which are represented in the significant similarity between the features of some diseases, which causes confusion for the classification algorithms when making a diagnosis. Solving these limitations in the future will require extracting features from various algorithms using traditional methods and combining them with deep feature maps extracted by CNN models, as well as applying hybrid methods between machine learning algorithms and deep learning models by using two blocks. In the first block, the deep features are extracted by CNN models. The second block is one of the machine learning algorithms that is fed with the output of the first block for classifying dermatology.