Abstract
The rapid spread of SARS-CoV-2 and the consequent global COVID-19 pandemic has prompted the public administrations of different countries to establish health procedures and protocols based on information generated through predictive techniques and models, which, in turn, are based on technology such as artificial intelligence (AI) and machine learning (ML). This article presents some AI tools and computational models used to collaborate in the control and detection of COVID-19 cases. In addition, the main features of the Epidempredict project regarding COVID-19 in Panama are presented. This initiative consists of the planning and design of a digital platform, with cloud-based technology, to manage the ingestion, analysis, visualization and exportation of data regarding the evolution of COVID-19 in Panama. The methodology for the design of predictive algorithms is based on a hybrid model that combines the dynamics associated with population data of an SIR model of differential equations and extrapolation with recurrent neural networks. The technological solution developed suggests that adjustments can be made to the rules implemented in the expert processes that are considered. Furthermore, the resulting information is displayed and explored through user-friendly dashboards, contributing to more meaningful decision-making processes.
1. Introduction
In December 2019, the spread of the SARS-CoV-2 virus began worldwide, which caused the COVID-19 pandemic [1]. This unprecedented crisis, which initially affected healthcare systems worldwide, has had an effect on every main sector of activity [2]. Therefore, a series of control measures have been taken in different countries to prevent high numbers of infections in the population [3,4].
One of the most widely used control initiatives is the establishment of restrictions on the mobility of individuals [5]. As a consequence, society is becoming increasingly interested in technological resources that facilitate the adoption of automated ways of efficiently managing the large volume of data currently being generated [6,7,8]. The global utilization of restrictions on movement has meant that while in the physical world, the activity of society is at a standstill, in the digital world, activity is increasing. For public administrations that manage healthcare systems, there is particularly high interest in the concept of having technological tools that contribute to the effective and efficient development of decision-making processes [9,10]. The current pandemic scenario poses challenges, as it requires the implementation of responsible and high quality public actions, through the adoption of actions under a decision-making process based on real data and information on the evolution of the disease and the levels of affectation that are being generated [11]. Initiatives taken under the influence of these conditions will benefit of the majority of global population. The context of the current healthcare crisis in which these types of public organizations operate, is associated with the so-called VUCA (volatile, uncertain, complex and ambiguous) environment [12,13]. For this scenario, the implementation of “disruptive” technologies allows for the maximization of the full potential of the latest advancements that are being applied to the intelligent management of large volumes of data (big data). These data inputs are being generated as a result of the growing digitization of organizational processes [14,15]. Furthermore, the implementation of solutions based on innovative technologies allows public organizations to deal with complex contexts in terms of decision making [15]. This implies a significant difference compared to conventional decision-making contexts. Given the exceptional nature of the crisis that is being experienced in these organizational environments, it is important to implement solutions that provide significant value and enable knowledge management, as these will be the organizations’ most valuable resource [16].
The integration of technological solutions based on technologies such as artificial intelligence (AI) and machine learning (ML) make it possible to create the necessary conditions to offer an efficient dataset, in real time that is available to all stakeholders [17]. Likewise, current computing advances provide great capabilities for the analysis and presentation of information and the intelligent exploitation of data generated from heterogeneous sources. Forms of visualization can be incorporated and aimed at non-technical audiences or personnel [18].
This paper is organized as follows: the Related Work section presents a review of scientific publications that propose technological solutions to address processes regarding the diagnosis and prediction of cases during the COVID-19 pandemic. The reviewed papers present solutions based on artificial intelligence and the application of methodologies that incorporate machine learning algorithms. Finally, in the Proposed Solution section, some of the most relevant attributes of the Epidempredict research project, developed in Panama to monitor the evolution of the COVID-19 pandemic, are presented.
2. Related Work
Artificial intelligence (AI) is a widely used term that encompasses a wide variety of areas and sub-areas of activity. In these areas, algorithms are designed and built to replicate human intelligence, facilitating decision making through the processing power of computer systems [19,20,21].
Recently, there has been an increase in the development of AI-based technological solutions in the healthcare sector. In this regard, the emergence of the COVID-19 pandemic has led to the development of initiatives involving the use of AI to achieve a more detailed analysis of the large volume and diversity of data being generated. On the other hand, the design of solutions based on machine learning and AI is contributing to the improvement of diagnostic and prevention systems, and significantly supporting the work performed by healthcare professionals [22].
On the other hand, AI and ML technologies are used to achieve improved accuracy in the predictions made about the occurrence of infectious and non-infectious diseases, such as COVID-19 [23]. One of the first expert systems designed for healthcare assistance was the MYCIN system [24]. Developed in 1976, it used 450 rules based on the criteria of a medical expert. The solution provided answers about which antibiotics could be used to treat a given bacterial infection. The system was used to support clinical decision making, facilitating the work of medical staff [25,26].
The application of technologies such as AI and ML in data management and decision making related to the spread of contagious diseases allows for the development of rapid diagnostic and screening processes [27,28]. However, one of the main differences between the current COVID-19 pandemic and previous events that represent a health risk lies in the differences in the efficient and effective management of data. The main cause for this is attributed to the significant increase in the volume of data generated. On the other hand, there is the need to use these data, in real time, as a means for timely decision making.
Computing advances currently provide great analytical capabilities, which is one of the reasons why the data analytics industry is booming. This is a situation that requires consideration, as not everyone who can access data knows how to correctly interpret it. In other words, most people who use specific datasets on a daily basis, which could add value to data, do not have advanced knowledge of data processing, which affects the obtainment of good-quality results [29].
Another situation that the COVID-19 pandemic has generated is that the available data provided by multiple sources, such as the WHO (World Health Organization) and others, are sometimes published without unifying criteria, either between countries or even within each country. In addition, different regions do not quantify the data using the same criteria with regard to their structuring. This fact, added to the particularities existing in each country or region in terms of treatment or measures adopted in the face of the pandemic, means that the analysis of the data must be tailor made. In this sense, there is a need for automated tools that allow different users, including those without great technical knowledge, to work with the specific data they know, merging different parameters, such as the dates of the measures adopted in a territory, the severity of the restrictions, the degree of compliance by citizens or the movements of the population in a territory with other data associated with the number of infected individuals, number of recovered individuals, number of hospitalized persons, or number of deaths [30].
The WHO highlights four fundamental aspects that must be managed in health situations of broad social impact, such as the current COVID-19 pandemic: determining what caused the disease; understanding the forms of transmission and the populations at risk; clearly substantiating the behavior of the infection (from the incubation stage to the level of mortality); and structuring epidemiological models that help to define the mechanisms to be adopted to prevent, control, and diagnose infection, as well as apply treatment [31].
The significant impact of the COVID-19 pandemic on different health systems has led to the development of new automated tools that allow health authorities to make timely decisions, facilitating a broader view of the impact of the disease. This paper presents a general exploration of the scientific literature. This exploration made it possible to determine some of the working approaches being used for the development of predictions and data processing in clinical settings. The existence of a strong integration of resources and techniques based on artificial intelligence (AI) and machine learning (ML) to increase the efficiency of the development of diagnostics and screening in patients infected with COVID-19 was identified. The advancements achieved in this area will strengthen the expert systems in place for healthcare, which will contribute to the structuring and design of screening for the identification and management of SARS-CoV-2 carriers. The aim is to strengthen the traditional approach to screening through the use of more rapid and accurate techniques [32].
Table 1 presents some studies that report the use of technology for radiographic imaging, such as computed tomography and X-rays, and includes studies that reference ML and AI as a technology for use in the structured screening of SARS-CoV-2 infected patients (based on Lalmuanawma, Hussain and Chhakchhuak, 2020) [26].

Table 1.
ML and AI research with regard to structured screening of SARS-CoV-2.
In the literature, references were found to studies conducted in different countries where the implementation of technological solutions focused on AI and ML are successfully contributing in the implementation of digital contact tracing systems. The application of information technologies considered to be disruptive represents a direct method of support to the development of more efficient screening processes. This allows the most appropriate measures to be taken to mitigate the effects of the disease in the population. The studies presented explore the use of technological solutions that collaborate with the prevention of the spread of pandemic diseases, such as Sars-Cov-2 and COVID-19. The authors present the tools as cost-effective technological solutions that significantly improve diagnostic processes compared to the results provided by traditional methods applied to the detection and follow-up of infected or ill patients [26]. On the other hand, these studies show that the main purpose of the application of expert systems is to obtain a solution that offers a rapid diagnosis, for which an increase in the precision of the values obtained by the selected procedures and methods is sought. In this sense, the rapid and early detection of positive cases reduces the levels of spread of the disease on the one hand and, on the other hand, grants physicians or other health personnel with more time to make more accurate diagnoses in patients, which saves their lives on many occasions. Using a more technical approach, the studies presented here employ a single classification algorithm; however, authors such as Lalmuanawma et al. [26] suggest the use of classification methods known as hybrids. In these studies, the use of these hybrid solutions in heterogeneous data regarding mammographic, demographic and other studies is considered possible. This is feasible due to the existence of significant factors in the data that shed light on the process of identifying and categorizing patients who are truly infected or highly likely to be infected, whether or not they exhibit symptoms.
The papers included in this general exploration of the scientific literature were classified by models (Table 2), frameworks (Table 3), algorithms (Table 4), and others (Table 5), such as architecture, systems or methods, as at least one study was identified in each of these categories. The studies represent new contributions to the design and implementation of automated models for data analysis through the use of disruptive resources and technologies with the aim of addressing the COVID 19 pandemic.

Table 2.
Models.

Table 3.
Frameworks.

Table 4.
Algorithms.

Table 5.
Others.
3. Proposed Solution: Guidance and Contributions
The integration of initiatives to the healthcare sector based on disruptive technologies, such as ML and AI, allow the creation of the conditions necessary to offer an efficient dataset in real time that is available to all stakeholders [16]. Current advancements in computing provide great opportunities for performing analysis, information presentation, and the intelligent exploitation of data generated from heterogeneous sources. Forms of visualization aimed at non-technical audiences or personnel can be incorporated [17].
The Epidempredict initiative regarding COVID-19 corresponds to a research and development project that aims to achieve the optimal management of different data inputs regarding the evolution of COVID-19 in Panama, using a set of technological resources and tools that facilitate the design and implementation of an innovative technological solution.
According to the considerations that must be taken into account regarding innovation, it is intended that the developed solution facilitates the transformation of data and information into knowledge that is of value to an organization. Especially today, in a society characterized by a clear focus on the digital, the perception that knowledge is the most important factor of production, in addition to land, labor and capital, can be revitalized [54].
The development of this project incorporated a series of intelligent models that allowed prediction and optimization adapted to current needs in relation to data and information on the evolution of the pandemic in Panama. The design and construction of models and algorithms favor the adoption of more agile and simple decision-making processes. Here, the process contemplated the use of models that are executed and applied to data collected from various sources, as well as the knowledge generated and stored in relational, non-relational and other databases that are captured in real time [18,55].
The procedure used for the development of the project was based on a set of daily data in which the evolution of COVID-19 during a certain period of time in Panama was recorded, shown in Figure 1: t ≤ t_max. These data were used to calculate the temporal variation of the variables S (number of people susceptible to the disease), I (number infected) and R (number recovered/dead) in that time range. These variables were introduced in the SIR model, whose differential equations are solved using a Runge–Kutta method, thus extracting the unknown parameters of the model [56]: Ø_fit(t) and µ _fit(t) to: t ≤ t_max. Then, an LSTM neural network was applied to extrapolate these parameters to higher time values, with which, after being implemented in the SIR model, it was possible to make predictions with regard to the evolution of the S, I, and R variables.
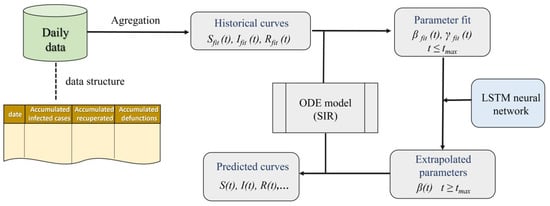
Figure 1.
Procedure used in the project.
Actions Applied to Input Variables and Results Obtained
The input variables used to implement the SIR model were the variation of the cumulative number of infected cases, the cumulative number of recovered persons and the cumulative number of deaths, taking 9 March 2020 as the starting date for the count. With these data, we could extract the variables S(t), I(t) and R(t), which were necessary for the implementation of the SIR model.
Figure 2, Figure 3 and Figure 4 present some visualizations obtained by using the initial data. The variables were obtained by solving the system of differential equations of the SIR model, that is, the parameters (t) and (t), as well as the basic re-productive number R0(t), which is no more than the ratio of Ø(t) to (t), with which the speed at which the disease spreads in the population was estimated. This ratio indicates the number of new infections expected from a single infection in a population where all individuals are susceptible, so that if its value is greater than 1, the disease is spreading, while if it is less than 1, the incidence of the disease is decreasing. From these output variables, we extrapolated the time series of coefficients with the LTSM neural network and made predictions regarding the evolution of the disease using the hybrid SIR model.
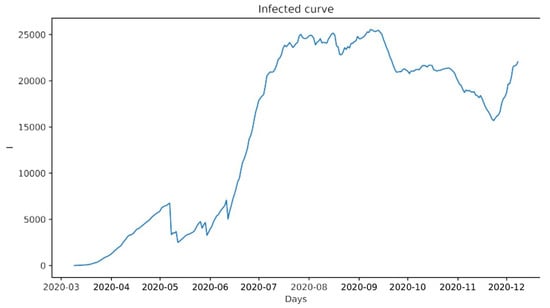
Figure 2.
Variation in the number of infected persons.

Figure 3.
Variation in the number of deaths per day.

Figure 4.
Variation of basic reproductive number R0.
For example, Figure 5 displays the results obtained by performing a 14-day forecast, taking the data collected from 9 September to 15 October 2020 as the input data. The results are represented in the following graphs, where the orange lines show the predictions with an error range and the blue lines show the actual data collected on those dates.
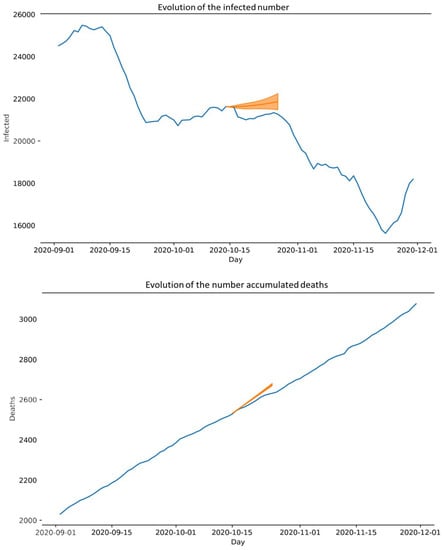
Figure 5.
Results obtained: (Above) Evolution of the infected number and (Below) Evolution of the number accumulated deaths.
A dashboard was designed to make all the project information visible via the website that was designed for the project. As Figure 6 shows, it uses a login with a permitted user so as not to expose sensitive health data. Once logged in, a dashboard breakdown view can be accessed, along with the defined sections, among which are a comparison of neighboring countries, a COVID-19 dashboard, COVID-19 open data, a document repository, and information regarding vaccines.
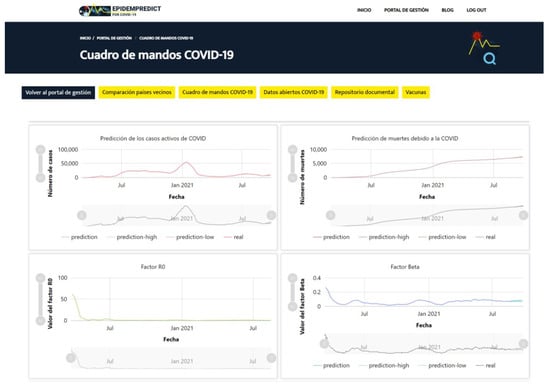
Figure 6.
Dashboard designed to make all the project information visible.
4. Discussion
Establishing the course of action and the effects that the COVID-19 pandemic will have in a given territory is one of the most commonly demanded requirements by health authorities worldwide. However, despite the successful development of digital tools and resources using disruptive technologies, such as artificial intelligence (AI), the dynamic attributes that characterize the data of the population under study and the diversity of restrictive measures adopted by health authorities make the prediction process a subject with different levels of complexity.
AI forms a part of everyday solutions used in the medical field, ranging from improving patient care and surgical techniques to predicting diagnoses. With regard to the COVID-19 pandemic, the scientific community has relied on AI to achieve the effective and efficient management of the large volume of data that has been generated, offering solutions aimed at diagnosing and predicting possible cases, which enabled authorities to make decisions based on valid and reliable data by establishing timely regulations adapted to the particular conditions of each territory.
The works included in the general exploration of the scientific literature conducted in this paper were classified by models, frameworks, algorithms, and others labels, such as architecture, systems or methods because research was identified that fit each of these categories. The studies included represent new contributions to the design and implementation of automated models for data analysis using disruptive resources and technologies with the goal of addressing the challenges caused by the COVID-19 pandemic.
In addition, the development of a platform called Epidempredict was proposed. This solution uses cloud storage technology to manage data regarding the evolution of COVID-19 in Panama. In this platform, a prediction system was designed and implemented based on a hybrid model that considers the ingestion and processing of data obtained from heterogeneous sources incorporating a SIR model, with differential equations and extrapolation processes based on recurrent neural networks. The input variables used to implement the SIR model correspond to the variation of the cumulative number of infected cases, the number of people recovered and the cumulative number of deaths. In order to process and analyze data and make predictions from the data, the data generated from the official registration of the first case of COVID-19 in Panama were used.
We established that the project is based on a hybrid solution, combining an expert system and LSTM within the SIR model. This allowed us to obtain explainable results, exposing the level of impact that the application of restrictive regulations has had on the fluctuation obtained for the coefficient. On the other hand, the expert rules integrated in the solution helped to obtain a prediction of the effect that the application of these measures has had on the dispersion index. In addition, explainable artificial intelligence was used to help understand the system’s interpretation of the data associated with the pandemic.
5. Conclusions
The hybrid model that was incorporated in the developed solution allows the population dynamics of the SIR model to be handled as well as those of the LSTM recurrent neural networks. Machine learning algorithms were incorporated in this model to predict the transmission rate of the virus in Panama. Recurrent neural networks have feedback paths between all of the elements that compose them. A single neuron is connected to subsequent neurons in the next layer, to past neurons in the previous layer, and to itself through vectors of varying weights that undergo alterations to achieve operation parameters or goals.
Furthermore, the model implemented in EpidemPredict for COVID-19 adapts the prediction generated by integrating a process of the identification and detection of changes in the structure of the data processed by the SIR model. In this sense, the adjustment made in the model allows for the generation of predictions that tolerate fluctuations of existing values in the key attributes of the data used and the integration of new data characteristics. This fact allows for the coefficients and results obtained from the implemented model to be adapted. In addition, the data are consumed and explored through dashboards that facilitate their use by non-specialized users. In short, the solution generated is a direct method of support for decision-making systems used by health authorities.
Author Contributions
Conceptualization: L.M., J.M.C.; investigation: Y.C., M.N.; methodology: I.S.-C.; writing—original draft: I.S.-C., Y.C.; writing—review and editing: L.M., J.M.C., V.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Secretaria Nacional de Ciencia, Tecnología e Innovación (SENACYT Panamá) under Grant number 48-2020-COVID19-045.
Data Availability Statement
EPIDEMPREDICT PLATFORM: https://epidempredict.utp.ac.pa/, accessed on 19 October 2021.
Acknowledgments
The authors Lilia Muñoz and Vladimir Villarreal are members of the National Research System (SNI).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Tanigawa, Y.; Rivas, M. Initial review and analysis of COVID-19 host genetics and associated phenotypes. Preprints 2020, 367. [Google Scholar] [CrossRef][Green Version]
- Trilla, A. Un mundo, una salud: La epidemia por el nuevo coronavirus COVID-19. Med. Clín. 2020, 154, 175. [Google Scholar] [CrossRef]
- Hattaf, K.; Yousfi, N. Mathematical Modeling in Virology. In Emerging and Reemerging Viral Pathogens; Elsevier: Amsterdam, The Netherlands, 2020; pp. 325–339. [Google Scholar]
- Wearing, H.J.; Rohani, P.; Keeling, M.J. Appropriate models for the management of infectious diseases. PLoS Med. 2005, 2, e174. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Lista, E.G.; Torres, M.S. Revisión sistemática de literatura sobre procesos de gestión de conocimiento. Rev. GTI 2014, 13, 45–67. [Google Scholar]
- Sáez, O.S. La responsabilidad patrimonial de la Administraciones Públicas en la gestión del Coronavirus. Diario La Ley 2020, 9617, 5. [Google Scholar]
- Peñas, S.L. COVID-19: Un terremoto para las cuentas públicas. Cuad. Inf. Econ. 2020, 276, 33–39. [Google Scholar]
- Moreno, O.M.C. VUCA World y lecciones de interdependencia COVID-19. GIGAPP Estud. Work. Pap. 2020, 7, 513–532. [Google Scholar]
- Berner, H.; Van Hemelryck, T. Sistemas de Información Social y Registros de Destinatarios de la Protección Social no Contributiva en América Latina: Avances y Desafíos Frente al COVID-19. 2020. Available online: https://repositorio.cepal.org/handle/11362/46452 (accessed on 19 October 2021).
- Belardo, M.B.; Herrero, M.B. La salud internacional frente al COVID-19: Entre los sistemas sanitarios y las políticas. Hamartia 2020, 7, 1–10. [Google Scholar]
- Mar Cornelio, O.; Gulín González, J.; Bron Fonseca, B.; Garcés Espinosa, J.V. Sistema de apoyo al diagnóstico médico de COVID-19 mediante mapa cognitivo difuso. Rev. Cuba. Salud Pública 2020, 46, e2459. [Google Scholar]
- Prieto-Castrillo, F.; Shokri Gazafroudi, A.; Prieto, J.; Corchado, J.M. An ising spin-based model to explore efficient flexibility in distributed power systems. Complexity 2018, 2018, 5905932. [Google Scholar] [CrossRef]
- Casado-Vara, R.; Chamoso, P.; De la Prieta, F.; Prieto, J.; Corchado, J.M. Non-linear adaptive closed-loop control system for improved efficiency in IoT-blockchain management. Inf. Fusion 2019, 49, 227–239. [Google Scholar] [CrossRef]
- Casado-Vara, R.; Martin-del Rey, A.; Affes, S.; Prieto, J.; Corchado, J.M. IoT network slicing on virtual layers of homogeneous data for improved algorithm operation in smart buildings. Future Gener. Comput. Syst. 2020, 102, 965–977. [Google Scholar] [CrossRef]
- de Freitas Barbosa, V.A.; Gomes, J.C.; de Santana, M.A.; Jeniffer, E.d.A.; de Souza, R.G.; de Souza, R.E.; dos Santos, W.P. Heg.IA: An intelligent system to support diagnosis of COVID-19 based on blood tests. Res. Biomed. Eng. 2021, 1–18. [Google Scholar] [CrossRef]
- Dhiman, C.; Vishwakarma, D.K. View-invariant deep architecture for human action recognition using two-stream motion and shape temporal dynamics. IEEE Trans. Image Process. 2020, 29, 3835–3844. [Google Scholar] [CrossRef]
- Mutasa, S. Understanding artificial intelligence based radiology studies: What is overfitting? Clin. Imaging 2020, 65, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Pathak, Y.; Shukla, P.K.; Tiwari, A.; Stalin, S.; Singh, S. Deep transfer learning based classification model for COVID-19 disease. IRBM 2020. [Google Scholar] [CrossRef]
- Agrebi, S.; Larbi, A. Use of artificial intelligence in infectious diseases. In Artificial Intelligence in Precision Health; Elsevier: Amsterdam, The Netherlands, 2020; pp. 415–438. [Google Scholar]
- Shortliffe, E.H. Computer-Based Medical Consultants: MYCIN; Elsevier: New York, NY, USA, 1976. [Google Scholar]
- Peiffer-Smadja, N.; Rawson, T.M.; Ahmad, R.; Buchard, A.; Pantelis, G.; Lescure, F.X.; Birgand, G.; Holmes, A.H. Machine learning for clinical decision support in infectious diseases: A narrative review of current applications. Clin. Microbiol. Infect. 2019, 26, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Lalmuanawma, S.; Hussain, J.; Chhakchhuak, L. Applications of machine learning and artificial intelligence for COVID-19 (SARS-CoV-2) pandemic: A review. Chaos Solitons Fractals 2020, 139, 110059. [Google Scholar] [CrossRef] [PubMed]
- Vaishya, R.; Javaid, M.; Khan, I.H.; Haleem, A. Artificial intelligence (AI) applications for COVID-19 pandemic. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 337–339. [Google Scholar] [CrossRef] [PubMed]
- Ai, T.; Yang, Z.; Hou, H.; Zhan, C.; Chen, C.; Lv, W.; Tao, Q.; Sun, Z.; Xia, L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases. Radiology 2020, 296, E32–E40. [Google Scholar] [CrossRef]
- Ávila-Tomás, J.F.; Mayer-Pujadas, M.A.; Quesada-Varela, V.J. La inteligencia artificial y sus aplicaciones en medicina II: Importancia actual y aplicaciones prácticas. Atención Primaria 2021, 53, 81. [Google Scholar] [CrossRef] [PubMed]
- Salado-Cid, R.; Romero, J.R. Lenguaje especıfico para el modelado de flujos de trabajo aplicados a ciencia de datos. XXI Jornadas de Ingeniería del Software y Bases de Datos 2020, 219, 227. [Google Scholar]
- Castro, J.L.A.; Villegas, O.R.T. COVID-19 y sus implicaciones sociales: Una mirada desde las ciencias computacionales. Educere 2021, 25, 147–164. [Google Scholar]
- Peris-Martínez, C.; Shaha, A.; Clarida, W.; Amelon, R.; Hernáez-Ortega, M.C.; Navea, A.; Morales-Olivas, J.; Dolz-Marco, R.; Pérez-Jordá, P.; Verbraak, F.; et al. Uso en la práctica clínica, de un método de cribado automatizado de retinopatía diabética derivable mediante un sistema de inteligencia artificial de diagnóstico. Archivos de la Sociedad Española de Oftalmología 2021, 96, 117–126. [Google Scholar] [CrossRef]
- Ardakani, A.A.; Kanafi, A.R.; Acharya, U.R.; Khadem, N.; Mohammadi, A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020, 121, 103795. [Google Scholar] [CrossRef]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef]
- Sun, L.; Song, F.; Shi, N.; Liu, F.; Li, S.; Li, P.; Zhang, W.; Jiang, X.; Zhang, Y.; Sun, L.; et al. Combination of four clinical indicators predicts the severe/critical symptom of patients infected COVID-19. J. Clin. Virol. 2020, 128, 104431. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, P.; Zhang, L.; Meng, W.; Li, J.; Tong, C.; Li, Y.; Cai, J.; Yang, Z.; Zhu, J.; et al. Rapid and accurate identification of COVID-19 infection through machine learning based on clinical available blood test results. MedRxiv 2020. [Google Scholar] [CrossRef]
- Hu, Z.; Ge, Q.; Li, S.; Boerwinkle, E.; Jin, L.; Xiong, M. Forecasting and evaluating multiple interventions for COVID-19 worldwide. Front. Artif. Intell. 2020, 3, 41. [Google Scholar] [CrossRef] [PubMed]
- Chenthamarakshan, V.; Das, P.; Padhi, I.; Strobelt, H.; Lim, K.W.; Hoover, B.; Hoffman, S.C.; Mojsilovic, A. Target-specific and selective drug design for COVID-19 using deep generative models. arXiv 2020, arXiv:2004.01215. [Google Scholar]
- Linda, W. A tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. J. Netw. Comput. Appl. 2020, 20, 1–12. [Google Scholar]
- Tang, Z.; Zhao, W.; Xie, X.; Zhong, Z.; Shi, F.; Liu, J.; Shen, D. Severity assessment of coronavirus disease 2019 (COVID-19) using quantitative features from chest CT images. arXiv 2020, arXiv:2003.11988. [Google Scholar]
- Chen, Y.C.; Lu, P.E.; Chang, C.S. A Time-dependent SIR model for COVID-19. arXiv 2020, arXiv:2003.00122. [Google Scholar]
- Ribeiro, M.H.D.M.; da Silva, R.G.; Mariani, V.C.; dos Santos Coelho, L. Short-term forecasting COVID-19 cumulative confirmed cases: Perspectives for Brazil. Chaos Solitons Fractals 2020, 135, 109853. [Google Scholar] [CrossRef]
- Khan, A.I.; Shah, J.L.; Bhat, M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020, 196, 105581. [Google Scholar] [CrossRef]
- Gupta, A.; Anjum; Gupta, S.; Katarya, R. InstaCovNet-19: A Deep learning classification model for the detection of COVID-19 patients using chest X-ray. Appl. Soft Comput. 2021, 99, 106859. [Google Scholar] [CrossRef]
- Abbas, A.; Abdelsamea, M.M.; Gaber, M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl. Intell. 2021, 51, 854–864. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Tian, T.; Huang, S.; Wan, F.; Li, J.; Li, S.; Yang, H.; Hong, L.; Wu, N.; Yuan, E.; et al. A data-driven drug repositioning framework discovered a potential therapeutic agent targeting COVID-19. Cold Spring Harb. Lab. 2020, 6, 165. [Google Scholar] [CrossRef]
- Imran, A.; Posokhova, I.; Qureshi, H.N.; Masood, U.; Riaz, M.S.; Ali, K.; John, C.N.; Hussain, M.I.; Nabeel, M. AI4COVID-19: AI enabled preliminary diagnosis for COVID-19 from cough samples via an app. Inform. Med. Unlocked 2020, 20, 100378. [Google Scholar] [CrossRef] [PubMed]
- Ahamad, M.M.; Aktar, S.; Rashed-Al-Mahfuz, M.; Uddin, S.; Liò, P.; Xu, H.; Summers, M.A.; Quinn, J.M.; Moni, M.A. A machine learning model to identify early stage symptoms of SARS-Cov-2 infected patients. Expert Syst. Appl. 2020, 160, 113661. [Google Scholar] [CrossRef]
- Yan, L.; Zhang, H.T.; Goncalves, J.; Xiao, Y.; Wang, M.; Guo, Y.; Sun, C.; Tang, X.; Jing, L.; Zhang, M.; et al. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020, 2, 283–288. [Google Scholar] [CrossRef]
- Chimmula, V.K.R.; Zhang, L. Time series forecasting of COVID-19 transmission in Canada using LSTM networks. Chaos Solitons Fractals 2020, 135, 109864. [Google Scholar] [CrossRef] [PubMed]
- Hassantabar, S.; Ahmadi, M.; Sharifi, A. Diagnosis and detection of infected tissue of COVID-19 patients based on lung X-ray image using convolutional neural network approaches. Chaos Solitons Fractals 2020, 140, 110170. [Google Scholar] [CrossRef] [PubMed]
- Afshar, P.; Heidarian, S.; Naderkhani, F.; Oikonomou, A.; Plataniotis, K.N.; Mohammadi, A. COVID-caps: A capsule network-based framework for identification of COVID-19 cases from x-ray images. Pattern Recognit. Lett. 2020, 138, 638–643. [Google Scholar] [CrossRef]
- Ye, Y.; Hou, S.; Fan, Y.; Qian, Y.; Zhang, Y.; Sun, S.; Peng, Q.; Laparo, K. α Satellite: An AI-driven System and Benchmark Datasets for Hierarchical Community-level Risk Assessment to Help Combat COVID-19. arXiv 2020, arXiv:2003.12232. [Google Scholar]
- Trujillo, L.L.; Trujillo, P.L.; Trujillo, F.L. Modelo de Gestión del Conocimiento para la Innovación. Adm. Y Organ. 2020, 23, 69–83. [Google Scholar]
- Diekmann, O.; Heesterbeek, J.A.P. Mathematical Epidemiology of Infectious Diseases: Model Building, Analysis and Interpretation; John Wiley & Sons: Hoboken, NJ, USA, 2000; Volume 5. [Google Scholar]
- Feng, L. SEIR Model Combined with LSTM and GRU for the Trend Analysis of COVID-19; Middle Tennessee State University: Murfreesboro, TN, USA, 2021. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).