Small-Scale Depthwise Separable Convolutional Neural Networks for Bacteria Classification
Abstract
:1. Introduction
- The DS-CNN was exploited to construct a compact network architecture for the automated recognition and classification of 33 bacteria species in the DIBaS dataset with reliable accuracy and less time consumption;
- As part of our methodology, we incorporated preprocessing and data augmentation strategies to improve the model’s input quality and achieve higher classification accuracy.
2. Related Works
3. Overview of the Depthwise Separable Convolutional Neural Network
3.1. DS-CNN Layer Primer
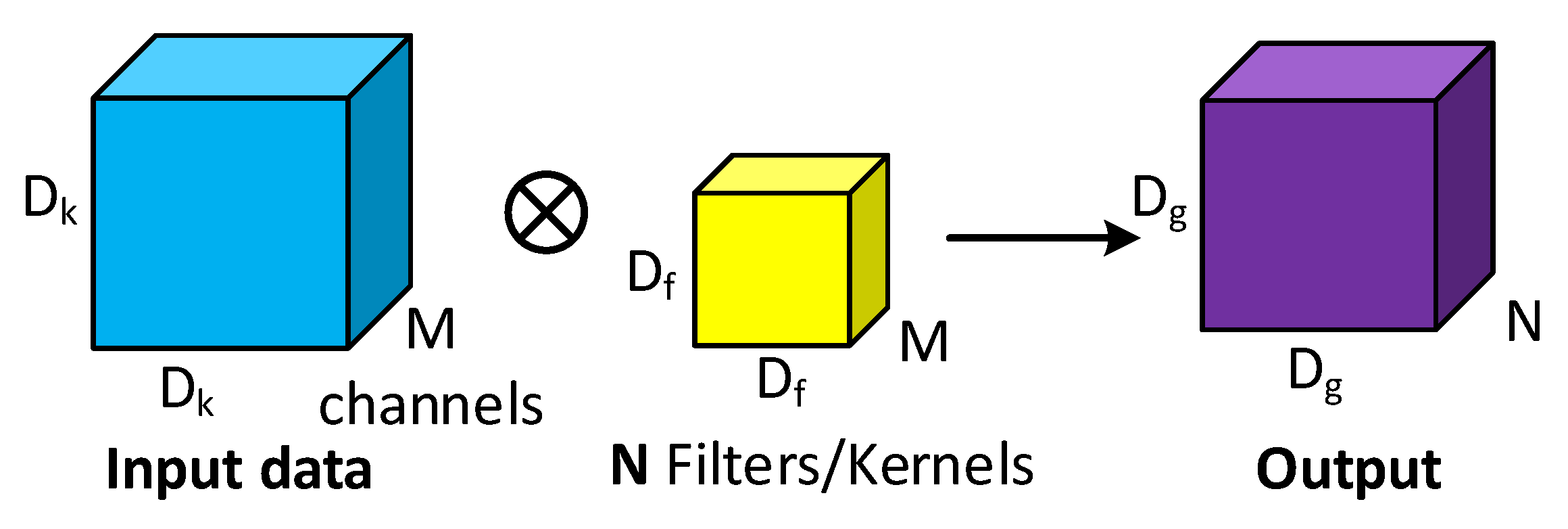
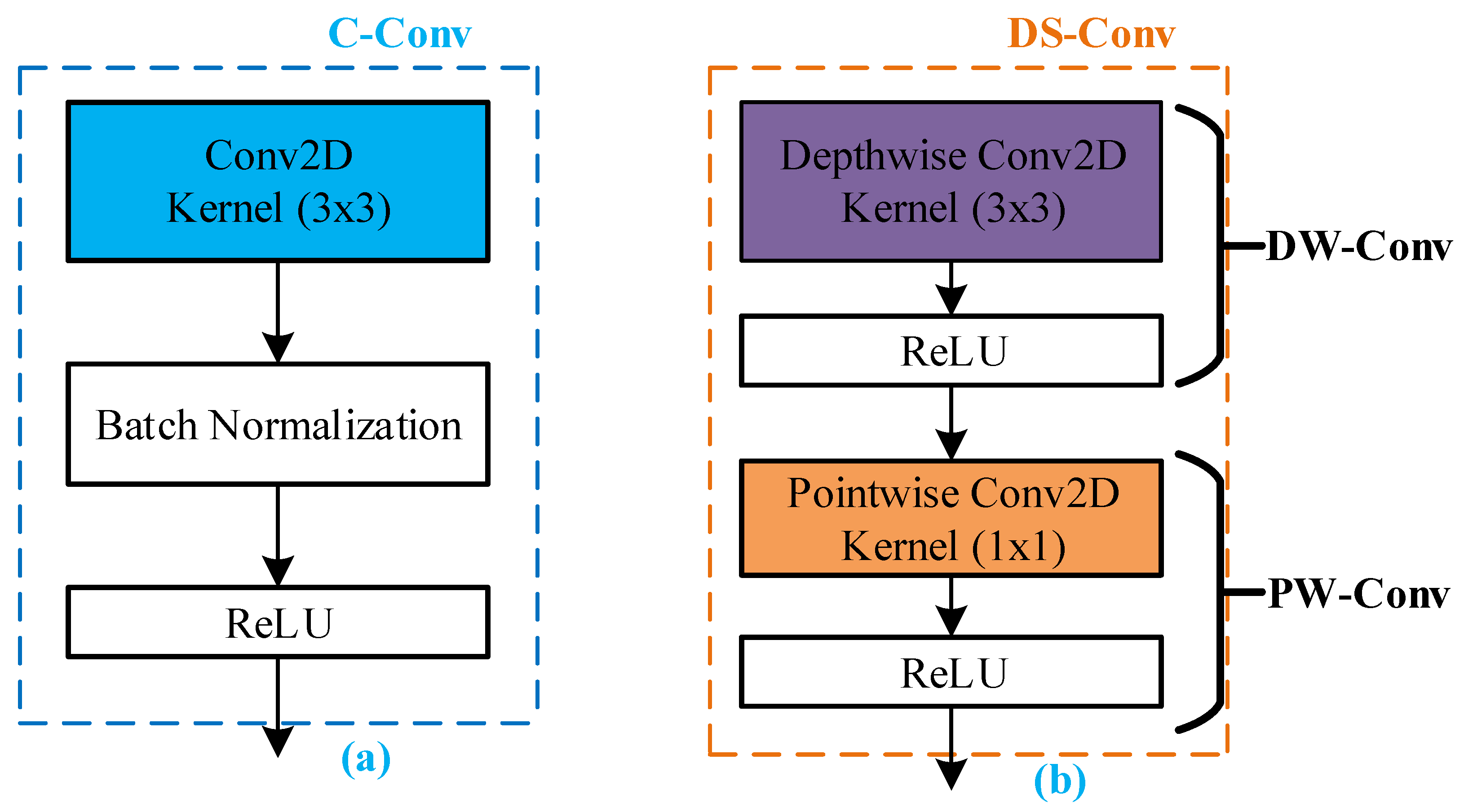
3.1.1. Conventional Convolution Block
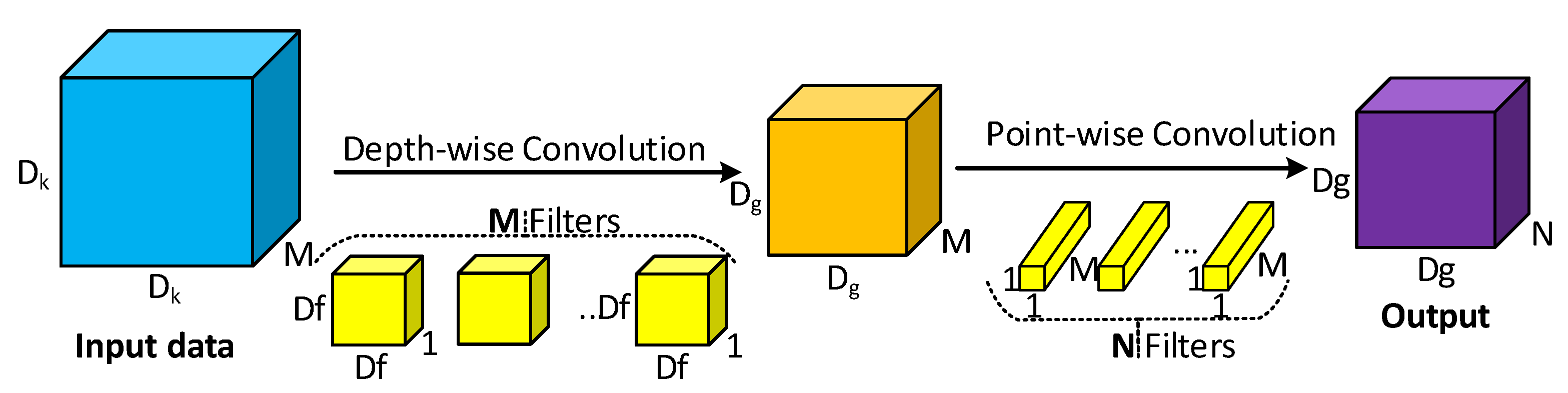
3.1.2. Depthwise Separable Convolution
3.1.3. Activation Functions
- Sigmoid function:
- Rectified linear unit (ReLU):where x is the input to the neuron.
3.1.4. Batch Normalization
3.1.5. Pooling Layer
3.1.6. Fully Connected Layer
3.1.7. Dropout
3.1.8. Classifier Layers
3.1.9. Learning Rate and Optimizers
3.2. The Proposed Architecture
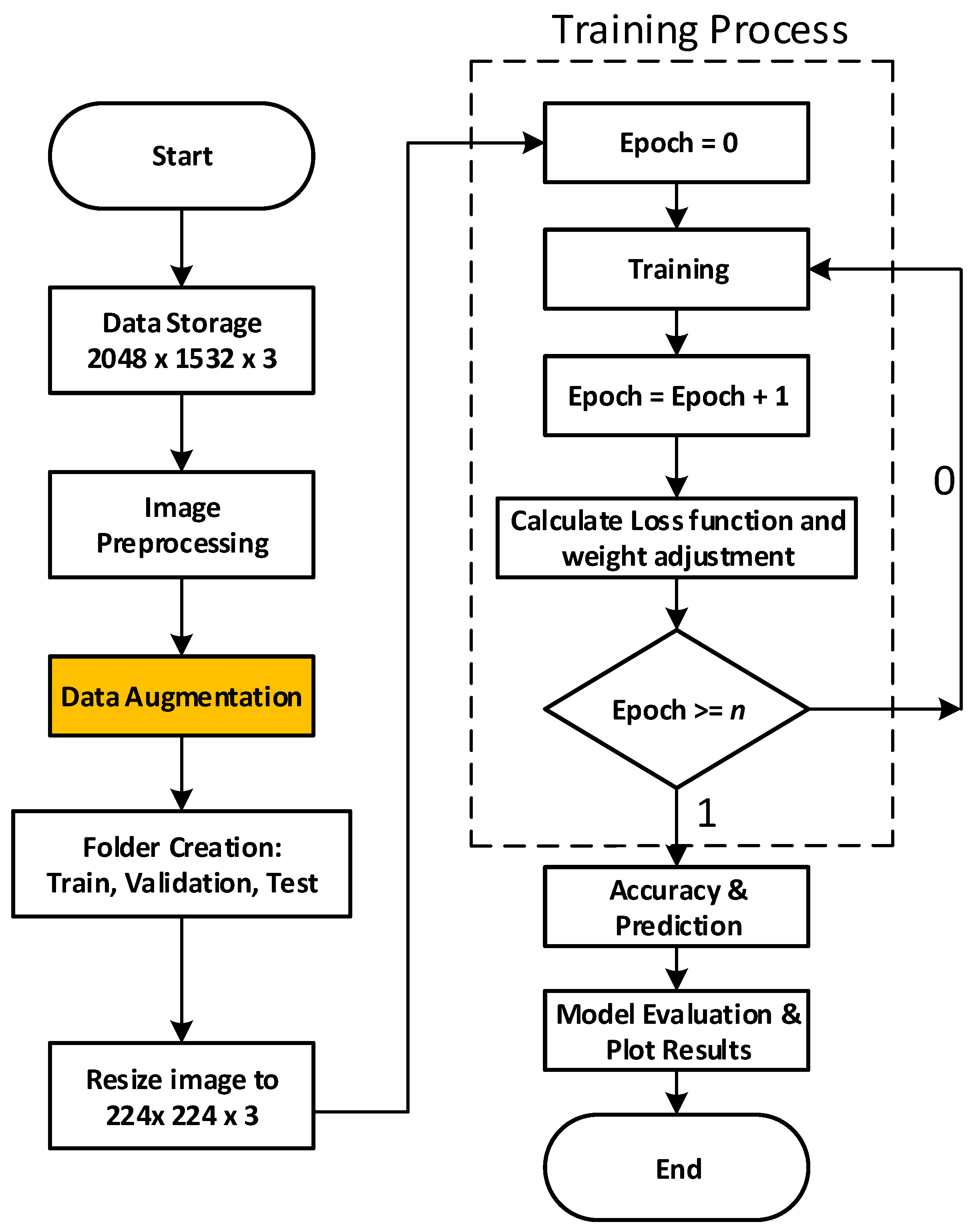
4. Materials and Methods
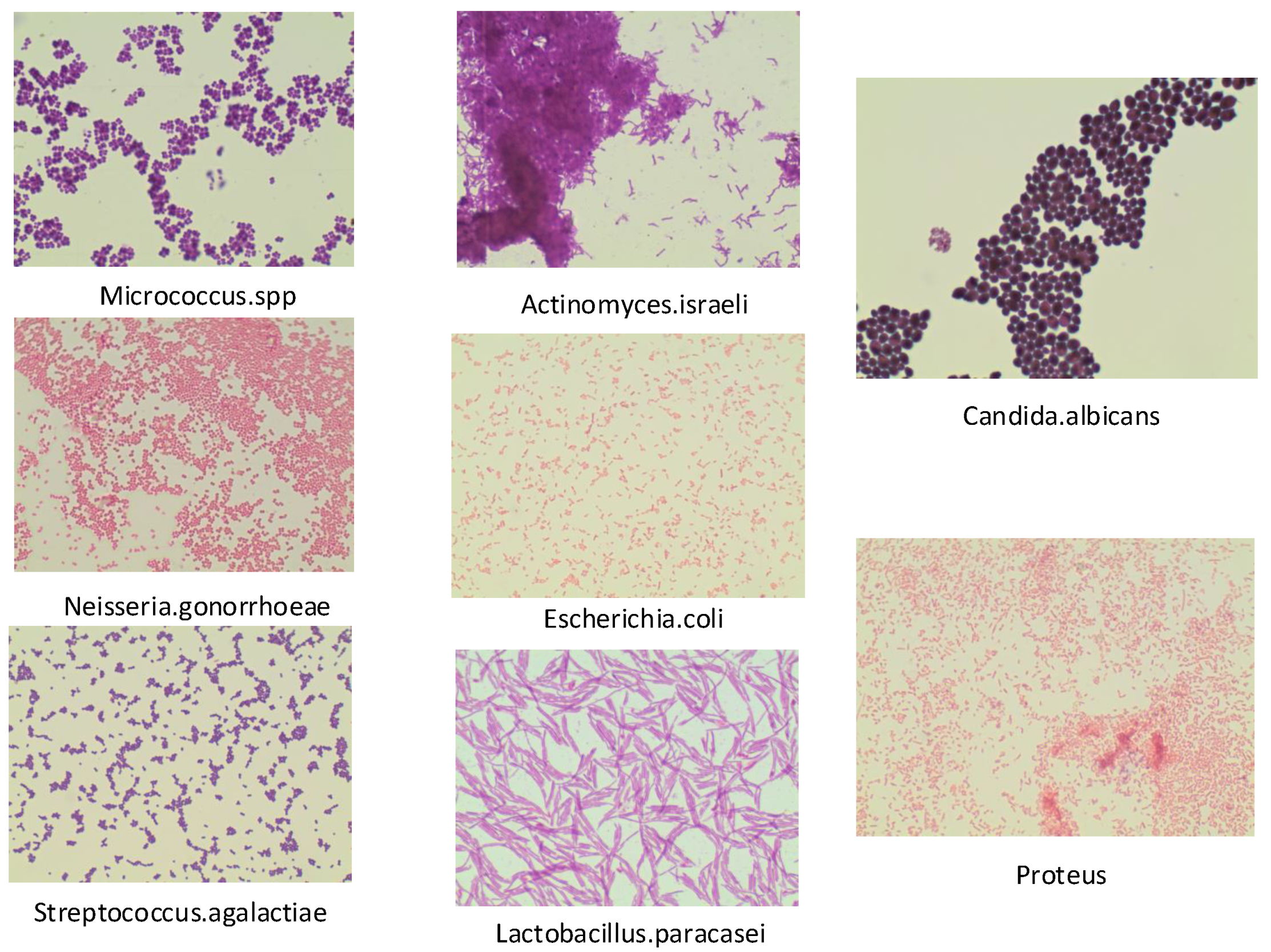
4.1. Dataset
4.2. Dataset Augmentation
- is a value in the range of to within which to rotate pictures randomly; was the random value selected;
- , and are thresholds (as a fraction of total width or height) within which to randomly shift images vertically or horizontally;
- is for randomly employing shearing transformations. It is 0.2;
- is for randomly zooming the picture sizes;
- is for randomly horizontally reversing the pixels of the image;
- is the strategy used for filling in newly created pixels, which can appear after a rotation or a width/height shift.
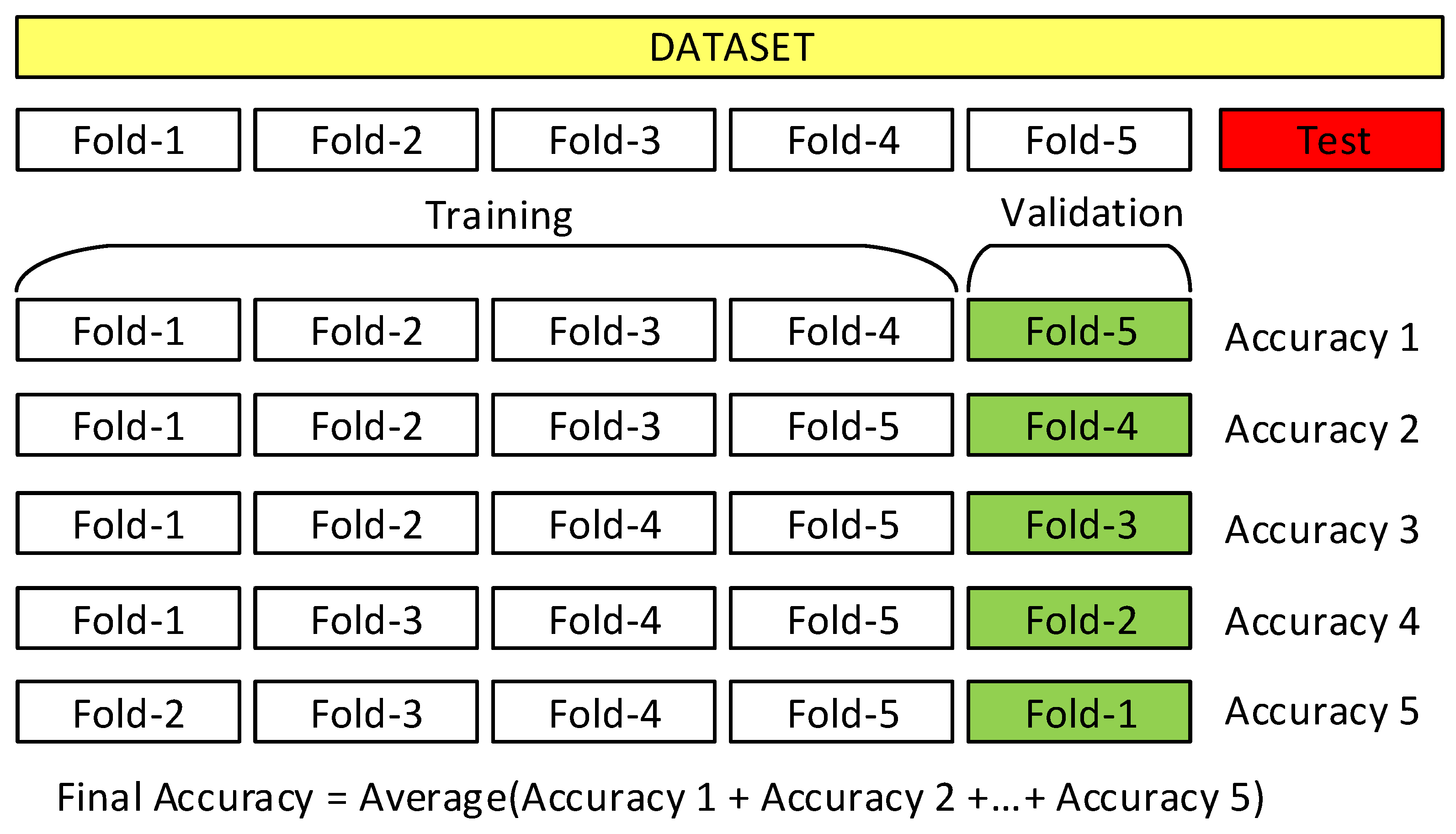
5. Experimental Setups
5.1. Training Strategies
5.2. Parameter Selection
6. Results and Discussion
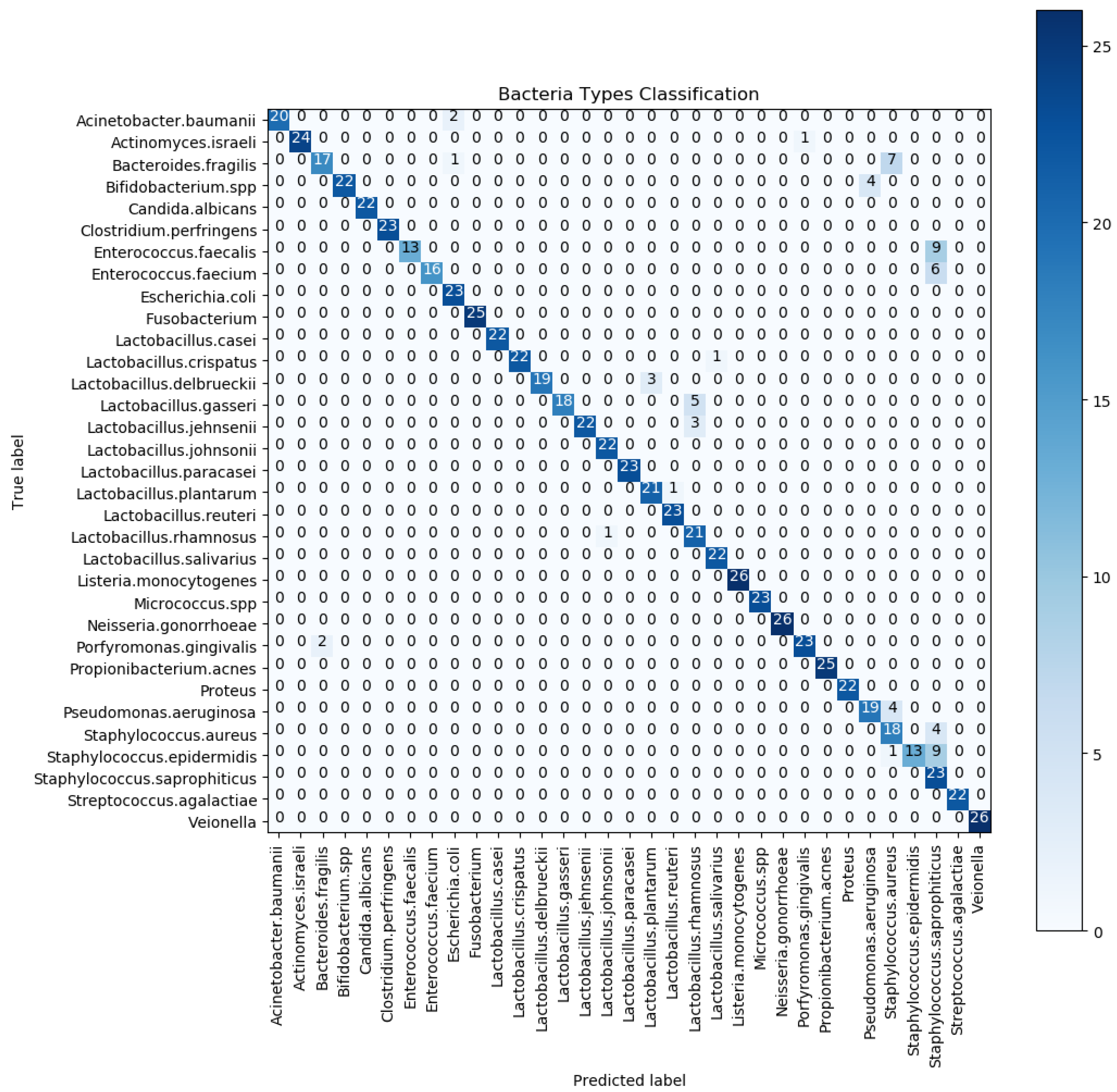
6.1. Statistical Analysis
- Accuracy is the most straightforward metric that the model evaluation process requires to quantify a model’s performance. Accuracy is the fraction of correct predictions and the overall number of forecasts. The formula for calculating accuracy is written by:
- Precision is an evaluation metric that describes a fraction of the true positive prediction and the total number of positive predictions. Precision refers to the frequency with which we are correct when the predicted value is positive:
- Sensitivity is the ratio of positive predictions to the total actual number of positives. Sensitivity is also referred to as the recall or true positive rate. Sensitivity means how often the forecast is correct when the real value is positive.
- Specificity is calculated by dividing the total number of negative predictions by the total number of actual negatives. Specificity is also understood as the true negative rate. The term “specificity” refers to the frequency with which a prediction is correct when the actual value is negative.
- The -score, alternatively called the balanced F-score or F-measure, can be calculated as a weighted average of the precision and sensitivity:
6.2. Comparison with Other Studies
7. Conclusions and Future Work
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| AUC | Area under the curve |
| BN | Batch normalization |
| C-CNNs | Conventional CNNs |
| CM | Confusion matrix |
| CV | Computer vision |
| DS-CNNs | Depthwise separable CNNs |
| DCNNs | Deep convolutional neural networks |
| DIBaS | Digital Images of Bacteria Species |
| FC | Fully connected |
| LSTM | Long short-term memory |
| MAC | Multiply–accumulate |
| GPU | Graphics processing unit |
| PR Curve | Precision-recall curve |
| ReLU | Rectified linear unit |
| RF | Random forest |
| ROC | Receiver operating characteristic |
References
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Simonyan, K.; Zisserman, A. Two-Stream Convolutional Networks for Action Recognition in Videos. arXiv 2014, arXiv:1406.2199. [Google Scholar]
- Collobert, R.; Weston, J.; Bottou, L.; Karlen, M.; Kavukcuoglu, K.; Kuksa, P. Natural Language Processing (Almost) from Scratch. J. Mach. Learn. Res. 2011, 12, 2493–2537. [Google Scholar]
- Zhang, T.; Kahn, G.; Levine, S.; Abbeel, P. Learning deep control policies for autonomous aerial vehicles with MPC-guided policy search. In Proceedings of the 2016 IEEE International Conference on Robotics and Automation (ICRA), Stockholm, Sweden, 16–21 June 2016; IEEE: Stockholm, Sweden, 2016; pp. 528–535. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Troyanskaya, O.G. Predicting effects of noncoding variants with deep learning–based sequence model. Nat. Methods 2015, 12, 931–934. [Google Scholar] [CrossRef] [Green Version]
- Alipanahi, B.; Delong, A.; Weirauch, M.T.; Frey, B.J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015, 33, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, B.; Shen, C.; Tan, M.; Liu, L.; Reid, I. Structured Binary Neural Networks for Accurate Image Classification and Semantic Segmentation. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 16–20 June 2019; pp. 413–422. [Google Scholar] [CrossRef] [Green Version]
- He, T.; Zhang, Z.; Zhang, H.; Zhang, Z.; Xie, J.; Li, M. Bag of Tricks for Image Classification with Convolutional Neural Networks. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 16–20 June 2019; pp. 558–567. [Google Scholar] [CrossRef] [Green Version]
- Nuriel, O.; Benaim, S.; Wolf, L. Permuted AdaIN: Reducing the Bias towards Global Statistics in Image Classification. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 19–25 June 2021. [Google Scholar]
- Nie, D.; Shank, E.A.; Jojic, V. A deep framework for bacterial image segmentation and classification. In Proceedings of the 6th ACM Conference on Bioinformatics, Computational Biology and Health Informatics, Atlanta, Georgia, 9–12 September 2015; pp. 306–314. [Google Scholar] [CrossRef]
- Ates, H.; Gerek, O.N. An image-processing based automated bacteria colony counter. In Proceedings of the 2009 24th International Symposium on Computer and Information Sciences, Guzelyurt, Cyprus, 14–16 September 2009; pp. 18–23. [Google Scholar] [CrossRef]
- Divya, S.; Dhivya, A. Human Eye Pupil Detection Technique Using Circular Hough Transform. Int. J. Adv. Res. Innov. 2019, 7, 3. [Google Scholar]
- Limare, N.; Lisani, J.L.; Morel, J.M.; Petro, A.B.; Sbert, C. Simplest Color Balance. Image Process. Line 2011, 1, 297–315. [Google Scholar] [CrossRef] [Green Version]
- Ganesan, P.; Sajiv, G. A comprehensive study of edge detection for image processing applications. In Proceedings of the 2017 International Conference on Innovations in Information, Embedded and Communication Systems (ICIIECS), Coimbatore, India, 17–18 March 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Xuan, L.; Hong, Z. An improved canny edge detection algorithm. In Proceedings of the 2017 8th IEEE International Conference on Software Engineering and Service Science (ICSESS), Beijing, China, 24–26 November 2017; pp. 275–278. [Google Scholar] [CrossRef]
- Hiremath, P.S.; Bannigidad, P. Automatic Classification of Bacterial Cells in Digital Microscopic Images. In Proceedings of the Second International Conference on Digital Image Processing, Singapore, 26–28 February 2010; 2010; p. 754613. [Google Scholar] [CrossRef]
- Ho, C.S.; Jean, N.; Hogan, C.A.; Blackmon, L.; Jeffrey, S.S.; Holodniy, M.; Banaei, N.; Saleh, A.A.E.; Ermon, S.; Dionne, J. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun. 2019, 10, 4927. [Google Scholar] [CrossRef]
- Kang, R.; Park, B.; Eady, M.; Ouyang, Q.; Chen, K. Single-cell classification of foodborne pathogens using hyperspectral microscope imaging coupled with deep learning frameworks. Sens. Actuators B Chem. 2020, 309, 127789. [Google Scholar] [CrossRef]
- Sajedi, H.; Mohammadipanah, F.; Pashaei, A. Image-processing based taxonomy analysis of bacterial macromorphology using machine-learning models. Multimed. Tools Appl. 2020, 79, 32711–32730. [Google Scholar] [CrossRef]
- Tamiev, D.; Furman, P.E.; Reuel, N.F. Automated classification of bacterial cell sub-populations with convolutional neural networks. PLoS ONE 2020, 15, e0241200. [Google Scholar] [CrossRef]
- Mhathesh, T.S.R.; Andrew, J.; Martin Sagayam, K.; Henesey, L. A 3D Convolutional Neural Network for Bacterial Image Classification. In Intelligence in Big Data Technologies—Beyond the Hype; Peter, J.D., Fernandes, S.L., Alavi, A.H., Eds.; Springer: Singapore, 2021; Volume 1167, pp. 419–431. [Google Scholar] [CrossRef]
- Korzeniewska, E.; Szczęsny, A.; Lipiński, P.; Dróżdż, T.; Kiełbasa, P.; Miernik, A. Prototype of a Textronic Sensor Created with a Physical Vacuum Deposition Process for Staphylococcus aureus Detection. Sensors 2020, 21, 183. [Google Scholar] [CrossRef]
- Zieliński, B.; Plichta, A.; Misztal, K.; Spurek, P.; Brzychczy-Włoch, M.; Ochońska, D. Deep learning approach to bacterial colony classification. PLoS ONE 2017, 12, e0184554. [Google Scholar] [CrossRef] [Green Version]
- Nasip, O.F.; Zengin, K. Deep Learning Based Bacteria Classification. In Proceedings of the 2018 2nd International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT), Ankara, Turkey, 19–21 October 2018; pp. 1–5. [Google Scholar] [CrossRef]
- Talo, M. An Automated Deep Learning Appoach for Bacterial Image Classification. In Proceedings of the International Conference on Advanced Technologies, Computer Engineering and Science (ICATCES), Karabuk, Turkey, 11–13 May 2019. [Google Scholar]
- Transfer Learning, CS231n Convolutional Neural Networks for Visual Recognition. Available online: https://cs231n.github.io/transfer-learning/ (accessed on 24 November 2021).
- Khalifa, N.E.M.; Taha, M.H.N.; Hassanien, A.E. Deep bacteria: Robust deep learning data augmentation design for limited bacterial colony dataset. Int. J. Reason.-Based Intell. Syst. 2019, 11, 9. [Google Scholar] [CrossRef]
- Plichta, A. Recognition of species and genera of bacteria by means of the product of weights of the classifiers. Int. J. Appl. Math. Comput. Sci. 2020. [Google Scholar] [CrossRef]
- Plichta, A. Methods of Classification of the Genera and Species of Bacteria Using Decision Tree. J. Telecommun. Inf. Technol. 2020, 4, 74–82. [Google Scholar] [CrossRef]
- Patel, S. Bacterial Colony Classification Using Atrous Convolution with Transfer Learning. Ann. Rom. Soc. Cell Biol. 2021, 25, 1428–1441. [Google Scholar]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef] [Green Version]
- Sifre, L. Rigid-Motion Scattering for Image Classification. Ph.D. Thesis, Ecole Polytechnique, CMAP, Palaiseau, France, 2014. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 22–25 July 2017; pp. 1800–1807. [Google Scholar] [CrossRef] [Green Version]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the 32nd International Conference on International Conference on Machine Learning, Lille, France, 6–11 July 2015; Volume 37, pp. 448–456. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- DIBaS—Krzysztof Paweł Misztal. Available online: http://misztal.edu.pl/software/databases/dibas/ (accessed on 24 November 2021).
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. arXiv 2015, arXiv:1409.0575. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Patras, I. Mirror, mirror on the wall, tell me, is the error small? In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 4685–4693. [Google Scholar] [CrossRef] [Green Version]
- Xie, S.; Tu, Z. Holistically-Nested Edge Detection. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 1395–1403. [Google Scholar] [CrossRef] [Green Version]
- Salamon, J.; Bello, J.P. Deep Convolutional Neural Networks and Data Augmentation for Environmental Sound Classification. IEEE Signal Process. Lett. 2017, 24, 279–283. [Google Scholar] [CrossRef]
- Eigen, D.; Fergus, R. Predicting Depth, Surface Normals and Semantic Labels with a Common Multi-Scale Convolutional Architecture. arXiv 2015, arXiv:1411.4734. [Google Scholar]
- Tutorials|TensorFlow Core. Available online: https://www.tensorflow.org/tutorials (accessed on 24 November 2021).
- Keras documentation: Keras API reference. Available online: https://keras.io/api/ (accessed on 24 November 2021).
- Le, N.Q.K.; Kha, Q.H.; Nguyen, V.H.; Chen, Y.C.; Cheng, S.J.; Chen, C.Y. Machine Learning-Based Radiomics Signatures for EGFR and KRAS Mutations Prediction in Non-Small-Cell Lung Cancer. Int. J. Mol. Sci. 2021, 22, 9254. [Google Scholar] [CrossRef]
- Le, N.Q.K.; Hung, T.N.K.; Do, D.T.; Lam, L.H.T.; Dang, L.H.; Huynh, T.T. Radiomics-based machine learning model for efficiently classifying transcriptome subtypes in glioblastoma patients from MRI. Comput. Biol. Med. 2021, 132, 104320. [Google Scholar] [CrossRef] [PubMed]
- Schneider, P.; Müller, D.; Kramer, F. Classification of Viral Pneumonia X-ray Images with the Aucmedi Framework. arXiv 2021, arXiv:2110.01017. [Google Scholar]
- Xie, Z.; Deng, X.; Shu, K. Prediction of Protein–Protein Interaction Sites Using Convolutional Neural Network and Improved Data Sets. Int. J. Mol. Sci. 2020, 21, 467. [Google Scholar] [CrossRef] [Green Version]
- Pei, Y.; Huang, Y.; Zou, Q.; Zhang, X.; Wang, S. Effects of Image Degradation and Degradation Removal to CNN-Based Image Classification. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 1239–1253. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Xie, D.; Zhang, Y.; Pu, S. All You Need Is a Few Shifts: Designing Efficient Convolutional Neural Networks for Image Classification. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 7234–7243. [Google Scholar] [CrossRef] [Green Version]


| Layer | Type | Filter Size | Output Shape | Parameters (M) | MACs (M) |
|---|---|---|---|---|---|
| Input | 224 × 224 × 3 | ||||
| Conv1 | Conv_1/Stride 2 | 3 × 3 × 3 × 64 | 112 × 112 × 64 | 0.002 | 21.68 |
| Batch Norm | - | 112 × 112 × 64 | 0.0003 | 0 | |
| Max-Pooling_1 | Pool 2 × 2 | 56 × 56 × 64 | 0 | 0 | |
| Conv2 | DW-Conv_2 | 3 × 3 × 1 × 64 | 56 × 56 × 64 | 0.0006 | 1.80 |
| PW-Conv_2 | 1 × 1 × 64 × 64 | 0.004 | 12.85 | ||
| Max-Pooling_2 | Pool 2 × 2 | 28 × 28 × 64 | 0 | 0 | |
| Conv3 | DW-Conv_3 | 3 × 3 × 1 × 64 | 28 × 28 × 84 | 0.0006 | 0.45 |
| PW-Conv_3 | 1 × 1 × 64 × 64 | 0.004 | 3.21 | ||
| Max Pooling_3 | Pool 2 × 2 | 14 × 14 × 64 | 0 | 0 | |
| FC | Fully Connected_1 | 256 | 14 × 14 × 256 | 3.21 | 0.03 |
| Dropout | dropout_1 | 0.25 | 0 | ||
| Classifier | Softmax | 33 | 256 × 33 | 0.008 | |
| Total | 3.23 | 40.02 | |||
| Models | Epochs = n | Dropout | Activations Functions | Optimizers | Batch Size |
|---|---|---|---|---|---|
| This work | 30–50 | 0.25–0.5 | ReLU | Adam | 16;32 |
| 30–50 | 0.25–0.5 | ReLU | RMSprop | 16;32 | |
| 30–50 | 0.25–0.5 | ReLU | Adamax | 16;32 | |
| 30–50 | 0.25–0.5 | ReLU | Nadam | 16;32 |
| Class | Classified Positive | Classified Negative |
|---|---|---|
| Positive | TP (True Positive) | FN (False Negative) |
| Negative | FP (False Positive) | TN (True Negative) |
| Models | Data Aug | Optimizers | Accuracy | Precision | Sensitivity | F1 |
|---|---|---|---|---|---|---|
| This work | Yes | Adam | 95.01 | 94.02 | 90.78 | 92.37 |
| Adamax | 96.08 | 94.27 | 93.22 | 93.74 | ||
| Nadam | 96.28 | 95.81 | 93.26 | 94.52 | ||
| RMSprop | 95.17 | 95.44 | 94.84 | 95.14 | ||
| This work | No | Adam | 86.24 | 91.11 | 81.63 | 86.11 |
| Adamax | 86.42 | 88.99 | 83.17 | 85.98 | ||
| Nadam | 83.95 | 85.69 | 79.26 | 82.35 | ||
| RMSprop | 81.48 | 83.09 | 75.66 | 79.21 |
| Works | Zielinski et al. [24] | Nasip et al. [25] | Khalifa et al. [28] | M. Talo [26] | S. Patel [31] | This Work |
|---|---|---|---|---|---|---|
| Year | 2017 | 2018 | 2019 | 2019 | 2021 | 2021 |
| CNN Algorithms | FV, SVM, RF CNNs | VGG-16 AlexNet | AlexNet | ResNet-50 | VGG-16 | DS-CNN |
| Layers | 8/8/16 | 16/8 | 8 | 50 | 16 | 5 |
| Data Augmentation | No | Yes | Yes | No | No | Yes |
| Transfer Learning | Yes | Yes | Yes | Yes | Yes | No |
| Number of Images | 660 | 35,600 | 6600 | 689 | 660 | 6600 |
| Image Dimensions | 224 × 224 × 3 227 × 227 × 3 | 224 × 224 × 3 227 × 227 × 3 | 227 × 227 × 3 | 224 × 224 × 3 | 224 × 224 × 3 | 224 × 224 × 3 |
| Parameters (M) | 58.42 98.95 134.4 | 134.4 | 58.42 | 23.69 | 134.4 | 3.23 |
| Accuracy (%) | 97.24 | 98.25 | 98.22 | 99.2 | 93.38 | 96.28 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mai, D.-T.; Ishibashi, K. Small-Scale Depthwise Separable Convolutional Neural Networks for Bacteria Classification. Electronics 2021, 10, 3005. https://doi.org/10.3390/electronics10233005
Mai D-T, Ishibashi K. Small-Scale Depthwise Separable Convolutional Neural Networks for Bacteria Classification. Electronics. 2021; 10(23):3005. https://doi.org/10.3390/electronics10233005
Chicago/Turabian StyleMai, Duc-Tho, and Koichiro Ishibashi. 2021. "Small-Scale Depthwise Separable Convolutional Neural Networks for Bacteria Classification" Electronics 10, no. 23: 3005. https://doi.org/10.3390/electronics10233005
APA StyleMai, D.-T., & Ishibashi, K. (2021). Small-Scale Depthwise Separable Convolutional Neural Networks for Bacteria Classification. Electronics, 10(23), 3005. https://doi.org/10.3390/electronics10233005






