Abstract
In the past years, deep neural networks (DNN) have become popular in many disciplines such as computer vision (CV), natural language processing (NLP), etc. The evolution of hardware has helped researchers to develop many powerful Deep Learning (DL) models to face numerous challenging problems. One of the most important challenges in the CV area is Medical Image Analysis in which DL models process medical images—such as magnetic resonance imaging (MRI), X-ray, computed tomography (CT), etc.—using convolutional neural networks (CNN) for diagnosis or detection of several diseases. The proper function of these models can significantly upgrade the health systems. However, recent studies have shown that CNN models are vulnerable under adversarial attacks with imperceptible perturbations. In this paper, we summarize existing methods for adversarial attacks, detections and defenses on medical imaging. Finally, we show that many attacks, which are undetectable by the human eye, can degrade the performance of the models, significantly. Nevertheless, some effective defense and attack detection methods keep the models safe to an extent. We end with a discussion on the current state-of-the-art and future challenges.
1. Introduction
Deep learning provides researchers, powerful models evolving science and technology. Convolutional neural networks (CNNs) are the most important type of DL models for image processing and analysis, as they are very effective in learning meaningful features. Some of the most representative disciplines that use DL for computer vision tasks are robotics [1], autonomous cars, biometrics [2,3], face recognition [4], image classification [5], etc.
Because of its success, DL has become a useful supportive tool for doctors through medical image analysis as it saves significant time from doctors’ tasks. In medical image analysis, DL algorithms analyze and process MRI, CT scans, X-ray, and skin images for cancer diagnosis, retinopathy detection, lung disease classification, brain tumors, etc. Although deep learning has a very high performance on vision tasks, some recent studies proved that it can be vulnerable to adversarial attacks [6] and stealth attacks [7]. In the first case adversarial examples are introduced by small perturbations in the input data while in the second case, small perturbations to the AI system itself are introduced. Szegedy et al. [8] shown that a very small perturbation on an image can drive the model to wrong decisions. The perturbation must be imperceptible to the human eye so that the images look the same. In Figure 1, we can see that the initial image has been correctly predicted as a panda while with a very small noise the model predicted the panda as a gibbon with high confidence. The first explanations about adversarial attacks were about nonlinearity and overfitting while, later, Goodfellow et al. [9] showed that the linearity of models is the reason for such vulnerability. Some other studies tried to explain this phenomenon. Schmid et al. [10] supported that lack of data and the non-well distributed true data are the consequence of adversarial examples. Ilyas et al. [11] claimed that the success of adversarial attacks is due to models’ abilities to generalize on a specific dataset and non-robust features.
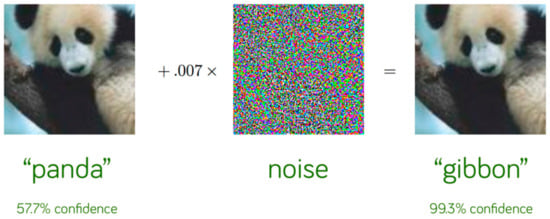
Figure 1.
Prediction before and after attack [9].
Adversarial attacks have raised concerns in the research community about the safety of deep neural networks and how we can trust our lives on them when they can be fooled easily. Adversarial examples can be created either we know the parameters of the DL model (white box attacks) or not (black box attacks) [12]. Usually, the noise that the attackers add in a clean image is not random but is computed by optimizing the input to maximize the prediction error. However, there are random noises too, which are implemented when the model’s parameters are unknown. Furthermore, there is a phenomenon that is called “adversarial transferability” and this means that adversarial examples which are created from one model can be effective on another model [13]. In addition to this, a study from Kurakin et al. [14] proved that adversarial examples are able to fool a model in the real-world when an adversarial example is printed as is shown in Figure 2.

Figure 2.
Adversarial attacks on printed out images [14].
There are two categories of defenses for decreasing the success rate of adversarial attacks, data level defense and algorithmic level defense. In the first category belong the adversarial training [8,9], preprocessing and postprocessing methods such as feature squeezing [15], magnet method [16]. In adversarial training, the model is trained with adversarial examples, which are correctly labeled. In the second category, some methods modify the model’s architecture, classifier and capacity [17]. However, these techniques are not always effective as most of them work with specific kinds of attacks either white box or black box. Moreover, many of them sacrifice accuracy on clean images.
At the same time, most doctors and researchers in the field of medicine deny trusting these models because they are treated as ‘black-boxes’ since we cannot explain how these models make a decision. This happens because a wrong decision in medicine has very high value as it is about human lives. Adversarial examples enhance the doctors’ view due to the efficacy of attacks proving that these models are not able to deal with real-world problems. Although adversarial examples seem unrealistic in medical image analysis, there are some serious motivations that we should take into consideration. For example, attackers can perturbate test reports in order to receive medical compensation [18]. Moreover, a wrong decision can cause dangerous effects on the patient’s life, needless costs and healthcare resources [19]. In addition, some malicious doctors can exploit these attacks so that they earn more income as they can manipulate the test reports leading to unnecessary surgeries.
- Until now, most existing studies have been done in natural images. Natural images have numerous differences from medical images and this is an important reason to study how adversarial attacks affect medical images. First of all, we lack big datasets with annotated labels due to the high cost and time consumption. In combination with the fact that the normal class is often overrepresented we result in slow convergence and overfitting. Another difference between these two types of images is that medical data often contain quantitative information while nature does not. Contrary to natural images, the orientation is usually not related to medical image analysis. In addition, there are various tasks in which the differences between the classes are very small. For example, an X-ray with early-stage pneumonia is quite similar to a normal one. Another difference is that natural images are generated from RGB cameras while most medical images are not. However, Finlayson et al. [20] showed that medical images can also be affected by adversarial examples. According to Ma et al. [21] medical DL models are more vulnerable than natural images models for two reasons: (1) the characteristic biological texture of medical images has many areas that can be easily fooled; and (2) modern DL models are quite deep as they are designed for natural images processing and this can lead to overparameterization in medical image analysis that increases vulnerability. However, attacks in medical images are detected more easily than in a natural image as adversarial features are linearly separated from normal features while in natural images adversarial examples are similar to normal. Even if adversarial attacks on medical imaging are an extreme case, robust machine learning (ML) focuses on these cases and according to Caliva et al. [22], this point of view is significant as medical image analysis hides many dangers and abnormalities which can be extreme cases as well.
- The field of adversarial attacks is relatively new and especially for medical applications. In Figure 3, we show the papers that have been done per year, on this field. We use app dimensions [23] tool to find how many papers have been done, by using as keywords “adversarial attack” and “medical”. We can see that the interest has been increased rapidly from 2018 to 2020. A short survey about adversarial attacks on the medical domain has been done by Sipola et al. [24]. However, it contains only a few studies about attacks by providing information about the consequences of these attacks, but without defense or detection mechanisms. Our paper contains much more studies about medical images and adversarial attacks. We also present not only attacks but also defenses, detections and new attacks designed for medical image analysis.
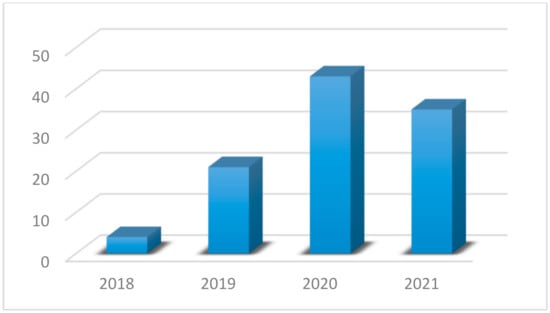 Figure 3. Papers per year from app dimensions tool.
Figure 3. Papers per year from app dimensions tool.
In this paper, our main contributions are: (1) to summarize for the first time in the literature, all works that have been done about adversarial attack, defense and attack detection methods in medical image analysis; (2) to bring to light the importance, the gaps and the challenges of this research field. There is no doubt that the resilience of the DL models in attacks is a key factor to increase the trustworthiness of the models and therefore their security issues should be a key research objective to enhance the integration of DL technology in real-world applications.
The rest of this paper is organized as follows. Section 2 presents the literature analysis conducted in this study. In Section 3, an overview of deep learning in medical image analysis is provided. Section 4 introduces general adversarial attacks applied to both natural and medical images. In Section 5 all attacks, defenses and detections in proposed for medical images are described. Section 6 pointing out some implementation aspects, while Section 7 discusses the current status and challenges. Finally, Section 8 concludes this study.
2. Literature Analysis
In this section, a statistical analysis of the literature is presented for identifying the current trends the research community is focused on. The analysis is based on the outcomes of provided by the search mentioned in the previous section using the app dimensions tool. As we can see from Figure 3, the first studies were presented in 2018, which means that until 2021 there are few works on this domain (103 in total). Most of these papers aim to prove that adversarial attacks affect medical images using existing attacks, while some studies propose new attacks for medical images or try to defend against these attacks (Figure 4a). From Figure 4a we can understand that so far researchers have focused on whether adversarial attacks can affect the models’ effectiveness. This is reasonable because the research area of adversarial attacks is new. However, more studies on defense methods will be expected in the next years. First papers tried to implement known attacks such as FGSM, PGD, etc. in order to test medical images under general attacks. Results have shown that these attacks are quite effective on all modalities but also, they are easily detected. Later, researchers tried to create custom attacks for medical images. Most of these attacks exploit features from several modalities. Two studies simulate the phenomenon of bias field which occur on medical images [25,26] to attack the models while others take advantage of noise that high tech medical imaging systems create. On the other hand, some studies tried to deploy these features in order to build more robust models. Mainly, adversarial training was the most used defense method, but it is not very robust under unknown attacks. Ensemble training is another method that is implemented as a defense method. However, there are studies that develop their own way to detect or defend against attacks which is more sophisticated.
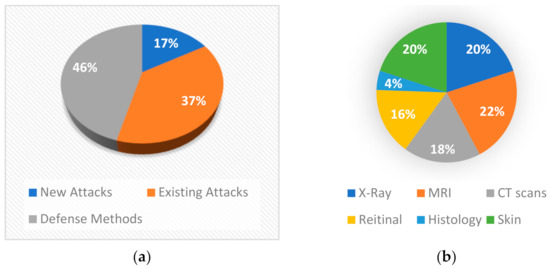
Figure 4.
(a) Methods that were analyzed in this paper and (b) Image modalities that were studied.
Furthermore, in Figure 4b it appears that most researchers experimented with MRI, X-rays, and dermoscopy images as they are the most frequently used because most medical free datasets contain these modalities. Moreover, CT-scans and fundoscopy were widely examined while histopathological and microscopy images were tested by only one paper. Also, as it is shown in Figure 5 the studies are mainly focusing on classification tasks and secondarily on segmentation tasks. In addition, only three studies are dealing with reconstruction tasks.
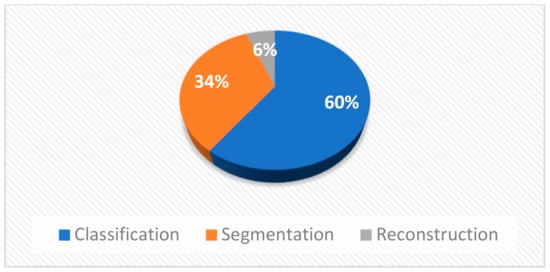
Figure 5.
Types of problems that were studied.
Moreover, most studies used pre-trained models for experiments as shown in Figure 6b. Transfer Learning has boosted deep learning as we can train easily very big models and that is why this method is widely used in medical image analysis. However, these models are designed for natural images, which are more complicated and therefore require more parameters. However, the models for medical images, need fewer parameters and according to Ma et al. [21], the overparameterization of these models could be an important reason for the significant reduction in accuracy. U-Net was the most used model for segmentation task as it is the state-of-the-art on this domain while ResNets was widely used for the task of classification. Experiments with custom models that are designed for medical applications, may help us to draw more safe conclusions. From the pre-trained models, DenseNets seem to be the most robust and as a consequence, the dense blocks enhance the model’s safety. Furthermore, gradient-based attacks are the most efficient—such as FGSM, PGD, I-FGSM, etc.—and that is why they are often used as shown in Figure 6a. Also, most of new attacks compared with these.
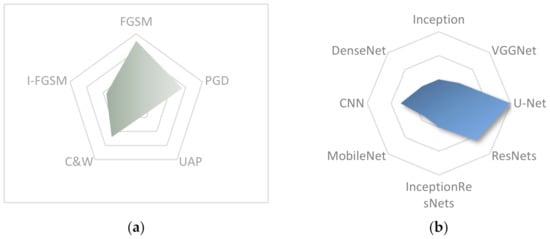
Figure 6.
Most often used attacks (a) and models (b) in adversarial medical imaging.
3. Medical Image Analysis
Medical image analysis aims at processing the human body through different image modalities for medical reasons like diagnosis, treatments, and health monitoring. The evolution of deep neural networks in the field of computer vision solves problems that classical image processing techniques performed poorly. These solutions have been widely applied in medical imaging because these networks have shown that they are the best choice for dealing with complex and high dimensional data such as medical images. The usage of computer vision in medicine is quite significant as it offers high rates of successful earlier diagnosis, which is crucial for reducing mortality rates. Furthermore, medical image analysis decreases medical errors and this is important, as a study from Daniel M. [27] has shown that medical errors are the third leading cause of death in the USA. An interesting study from Frank R. [28] has shown that the rise of medical imaging increases human life expectancy. Another aspect of this view is presented by Beinfeld et al. [29]. They claimed that spending $385 on medical imaging leads to saving approximately $3000. The most used image modalities are MRI, CT scans, ultrasound (US), and X-ray. However, due to the difficulty of acquiring medical images, the datasets are smaller when compared to other computer vision tasks and as a consequence transfer learning [30] method is often used. In addition, there are several tasks on medical image analysis, which deep learning deals with, with the most important being classification or diagnosis, detection and segmentation.
3.1. Classification—Diagnosis
A major category of applying deep learning in medical image analysis is classification or computer-aided-diagnosis (CAD) in which images are inputs and the DL models classify the images into several classes. Usually, models predict if a patient has a disease or not. One of the first works was done by Lo et al. [31] in 1995. They used a CNN with two hidden layers in order to diagnose whether an X-ray image has lung nodules or not. Another common image modality in medical imaging is a chest X-ray. Rajpurkar et al. [32] modified a DenseNet 121 model to classify a chest X-ray into 14 diseases. The model is called CheXNet. Diabetic Retinopathy (DR) is also a well-known diagnose method for DL models. Korolev et al. [33] evaluated their model, which is based on VGGNet [34] and ResNet [35] architectures for Alzheimer diagnosis.
3.2. Detection
Detection is an additional important target of medical image analysis. Accurate and fast detection of anatomical or pathological object localization such as organs and landmarks is quite significant for image registration and segmentation tasks [36,37]. Payer et al. [38] used a CNN end-to-end framework for anatomical landmark extraction from hand X-rays and MRIs. As a result, 37 landmarks were detected from X-rays while 28 from MRIs. LUNA16 [39] challenge is created to boost pulmonary nodules detection in CT scans as it is crucial for diagnosis of pulmonary cancer [40]. Platania et al. [41] applied CNNs for breast cancer detection using mammography images. An interesting study for COVID-19 detection was done by Horry et al. [42], (p. 19). They investigated transfer learning with state-of-the-art DL models concluding that VGGNet was the more stable and robust.
3.3. Segmentation
Segmentation in medical imaging refers to extracting specific parts of a medical image such as cells, tumors, organs to be analyzed in detail [36]. In addition, the segmentation of these parts allows us to analyze clinical parameters like volume and shape [20]. Ronneberger et al. proposed the U-Net [43] model, which is one of the most widely used DL models for biomedical image segmentation. Li et al. [44] proposed the H-DenseUNet, which is a hybrid densely connected U-Net for liver tumor segmentation from CT scans. Milletari et al. [45] proposed a 3D variation of the U-Net called V-Net. V-Net is responsible for 3D image segmentation using 3D convolutional layers. Moreover, Drozdzal et al. [46] proposed a combination of U-Net with ResNet skip-connections. In 2019, Jin et al. [47] proposed a deformable U-Net called DUNet for retinal vessels segmentation.
4. General Adversarial Examples
An adversarial example is an input sample in which it has been added an imperceptible noise so that it can be misclassified. A characteristic example is presented in Figure 1 where an attack has been applied to a deep learning model [9] leading to a wrong classification with high confidence. Szegedy et al. [8] were the first authors who investigated adversarial examples and they concluded that the success of this attack is due to the lack of generalization in the low probability space of data. However, some later works [9,17] have shown that even linear models are vulnerable too and an increase to the model’s capacity improves its robustness to these attacks. According to Hu et al. [48], it is important to study why adversarial examples exist and to better understand deep learning models in order to create more robust models. Attacks can be divided into three categories depending on the knowledge of adversaries. The first category is the white-box attack in which adversaries know everything about the model such as architecture and parameters. The second category is named grey-box attack and the adversaries know the structure of the model but not the parameters. Lastly, in the third category, adversaries know nothing about the model’s structure and parameters. In addition to this, there are targeted and untargeted attacks. In the former, attackers want to misclassify the input sample in a specific class, while in the latter they just want the sample data to be misclassified. There are numerous adversarial attacks and defenses [49] but none of these defenses is a panacea for all types of attacks.
4.1. Adversarial Attacks
In this section, we describe some of the most known adversarial attacks proposed for natural images and have been also applied to medical images.
FGSM (fast gradient sign method) [9] was the first proposed adversarial attack. FGSM is a white-box attack and produces adversarial examples for computer vision systems. This method extracts the adversarial gradient and decreases or increases the value of pixels so that the loss function increases. It perturbs a clean sample for a one-step update along the direction of gradient descend. This attack is formulated as
where is the input image, y is the label and θ represents the weights of the model. Moreover, is the magnitude of perturbation, J(θ,,y) is the gradient loss, sign (·) is the sign function and ∇x(·) is the gradient w.r.t. .
BIM (basic iterative method) or I-FGSM [13] is an iterative and improving method of FGSM. It performs FGSM with a value ϵ and updates its value with a small perturbation for T iterations until the image is misclassified. This method is formulated as
where αΤ = and the α is the magnitude of the perturbation for each iteration.
PGD (projected gradient descent) [17] is a generalization of BIM but without the constraint αΤ = ϵ. Perturbations are constrained by projecting adversarial samples from each iteration into ϵ − or ϵ − neighbor of the clean image.
C&W (Carlini & Wagner) [50] is another state-of-the-art attack that consists of three methods, and , which minimize , and norm respectively in order to compute the perturbation’s value.
JSMA (Jacobian-based saliency map attack) [51] is an iterative method that affects the value of a few pixels and it changes the value of one pixel in every iteration while the rest are unchanged. In this way, the saliency map is computed. Then the region with the most effective perturbation is selected and this region is perturbed in a clean image.
UAP (universal adversarial perturbation) [52] is an attack that creates perturbation for all images in a dataset trying to find the optimal perturbation that misclassifies most of the data points.
DAG (dense adversary generation) [53] is a black-box method that creates adversarial samples for object detection and semantic segmentation tasks.
4.2. Adversarial Defenses
In this section, some of the state-of-the-art defenses that are used to mitigate the phenomenon of adversarial attacks are discussed.
Adversarial training is one of the most widely used defenses in which a model is trained with adversarial samples so that can be more robust. This method is a min-max game that is formulated as
where is the DNN function, is the adversarial example of , is the loss function on the adversarial example , and is the maximum perturbation constraint.
Ensemble adversarial training is another effective method, which is developed for black-box attacks. Adversarial training is an effective method but the used individual models are vulnerable to black-box attacks as they can defend only attacks in which they are trained. Tramèr et al. [54] implemented ensemble adversarial training to compromise this phenomenon. They trained neural networks with adversarial samples from several methods such as FGSM and PGD so that the model has diversity on training samples.
There are numerous defense methods [49] such as randomization, which aims to randomize the adversarial samples [55]. Another method is denoising that tries to remove the perturbations from an input [15]. Some other are the weight-sparse DNNs [56], KNN-based defenses [57], Bayesian model-based defenses [58], and consistency-based defenses [59]. However, there are also detection methods that detect an adversarial sample and reject it before entering the model as input [60,61].
5. Adversarial Medical Image Analysis
Although several works have been done on adversarial examples for natural images, in medical images there are much fewer. Many researchers believe that adversarial examples are too difficult to occur in medical images. However, Finlayson et al. [62] presented some hypothetical scenarios in which bad actors could create adversarial examples. A characteristic example is a clinic that could perturb the medical images to lead all cases in surgery. Additionally, the U.S. Food and Drug Administration approved the first computer vision algorithm that can be deployed for medical diagnosis in Diabetic Retinopathy (DR) without the input of a human clinician [63]. In cases like these, we have to be sure about the accuracy of the algorithms and we must deal with adversarial examples as they can cause disastrous results. In this section, we present adversarial attacks, detections, and defenses that are applied on medical image analysis according to image modalities. Moreover, some works propose custom attacks and defenses.
5.1. Existing Adversarial Attacks on Medical Images
Paschali et al. [18] studied the effects of adversarial attacks on brain segmentation and skin lesion classification. For the classification task, InceptionV3, InceptionV4 [64], and MobileNet [65] models have been used, while SegNet [66], U-Net and DenseNet [67] were used for segmentation task. Experiments showed that InceptionV3 and DenseNet were the most robust models for classification and segmentation tasks respectively. The authors demonstrated that the robustness of a model is correlated with its depth for classification while for segmentation, dense blocks and skip connections increase its efficacy. The adversarial samples were imperceptibly as the SSIM was 0.97–0.99. Wetstein et al. [68] studied the factors that affect the efficacy of adversarial attacks. Their results show that the value of perturbation is correlated with the efficacy and perceptibility of the attacks. In addition, pre-training models enhance the adversarial transferability and finally, the performance of an attack can be reduced when there is inequality of data/model in target and attacker. Finlayson et al. [62] used PGD white and black box attacks on fundoscopy, dermoscopy, and chest X-ray images, using a pre-trained ResNet50 model. The accuracy of the model was dramatically decreased in both cases.
MRI images for brain tumor segmentation provide four different modalities (T1, T2, T1ce, and FLAIR) with different intensities in order for the brain tumor to be detected and labeled easily. Cheng et al. [69] investigated the effects of adversarial examples when they are applied on each modality and in all modalities simultaneously. Experiments were carried out with an ensemble U-Net model and MICCAI BraTS 2019 [70] dataset. For the generation of adversarial examples, they used universal random perturbation, which is similar to [52]. The results showed that simultaneous perturbation decreases the accuracy of the model, significantly, while when only one modality is perturbed, the accuracy is reduced slightly. Adversarial examples for age prediction from brain MRI have been applied by Li et al. [71]. They generated universal adversarial perturbation with L0, L2, and L∞ norms for the magnitude of perturbation value. In addition, they used two different architectures, a DNN and a hybrid DNN model that is combined with segmentation techniques and they showed that the hybrid model is much more robust than a conventional DNN on adversarial attacks. Huq et al. [72] analyzed adversarial attacks for skin cancer recognition. They experimented with VGG16 and MobileNet on the HAM10000 dataset, trying to classify an image into seven categories. After attacking with white-box attacks FGSM and PGD, accuracy was decreased significantly. They proposed adversarial training because it offers robustness and especially adversarial training with the PGD method, which resulted in a 1% accuracy reduction. Yilmaz et al. [73] applied FGSM attack on mammographic images. They used ‘Digital Database for Screening Mammography’ (DDSM) which contains two categories, normal and cancerous. The accuracy decreased up to 30% while the SSIM fell below 0.2. Pal et al. [74] examined the classification accuracy of COVID-19 from X-rays and CT-scans. They used FGSM attack to create adversarial samples and tested them on VGG-16 and InceptionV3 models. The results have shown that these models are vulnerable as the accuracy has decreased up to 90% in VGG-16 and up to 63% in InceptionV3.
Bortsova et al. [75] experimented with targeted PGD attack in X-rays for segmentation tasks. They added imperceptible noise in images and the model segmented the heart symbol instead of the heart Figure 7. The authors try to implement FGSM in the same experiment with no success. Also, they applied untargeted PGD attack, white-box and black-box which significantly decrease the average IoU of the model.
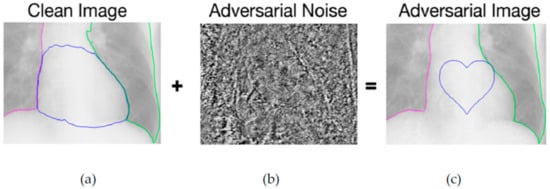
Figure 7.
(a) Prediction of a normal image. (b) Noise that added to image. (c) Prediction of adversarial image.
An interesting study was done by Anand et al. [76]. They compared the robustness of biomedical image analysis between transfer learning (TL) and self-supervised learning (SSL). Chest X-ray [77] for the pneumonia detection dataset and MRI RVSC [78] for the cardiac segmentation dataset were tested. For TL, a pre-trained model from ImageNet was used while SSL was done by Jigsaw puzzle task [79]. They experimented with PGD and FGSM attacks and VGG11 and U-Net models. The results showed that SSL outperforms TL as it learns more robust features. The authors propose SSL in combination with adversarial training as the default approach for better performance in small, labeled datasets and adversarial examples.
A study about adversarial attacks on NLST dataset (CT scans) for malignancy prediction of lung nodules was done by Rahul et al. [80]. They created adversarial samples with white-box attack FGSM and black-box One-pixel attack. Also, a custom model [81] was trained with three different architectures. After the FGSM attack, they received a 28–36% reduction in accuracy. However, in the black-box attack, the model was much more robust as the reduction was only 2–3%. For dealing with these attacks, they proposed an ensemble training approach where each CNN architecture was trained with seven different initializations minimizing significantly the attack accuracy.
The vulnerability of brain tumor classification on adversarial attacks was studied by Kotia et al. [82]. They applied three different white-box attacks, noise-based attack, FGSM and virtual adversarial training (VAT) [83]. The test of these attacks was done on the CE-MRI data set [84]. FGSM was the most effective attack as it decreased the accuracy by 69%, while VAT and noise-based, by 34% and 24% respectively. Adversarial training with FGSM and noise-based adversarial samples, showed very good results, as the training accuracy was almost the same as test accuracy on images under attack. However, adversarial training with VAT was not so effective, as the adversarial accuracy was decreased by 14% from training accuracy.
Shah et al. [85] studied the effect of adversarial examples on retinal images. They examined image-based (CNN-0 [86], CNN-1 [87]) and hybrid-lesion-based [88] algorithms for medical image analysis in order to diagnose diabetic retinopathy. CNN-0 and I-FGSM were used to generate adversarial images while CNN-1 and hybrid-lesion-based models were tested on them. The results have shown that CNN models are quite vulnerable and hybrid-lesion-based models more robust, as they have a 45% and 0.6% reduction of accuracy, respectively. Kovalev et al. [89] have studied the connection of control parameters and the size of image datasets with the efficacy of adversarial attacks. The adversarial samples were generated with white-box attack PGD. They chose two modalities for experiments, chest X-ray and histology for eight different classification tasks and the Inception V3 model. The experimental results showed that histology images are less vulnerable than X-rays. Moreover, an interesting observation was that adversarial attack accuracy was low if the original image was classified with high confidence. Additionally, as expected, the bigger value of perturbation leads to the higher success of attack except for X-ray images of the aorta with interior rotation. Finally, they showed that the size of the training set does not affect the success of the attacks.
The segmentation of the pancreas is very challenging due to its small size, abnormalities, texture, and localization. Li et al. [90] proposed a 3D deep coarse-to-fine framework for facing this challenge using NIH [91] and JHMI [92] datasets. The name of the model is ResDSN F2C and it is inspired by V-Net, U-Net, and VoxResNet. However, this framework is vulnerable to adversarial attacks as FGSM and I-FGSM cause an important reduction in accuracy (85.83%). The authors proposed adversarial training for this model to alleviate this phenomenon, as this technique decreased the accuracy only by 13.11%. Adversarial attacks on dermatoscopic images were done by Allyn et al. [93]. They perturbed the test set from the HAM10000 dataset [94] and tested it on DenseNet201. The overall accuracy decreased by 17%.
Hirano et al. [95] investigated universal adversarial attacks on DNNs for skin cancer diabetic retinopathy and pneumonia classification. They experimented in both targeted and untargeted attacks with several models such as VGG16, VGG19, InceptionResNetV2, DenseNet169, DenseNet121, and ResNet50. They discovered that adversarial training was not efficient in most cases and especially in untargeted attacks. At the same time, the transferability rate was low in non-targeted attacks. Finally, the authors claim that VGG16 and VGG19 seem to be more robust than the other models. One more study from Hirano et al. [96] proved that universal adversarial perturbation on COVIDNet [97] for the detection of COVID-19 from chest X-rays, is efficient and especially for non-targeted attacks. The dataset that they used was COVIDx. Attacks were both targeted and untargeted while adversarial retraining was applied as a defense method.
Ma et al. [21] examined the robustness of ResNet50 model on three datasets, chest X-ray [98], ISIC [99], and fundoscopy [100]. They applied four state-of-the-art white-box attacks, FGSM, PGD, C&W, and BIM, focusing on untargeted settings. Also, the value of the perturbation was computed with L∞ norm and when perturbation was = 1, the strongest attacks C&W, BIM, and PGD had almost everywhere 100% attack accuracy. Dermoscopy images were a little bit more robust than the other datasets but with no important deviation. For multiclass X-ray images with three and four classes, the attacks were efficient with = 0.3. They also applied four detectors, KD [101], LID [60], deep features and quantized deep features-based detectors for detecting adversarial samples, which had very high detection accuracy.
In Table 1 existing attacks implemented for medical images are summarized. The performance degradation column has shown that some attacks can dramatically reduce model’s accuracy. These attacks were tested only in classification and segmentation tasks. FGSM and PGD were the most used methods and PGD seems to be the most efficient. Moreover, the most of experiments were carried out in MRI, Dermoscopy and X-ray images. It is worth noting that the “dash” symbol in Table 1 implies that the authors did not provide results in the form of a percentage error.

Table 1.
Overview of existing adversarial attacks on medical images.
5.2. Adversarial Attacks for Medical Images
Byra et al. [102] proposed an attack method on ultra-sound (US) images for fatty liver. US images are reconstructed from radio-frequency signals, and authors applied a zeroth-order optimization attack [103] on the reconstruction method. The experiments were performed with the InceptionResNetV2 model and the attack achieved a 48% reduction in the model’s accuracy. Ozbulak et al. [104] proposed a targeted attack for medical image segmentation, which is named adaptive segmentation mask attack (ASMA). The proposed attack creates imperceptible samples for most parts and offers high intersection-over-union (IoU) degradation. For the experiments, they used the U-Net model because it is one of the most known models for medical image segmentation. Glaucoma optic disk segmentation [105] and ISIC skin lesion segmentation [106] datasets were used.
Chen et al. [107] proposed a method for generating adversarial examples to attack medical image segmentation. The adversarial examples are generated by geometrical deformations to model anatomical and intensity variations. They examined the efficiency of these examples by attacking a U-Net model for organ segmentation from abdominal CT scans. They achieved a significant reduction in terms of the Dice score metric for all organs. However, kidneys and pancreas are more difficult to attack than the liver and spleen and they required a higher value of perturbation.
Tian et al. [25] investigated the phenomenon of bias field, which can be caused by the wrong acquisition of a medical image, and it can affect the efficacy of a DNN, as shown in Figure 8. The authors, inspired by the adversarial attacks created an adversarial-smooth bias field attack to fool a model. Their experiments were carried out on a chest X-ray dataset with fine-tuning the ResNet50, MobileNet, and DenseNet121 models. They examined white-box attacks and the transferability of this attack. The proposed attack had higher attack accuracy on transferability than other state-of-the-art white-box attacks.
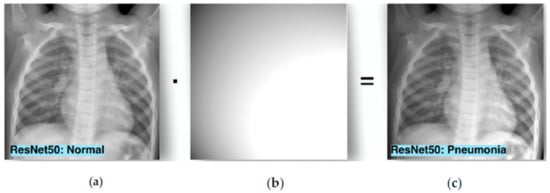
Figure 8.
(a) A clean image, (b) the bias field noise, and (c) the diagnosis after implementation of bias field noise.
A very interesting study was done by Kugler et al. [108] who investigated physical attacks on skin images. They used the HAM10000 dataset for training and the PADv1 dataset for attacking. The perturbations, in this case, were dots and lines with pen or acrylic (Figure 9). The model that they trained were ResNet, InceptionV3, InceptionResNetV2, Xception, and MobileNet. In contrast with digital attacks, physical attacks have a small difference in confidence compared to clean images. The most robust networks were Xception and InceptionResNet. Finally, the authors claimed that the attacks’ consequences are not statistical outliers but are related to the architectures and training procedures.
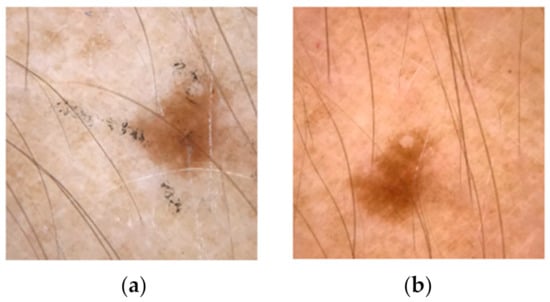
Figure 9.
The (a) image has some lines with a pen and the (b) image is clean.
Yao et al. [109] proposed a hierarchical feature constraint (HFC) method that can be added to any attack. Adversarial attacks are detected easier in medical images than in natural images, that is why this method aims to hide the adversarial features in order for them to not be easily detected. The experiments were performed on X-ray and Fundoscopy images with ResNet50 and VGG16.
Shao et al. [110] proposed a white-box targeted segmentation attack based on multi-scale gradients. This method combines adaptive segmentation mask and feature space perturbation in order to create a multi-scale attack (MSA). The authors, use not only the gradient of the last layer but also the gradient of the middle layer in order for perturbation to be small. Glaucoma optic disc segmentation dataset [111] and ISIC skin lesion segmentation dataset were used for experiments. The attack was performed on U-Net, R2U-Net [112], Attention U-Net [113], and Attention R2U-Net. The results have shown that the proposed method presents very high IoU with the targeted mask with imperceptible noise.
Qi et al. [114] proposed a new medical attack termed ‘stabilized medical image attack’ (SMIA). This method uses an objective function that consists of a loss deviation term and a loss stabilization term. The loss deviation term augments the discrepancy between the prediction of a perturbed image and its ground truth label. In addition to that, the loss stabilization term ensures similar behavior of CNN predictions of this example and its smoothed input. The experiment was conducted with fundus, endoscopic and CT scans datasets. ResNet, U-Net, and two other models [115,116] have been used. The accuracy has decreased up to 27%.
Table 2 shows all the attacks that have been created exclusively for medical images. Some of these methods use adversarial attacks in order to make medical models more robust, while others aim to decrease the efficacy of medical models. If we compare Table 2 with Table 1 we can conclude that medical adversarial attacks are not as strong as ordinary attacks.

Table 2.
Overview of medical adversarial attacks.
5.3. Defenses—Attack Detection
Wu et al. [117] have studied the classification of diabetic retinopathy with adversarial training. They used ResNet32 with the PGD method for generating adversarial samples. Adversarial training improved significantly the model’s efficacy under attack. He et al. [118] proposed a non-local context encoding network (NLCEN), which defends against adversarial attacks on medical image segmentation using contextual information of biomedical images. This network is based on ResNet and feature pyramid network (FPN) in combination with non-local context encoder. The experiments have been performed in JSRT and ISBI datasets with Iterative FGSM attack. The model requires 2 and 4 h of training and testing for JSRT and ISBI, respectively. NLCEN has been compared with state-of-the-art models such as, U-Net, InvertNet [119], SLSDeep [120], NWCN [121], and CDNN [122] by presenting the best accuracy of all. Furthermore, this method retains high accuracy even under attacks with big values of perturbation. Taghanaki et al. [123] studied adversarial examples on chest X-ray by trying to implement average pooling instead of max pooling. They used InceptionResNetV2 and Nasnet Large with 10 different attacks, which are divided into three categories, gradient-based, score-based, and decision-based attacks. The results showed that gradient-based attacks, fooled efficiently the models even with average pooling, but it provides an improvement in score-based and decision-based attacks.
The phenomenon of adversarial examples has not only negative aspects. Ren et al. [124] applied adversarial defense to deal with small datasets in Brain MRI segmentation. Due to small datasets, especially in 3D MRI, the task of segmentation is very challenging. However, the authors of this study proved that data augmentation with adversarial examples can improve the model’s robustness. They created adversarial samples with FGSM on a Cascade Anisotropic CNN [125]. Moreover, some studies applied adversarial attacks in the training procedure to create models that are more robust in general. Pervin et al. [126] used FGSM and inverse FGSM methods as a data augmentation approach for colon cancer segmentation in order to avoid overfitting. Inverse FGSM was used first time for adversarial training providing encouraging results. The authors achieved a 9% improvement in terms of IoU. Liu et al. [127] investigated the effect of adversarial training on Lung nodules from CT scans. The training was done with three 3DResUNets and data were collected from LUNA [38] and NLST cohort [128]. The PGD attack was used in order for them to find the patterns that lead to misclassification with high confidence and then they trained the network with these samples. The authors propose adversarial data augmentation to decrease the vulnerability of nodule detection against some unexpected noise and underrepresented properties of nodules. Vatian et al. [129] presented a very interesting work about the adversarial examples as ‘natural’ adversarial attacks. They have experimented with CT scans for lung cancer screening [130] and Brain MRI [131] using a CNN structure. They showed that in high-tech medical imaging systems a noise, which behaves as a ‘natural’ adversarial example, may occur. Three methods were applied to defend against these attacks. Adversarial training with FGSM and JSMA was the most effective defending methods, while the other two defense methods were data augmentation with Gaussian noise and replacing layers’ activation functions with Bounded ReLU. One more study about the bias field phenomenon has been done by Chen et al. [26]. They proposed an adversarial data augmentation method for segmentation, by modeling the intensity inhomogeneities (bias field), which is often appeared in MRI images. This method improves the efficacy and robustness of the models. Moreover, it can be applied in supervised and semi-supervised learning models. Experiments have been performed with the U-Net model and the ACDC dataset [132] for cardiac image segmentation.
Two interesting studies were carried out by Caliva et al. [22] and Cheng et al. [133]. They tried to mitigate the problem of false negatives in MRI reconstruction because the Fast-MRI challenge, has shown that even top models were not able to reconstruct some small abnormalities. They proposed false negative attack feature (FNAF) for robust training. FNAF applies adversarial attack in order to detect false negatives and then it improves the model’s generalization through robust training. The experiments were performed with U-Net and I-RIM [134] models, showing that FNAF with U-Net can significantly improve the efficacy of the model. The reconstructed images were evaluated with the SSIM metric which was 0.7197 ± 0.2613. Park et al. [135] proposed a defense mechanism for adversarial attacks on medical image segmentation. This method needs no knowledge about the model’s architecture or training examples. This mechanism converts an image in the frequency domain with discrete Fourier transform, as it helps to discrete clean images from adversarial images as shown in Figure 10. For the experiments, they used the OASIS [136] dataset, the SegNet, U-Net, DenseNet models, and DAG adversarial attack for crafting adversarial examples. DenseNet was the most robust network. This methodology does not increase the parameters of a model but increases the execution time because of the transformation of the image in the frequency domain.
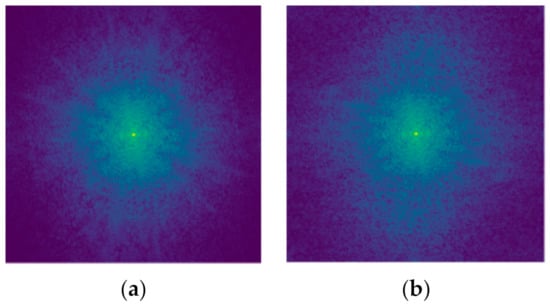
Figure 10.
The (a) image is the frequency domain of a clean image and the (b) is the frequency domain of an adversarial image.
Taghanaki et al. [137] proposed a defense method for classification, segmentation and object detection. In order for them to deal with the linearity of deep learning models, they proposed a non-linear radial basis convolutional feature mapping with distance functions, which is based on Mahalanobis distance. Chest X-ray 14 dataset was used for classification task with InceptionResNetV2 model, while ISIC (skin images) dataset with U-Net and V-Net for segmentation task. The proposed feature mapping increased the accuracy for classification and segmentation on both legitimate images and adversarial examples. This method does not increase the complexity of the model as it only changes the activation function. Another study about Mahalanobis distance was done by Anisie Uwimana and Ransalu Senanayake [138]. They proposed Mahalanobis distance in order to detect adversarial samples and out-of-distribution data (OOD). For the experiments, a malaria dataset was used for training while FGSM, BIM, C & W, and DeepFool methods were used for attacking. Also, a different malaria dataset was examined as the out-of-distribution data. This methodology presents up to 99.95 detection accuracy.
Daza et al. [139] introduced a benchmark for evaluation of adversarial robustness which is an extension of AutoAttack approach [140] and evaluate volumetric segmentation models in the medical domain. It contains four attacks APGD-CE, APGD-DLR, FAB-T, and Square Attack. Additionally, the authors proposed a model termed ROG for medical segmentation decathlon (MSD). This model is competitive with state-of-the-art models in clean images while outperforms them significantly in perturbated images. ROG model has a lattice architecture for general medical segmentation maintains high resolution features while also taking advantage of multiple image scales.
Another study tried to solve the problem of limited angle tomography that can cause problems in CT reconstruction because of missing data that lead to misinterpretation of the images. Huang et al. [141] proposed robust adversarial training in order to face this phenomenon. They applied poison noise as a perturbation on images for training because this noise is common in CT scans. The experiments have been performed with the U-Net model and AAPM Low-Dose CT Grand Challenge data. The results showed that retraining with poison noise is quite significant for limited angle reconstruction; however, it is not adequate for non-local adversarial examples. Xue et al. [142] proposed a defense mechanism to make models’ diagnosis more robust, by adding an auto-encoder on the CNN structure. This method can be combined with several models or with other defense methods. The method was tested on X-ray and fundoscopy images, under FGSM, IFGSM, and C&W attacks. However, embedding an auto-encoder into a CNN increases the complexity of the model. Tripathi and Mishra [143] proposed a fuzzy unique image transformation (FUIT) to defend against adversarial examples on diagnosis models for COVID-19. This technique models the pixels of an image into an interval and then the images are provided for training. The results showed that this method is effective on six non-targeted attacks (FGSM, BIM, PGD, PGD-r, Deep Fool, C&W). This method does not affect the model’s architecture but in testing, it requires the transformation of images taking more time. Another effective defense method for medical image segmentation was proposed by Liu et al. [144] by creating a low-cost frequency refinement approach. The experiments were carried out on skin and fundoscopy images under ASMA attack. This method has no impact on the model’s complexity as it only needs the frequency refinement of images.
Xu et al. [145] examined the robustness of three pre-trained deep diagnostic models. IPMI2019-AttnMel for Melanoma detection, InceptionV3 for diabetic retinopathy detection and CheXNet for classification of 14 types of diseases on ChestX-ray. They experimented with PGD and GAP (generative adversarial perturbations) attacks. Both attacks dramatically decrease model’s accuracy with PGD attacking even with 100% accuracy. The authors proposed two defenses in order to deal with attacks. The first is multi-perturbations adversarial training (MPAdvT) which trains the models with several perturbation levels and iteration steps during the training process. In adversarial training process, all samples are treated equally while according to Wang et al. [146] the perturbation on misclassified examples is more important for the robustness of the model and the minimization techniques are more significant than maximization in the natural image field. The second defense method, misclassification-aware adversarial Ttraining (MAAdvT) is based on these observations. The authors add a misclassification aware regularization to adversarial loss. They use Kullback–Leibler (KL) divergence in order for the classifier to be stable against misclassified adversarial examples. Both defense methods present better results than standard adversarial training.
Li et al. [147] proposed an attack detection method that rejects adversarial examples before a classification task. This method can be implemented in any medical image model without changing its architecture. When convolutions and pooling operations are applied to the adversarial sample then the noise getting worse. Because of that, an adversarial sample has a different feature map from a clean image. In this way, the detector extracts a feature map of an image and detects an adversarial sample. The experiments were carried out on the Chest X-ray 14 dataset and with the DenseNet121 model. Moreover, a significant advantage is that this method does not require knowledge of the attack model. Several experiments have been done with state-of-the-art attacks such as FGSM, BIM, PGD, and MIM, presenting encouraging results. Another detection method has been implemented by Li et al. [148] in order to minimize the adversarial risk. They proposed a robust medical imaging framework based on semi-supervised adversarial training (SSAT) and unsupervised adversarial detection (UAD). The experiments have been done with ResNet18 and the OCT image dataset [149]. The experimental results showed that SSAT decreases significantly the vulnerability of the model on adversarial samples while the UAD method rejects most adversarial examples. This method increases the model’s complexity as it uses the SSAT module as a complement to an existing model in order to find the robust features.
Some of the above methods use adversarial training [22,117,133,141] or the usage of average pooling [123] instead of max pooling as a defensive tactic, which do not affects the complexity of the models. Moreover, some studies like [26,124,127,129] applied adversarial learning in order to improve the performance of models not only in attacks but in general.
Table 3 summarizes all the defense and attack detection methods with the corresponding tasks, modalities and models. We observe that some methods provide significant protection against attacks while others simply reduce the success rate of an attack. At the same time, attack detection methods, detect adversarial samples with very high accuracy. For example, the studies [60,101] detect adversarial samples in several images and implemented in medical adversarial samples in [21].

Table 3.
Overview of defense and attack detection methods.
5.4. Benefits of Adversarially Robust Models
Creating models which are robust to adversarial attacks is crucial and especially in the medical domain. However, some studies have shown that adversarially robust models have some additional advantages. Lee et al. [150] proposed adversarial vertex mixup in order to overcome poor adversarial generalization. This method improves the robust generalization and decreases the trade-off between standard accuracy and adversarial robustness. Liu et al. [151] proposed a new framework, termed Neural SDE which incorporates several regularization mechanisms based on random noise injection. This framework creates more robust models as achieves better generalization and is resistant to adversarial and non-adversarial perturbations. Another interesting study [152], proposes adversarial robustness-based adaptive label smoothing (AR-AdaLS) which incorporates the correlations of adversarial robustness and uncertainty. The authors found that taking into account the adversarial robustness of the data within distribution, improves the calibration and stability of the model even under distributional shifts. Yi et al. [153] showed that adversarially trained models lead to an improved generalization on out-of-distribution data and this is quite important in medical image analysis. Adversarial learning not only improves adversarial accuracy, but also improves the models’ efficiency under various circumstances making them more robust in real life problems.
6. Implementation Aspects
In this section some implementation aspects of dealing with the attack, defense, and attack detection methods are presented. Furthermore, source code links for some of the described studies and dataset links that have been used are provided.
6.1. Open-Source Libraries
Some open-source libraries are available helping to create adversarial attacks and defenses. In this way, we can implement novel attacking or defending methods and to study their robustness performance using these implemented libraries. CleverHans [154] is one of the most known Python libraries that creates adversarial examples with state-of-the-art attacks. Another well-known python library for attacks is Foolbox [155] and runs easily adversarial attacks in PyTorch, TensorFlow, and JAX. Adversarial Robustness Toolbox (ART) [156] is also a Python library and provides developers with adversarial attacks in order to test their models. Advbox Family [157] is an open-source toolbox that supports Python and provides adversarial attacks and defenses. Another Python toolbox for adversarial robustness research is AdverTorch [158], which is implemented in PyTorch and generates adversarial perturbations and defending against adversarial examples. Finally, DEEPSEC [159] is a uniform platform for security analysis of deep learning models and provides state-of-the-art adversarial attacks, defenses and relative utility metrics of them. Table 4 summarizes the main characteristics of the discussed open-source libraries.

Table 4.
Main characteristics of the available open-source libraries.
6.2. Source Codes and Datasets
Apart from the available open-source libraries discussed in the previous section, additional source codes that implement novel attack, defense, and attack detection methods are provided by the authors mainly via a GitHub repository. Table 5 presents some information regarding the software given by some authors of the papers analyzed in this study.

Table 5.
Source codes in GitHub.
It is worth noting that for comparison purposes between the attack and defense methods some certain datasets with images of different modalities and sizes are used to produce adversarial samples. Table 6 illustrates the main characteristics of the most used datasets by the studies analyzed in the previous sections.

Table 6.
Main characteristics of the datasets commonly used in medical adversarial deep learning.
7. Discussion
In the last years, the ability to generate adversarial examples have raised questions about the safety of deep learning models as they can easily fool them. This phenomenon can cause serious problems especially in the medical domain. A mistake in medicine could lead to death, so we have to be sure for the DL models that they work properly and no one can intentionally degrade their prediction accuracy. However, the majority of surveys, show that we can imperceptibly process a medical image in order to be misclassified. In addition to that, some studies have shown that a mistake during image acquisition can cause a kind of noise that behaves like an attack. All of these problems need to be addressed towards implementing DL-based systems in hospitals and clinics and help doctors make decisions more easily, quickly, and accurately.
We observe that existing attacks have differences from medical imaging attacks. Some modalities, create specific features in images and researchers exploit them in order to attack images. For example [25,26] deploy the phenomenon of bias field which can be caused by the wrong acquisition of a medical image and it can reduce the efficacy of the model. Also, Byra et al. [102] created an attack based on how ultrasound images were created. Furthermore, high-tech medical imaging systems create a specific noise on images and the authors of [129], used this noise in order to create the attack. We can easily understand that medical attacks use the peculiarities of medical images to harm or reinforce the models.
An interesting study [18] has shown that the robustness of a model is correlated with its depth for classification while dense blocks and residual connections for segmentation tasks. Additionally, the authors from the study [68] demonstrated that pre-trained models increase the adversarial transferability and inequality of data/model decreases attack’s efficacy. On the other hand, Hirano et al. [95] discovered that the transferability rate is low on non-targeted attacks. Also, two interesting observations have been done by Kovalev et al. [89]. Firstly, they claimed that adversarial accuracy is low when an image is classified with high confidence and secondly, they showed that the size of the dataset does not affect the adversarial accuracy. Transfer learning with big pre-trained models is a powerful method and it is widely used in medical image analysis. However, Ma et al. [21] claimed that these models have been made for natural images which are more complicated than medical images. This means that these models are overparameterized for medical images and this could be an important reason for the high adversarial accuracy of medical image analysis. Creating models which will be tailor-made for each imaging modality could lead to more robust predictions.
Adversarial training is widely used as a defense method due to its simplicity. Nevertheless, it does not provide safety to the models when they are attacked by a black-box attack. From the papers read, we see that PGD attack usually cause the biggest degradation in accuracy. Maybe, an adversarial training with PGD adversarial samples could lead to a smaller degradation not only under PGD attack but also in other attacks. The gradient based attacks are the most known and efficient and that is why most studies test their data with them. Ensemble training with the most efficient attacks like PGD, FGSM, and C&W could lead to a quite robust model. However, this method requires cost and time to craft all adversarial samples from these methods and this could be prohibitive sometimes. Furthermore, NLCEN [118] is a network that presents better results than state of the art models and it could be a good choice for medical imaging problems as it retains high accuracy even under big values of perturbation. According to Taghanaki et al. [123], average pooling does not improve adversarial accuracy from gradient based attacks. Adversarial attacks have not only negative aspects. FNAF attack [22,133] can improve accuracy in MRI reconstruction. Small datasets in brain MRI can be enlarged with adversarial samples, making the model more robust [124]. Also, Pervin et al. [126] created adversarial examples in order to augment their datasets and not to attack. Following this approach, we avoid overfitting and we build robust models at the same time. Another significant advantage of adversarial training is that they can deal with some unexpected noises and underrepresented properties of nodules [127]. An interesting defense method presented by Taghanaki et al. [137] provides increased accuracy on classification and segmentation tasks in normal and adversarial examples by changing the activation functions. According to Huang et al. [141], poison noise improves the efficiency in CT reconstruction. From these methodologies, we can build models which cope with adversarial attacks but we can build stronger models in general at the same time. This is quite important because through adversarial robustness we can develop and explore new techniques in order to build more reliable deep learning models in medicine.
According to [89], histology images tend to be more robust than X-ray images, due to their structures. However, this phenomenon needs to be further investigated to reach a safe conclusion about its robustness under several attacks and compared to other modalities. Also, self-supervised learning (SSL) with adversarial learning could lead to more powerful models as according to [76] it learns more robust features than a pre-trained model. Nevertheless, more studies on SSL are mandatory in order to enhance or not this case. Xu et al. [145] claimed that models with attention modules have better accuracy but they are more vulnerable to adversarial attacks. Moreover, they observe that if a model has many layers, it is more vulnerable and that is why we should consider the trade-off between accuracy and robustness. In addition to that, the authors observing the saliency maps of models point out that we should use regularization in order to smooth the loss function for more efficient defense. Furthermore, two studies ([71] and [85]) compared convolutional neural networks with hybrid CNN. Both studies proved that hybrid models present better results. This could be another way to encounter adversarial attacks and to create robust models.
On the other hand, adversarial samples can improve the generalizability of the models. Data augmentation with adversarial training can deal with some kind of noises that often occur in medical images. Hence, by trying to defend against adversarial attacks, we build more robust models that can work properly in many cases.
8. Conclusions
Deep learning has dramatically improved medical image analysis and it has become a crucial tool for doctors and hospitals. Nevertheless, adversarial attacks impede the proper functions of deep learning models and they create serious dangers for patients. In this paper, we summarize studies that investigate adversarial examples on medical image analysis. We conducted an inquiry into existing adversarial attacks, new attacks for medical images, and detection/defense mechanisms that are applied on medical image analysis. The phenomenon of adversarial attacks, is a new field in deep learning and especially in medical imaging. However, the studies have shown that these attacks are able to fool medical imaging models too.
Furthermore, some studies proposed new attacks, which are designed exclusively for the field of medical image analysis by presenting very high accuracy. On the other hand, some studies show how to face this phenomenon by detecting perturbed images, or by defending against these attacks. We believe that research community need to focus on tackling adversarial attacks so that the DL technology to be integrated in real world problems. As a future work we would like to compare all these attacks on a common database in order to conclude, which attack is the strongest and to evaluate the detection/defense methods. Also, we would like to examine all modalities under attack, in order to conclude which is the more robust and why. In addition, we want to study the robustness of deep learning models that have been built exclusively for medical images. Studying these cases, we will make safe assumptions on how to create safer models in the medical imaging domain.
Author Contributions
Conceptualization, G.A.P.; Methodology G.A.P. and K.D.A.; Investigation, K.D.A.; Resources, K.D.A.; Data curation, K.D.A.; Writing—original draft preparation, K.D.A.; Writing—review and editing, G.A.P.; Visualization, K.D.A.; Supervision, G.A.P.; Project administration, G.A.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
This work was supported by the MPhil program “Advanced Technologies in Informatics and Computers”, hosted by the Department of Computer Science, International Hellenic University, Kavala, Greece.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Geiger, A.; Lenz, P.; Stiller, C.; Urtasun, R. Vision meets robotics: The KITTI dataset. Int. J. Robot. Res. 2013, 32, 1231–1237. [Google Scholar] [CrossRef]
- Apostolidis, K.; Amanatidis, P.; Papakostas, G. Performance Evaluation of Convolutional Neural Networks for Gait Recognition. In Proceedings of the 24th Pan-Hellenic Conference on Informatics, Athens Greece, 20–22 November 2020; pp. 61–63. [Google Scholar] [CrossRef]
- Sidiropoulos, G.K.; Kiratsa, P.; Chatzipetrou, P.; Papakostas, G.A. Feature Extraction for Finger-Vein-Based Identity Recognition. J. Imaging 2021, 7, 89. [Google Scholar] [CrossRef] [PubMed]
- Filippidou, F.P.; Papakostas, G.A. Single Sample Face Recognition Using Convolutional Neural Networks for Automated Attendance Systems. In Proceedings of the 2020 Fourth International Conference on Intelligent Computing in Data Sciences (ICDS), Fez, Morocco, 21–23 October 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Shankar, K.; Zhang, Y.; Liu, Y.; Wu, L.; Chen, C.-H. Hyperparameter Tuning Deep Learning for Diabetic Retinopathy Fundus Image Classification. IEEE Access 2020, 8, 118164–118173. [Google Scholar] [CrossRef]
- Maliamanis, T.; Papakostas, G.A. Machine Learning Vulnerability in Medical Imaging. In Machine Learning, Big Data, and IoT for Medical Informatics, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2021; Available online: https://www.elsevier.com/books/machine-learning-big-data-and-iot-for-medical-informatics/xhafa/978-0-12-821777-1 (accessed on 4 June 2021).
- Tyukin, I.Y.; Higham, D.J.; Gorban, A.N. On Adversarial Examples and Stealth Attacks in Artificial Intelligence Systems. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Szegedy, C.; Zaremba, W.; Sutskever, I.; Bruna, J.; Erhan, D.; Goodfellow, I.; Fergus, R. Intriguing Properties of Neural Networks. arXiv 2014, arXiv:1312.6199. Available online: http://arxiv.org/abs/1312.6199 (accessed on 4 June 2021).
- Goodfellow, I.J.; Shlens, J.; Szegedy, C. Explaining and Harnessing Adversarial Examples. arXiv 2015, arXiv:1412.6572. Available online: http://arxiv.org/abs/1412.6572 (accessed on 4 June 2021).
- Schmidt, L.; Santurkar, S.; Tsipras, D.; Talwar, K.; Mądry, A. Adversarially Robust Generalization Requires More Data. arXiv 2018, arXiv:1804.11285. Available online: http://arxiv.org/abs/1804.11285 (accessed on 4 June 2021).
- Ilyas, A.; Santurkar, S.; Tsipras, D.; Engstrom, L.; Tran, B.; Madry, A. Adversarial Examples Are Not Bugs, They Are Features. arXiv 2019, arXiv:1905.02175. Available online: http://arxiv.org/abs/1905.02175 (accessed on 4 June 2021).
- Maliamanis, T.; Papakostas, G. Adversarial computer vision: A current snapshot. In Proceedings of the Twelfth International Conference on Machine Vision (ICMV 2019), Amsterdam, The Netherlands, 31 January 2020; p. 121. [Google Scholar] [CrossRef]
- Papernot, N.; McDaniel, P.; Goodfellow, I. Transferability in Machine Learning: From Phenomena to Black-Box Attacks using Adversarial Samples. arXiv 2016, arXiv:1605.07277. Available online: http://arxiv.org/abs/1605.07277 (accessed on 4 June 2021).
- Kurakin, A.; Goodfellow, I.; Bengio, S. Adversarial Examples in the Physical World. arXiv 2017, arXiv:1607.02533. Available online: http://arxiv.org/abs/1607.02533 (accessed on 4 June 2021).
- Xu, W.; Evans, D.; Qi, Y. Feature Squeezing: Detecting Adversarial Examples in Deep Neural Networks. In Proceedings of the 2018 Network and Distributed System Security Symposium, San Diego, CA, USA, 18–21 February 2018. [Google Scholar] [CrossRef]
- Meng, D.; Chen, H. MagNet: A Two-Pronged Defense against Adversarial Examples. arXiv 2017, arXiv:1705.09064. Available online: http://arxiv.org/abs/1705.09064 (accessed on 4 June 2021).
- Madry, A.; Makelov, A.; Schmidt, L.; Tsipras, D.; Vladu, A. Towards Deep Learning Models Resistant to Adversarial Attacks. arXiv 2019, arXiv:1706.06083. Available online: http://arxiv.org/abs/1706.06083 (accessed on 4 June 2021).
- Paschali, M.; Conjeti, S.; Navarro, F.; Navab, N. Generalizability vs. Robustness: Investigating Medical Imaging Networks Using Adversarial Examples. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 11070, pp. 493–501. [Google Scholar] [CrossRef]
- Mangaokar, N.; Pu, J.; Bhattacharya, P.; Reddy, C.K.; Viswanath, B. Jekyll: Attacking Medical Image Diagnostics using Deep Generative Models. In Proceedings of the 2020 IEEE European Symposium on Security and Privacy (EuroS&P), Genoa, Italy, 7–11 September 2020; pp. 139–157. [Google Scholar] [CrossRef]
- Finlayson, S.G.; Bowers, J.D.; Ito, J.; Zittrain, J.L.; Beam, A.L.; Kohane, I.S. Adversarial attacks on medical machine learning. Science 2019, 363, 1287–1289. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Niu, Y.; Gu, L.; Wang, Y.; Zhao, Y.; Bailey, J.; Lu, F. Understanding Adversarial Attacks on Deep Learning Based Medical Image Analysis Systems. arXiv 2020, arXiv:1907.10456. Available online: http://arxiv.org/abs/1907.10456 (accessed on 4 June 2021).
- Calivá, F.; Cheng, K.; Shah, R.; Pedoia, V. Adversarial Robust Training of Deep Learning MRI Reconstruction Models. arXiv 2021, arXiv:2011.00070. Available online: http://arxiv.org/abs/2011.00070 (accessed on 4 June 2021).
- Dimensions. Available online: https://app.dimensions.ai/discover/publication (accessed on 9 August 2021).
- Sipola, T.; Puuska, S.; Kokkonen, T. Model Fooling Attacks Against Medical Imaging: A Short Survey. ISIJ 2020, 46, 215–224. [Google Scholar] [CrossRef]
- Tian, B.; Guo, Q.; Juefei-Xu, F.; Chan, W.L.; Cheng, Y.; Li, X.; Xie, X.; Qin, S. Bias Field Poses a Threat to DNN-based X-Ray Recognition. arXiv 2021, arXiv:2009.09247. Available online: http://arxiv.org/abs/2009.09247 (accessed on 4 June 2021).
- Chen, C.; Qin, C.; Qiu, H.; Ouyang, C.; Wang, S.; Chen, L.; Tarroni, G.; Bai, W.; Rueckert, D. Realistic Adversarial Data Augmentation for MR Image Segmentation. arXiv 2020, arXiv:2006.13322. Available online: http://arxiv.org/abs/2006.13322 (accessed on 4 June 2021).
- Makary, M.A.; Daniel, M. Medical error—the third leading cause of death in the US. BMJ 2016, i2139. [Google Scholar] [CrossRef] [PubMed]
- Lichtenberg, F.R. The quality of medical care, behavioral risk factors, and longevity growth. Int. J. Health Care Financ. Econ. 2011, 11, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Beinfeld, M.T.; Gazelle, G.S. Diagnostic Imaging Costs: Are They Driving Up the Costs of Hospital Care? Radiology 2005, 235, 934–939. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, F.; Qi, Z.; Duan, K.; Xi, D.; Zhu, Y.; Zhu, H.; Xiong, H.; He, Q. A Comprehensive Survey on Transfer Learning. arXiv 2020, arXiv:1911.02685. Available online: http://arxiv.org/abs/1911.02685 (accessed on 4 June 2021). [CrossRef]
- Lo, S.-C.B.; Lou, S.-L.A.; Lin, J.; Freedman, M.T.; Chien, M.V.; Mun, S.K. Artificial convolution neural network techniques and applications for lung nodule detection. IEEE Trans. Med. Imaging 1995, 14, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv 2017, arXiv:1711.05225. Available online: http://arxiv.org/abs/1711.05225 (accessed on 4 June 2021).
- Korolev, S.; Safiullin, A.; Belyaev, M.; Dodonova, Y. Residual and Plain Convolutional Neural Networks for 3D Brain MRI Classification. arXiv 2017, arXiv:1701.06643. Available online: http://arxiv.org/abs/1701.06643 (accessed on 4 June 2021).
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. Available online: http://arxiv.org/abs/1409.1556 (accessed on 4 June 2021).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Chen, Y.-W.; Jain, L.C. (Eds.) Deep Learning in Healthcare: Paradigms and Applications; Springer International Publishing: Cham, Switzerland, 2020; Volume 171. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A Survey on Deep Learning in Medical Image Analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Payer, C.; Štern, D.; Bischof, H.; Urschler, M. Regressing Heatmaps for Multiple Landmark Localization Using CNNs. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016; Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W., Eds.; Springer International Publishing: Cham, Switzerland, 2016; Volume 9901, pp. 230–238. [Google Scholar] [CrossRef]
- Setio, A.A.A.; Traverso, A.; de Bel, T.; Berens, M.S.N.; van den Bogaard, C.; Cerello, P.; Chen, H.; Dou, Q.; Fantacci, M.E.; Geurts, B.; et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med. Image Anal. 2017, 42, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Setio, A.A.A.; Ciompi, F.; Litjens, G.; Gerke, P.; Jacobs, C.; van Riel, S.J.; Wille, M.M.W.; Naqibullah, M.; Sanchez, C.I.; van Ginneken, B. Pulmonary Nodule Detection in CT Images: False Positive Reduction Using Multi-View Convolutional Networks. IEEE Trans. Med. Imaging 2016, 35, 1160–1169. [Google Scholar] [CrossRef]
- Platania, R.; Shams, S.; Yang, S.; Zhang, J.; Lee, K.; Park, S.-J. Automated Breast Cancer Diagnosis Using Deep Learning and Region of Interest Detection (BC-DROID). In Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, Boston, MA, USA, 20–23 August 2017; pp. 536–543. [Google Scholar] [CrossRef]
- Horry, M.J.; Chakraborty, S.; Paul, M.; Ulhaq, A.; Pradhan, B.; Saha, M.; Shukla, N. COVID-19 Detection Through Transfer Learning Using Multimodal Imaging Data. IEEE Access 2020, 8, 149808–149824. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. Available online: http://arxiv.org/abs/1505.04597 (accessed on 4 June 2021).
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.-W.; Heng, P.A. H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation from CT Volumes. arXiv 2018, arXiv:1709.07330. Available online: http://arxiv.org/abs/1709.07330 (accessed on 4 June 2021). [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. arXiv 2016, arXiv:1606.04797. Available online: http://arxiv.org/abs/1606.04797 (accessed on 4 June 2021).
- Drozdzal, M.; Vorontsov, E.; Chartrand, G.; Kadoury, S.; Pal, C. The Importance of Skip Connections in Biomedical Image Segmentation. arXiv 2016, arXiv:1608.04117. Available online: http://arxiv.org/abs/1608.04117 (accessed on 4 June 2021).
- Jin, Q.; Meng, Z.; Pham, T.D.; Chen, Q.; Wei, L.; Su, R. DUNet: A deformable network for retinal vessel segmentation. Knowl. Based Syst. 2019, 178, 149–162. [Google Scholar] [CrossRef]
- Xu, H.; Ma, Y.; Liu, H.-C.; Deb, D.; Liu, H.; Tang, J.-L.; Jain, A.K. Adversarial Attacks and Defenses in Images, Graphs and Text: A Review. Int. J. Autom. Comput. 2020, 17, 151–178. [Google Scholar] [CrossRef]
- Ren, K.; Zheng, T.; Qin, Z.; Liu, X. Adversarial Attacks and Defenses in Deep Learning. Engineering 2020, 6, 346–360. [Google Scholar] [CrossRef]
- Carlini, N.; Wagner, D. Towards Evaluating the Robustness of Neural Networks. arXiv 2017, arXiv:1608.04644. Available online: http://arxiv.org/abs/1608.04644 (accessed on 4 June 2021).
- Papernot, N.; McDaniel, P.; Jha, S.; Fredrikson, M.; Celik, Z.B.; Swami, A. The Limitations of Deep Learning in Adversarial Settings. arXiv 2015, arXiv:1511.07528. Available online: http://arxiv.org/abs/1511.07528 (accessed on 4 June 2021).
- Moosavi-Dezfooli, S.-M.; Fawzi, A.; Fawzi, O.; Frossard, P. Universal Adversarial Perturbations. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 86–94. [Google Scholar] [CrossRef]
- Xie, C.; Wang, J.; Zhang, Z.; Zhou, Y.; Xie, L.; Yuille, A. Adversarial Examples for Semantic Segmentation and Object Detection. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 1378–1387. [Google Scholar] [CrossRef]
- Tramèr, F.; Kurakin, A.; Papernot, N.; Goodfellow, I.; Boneh, D.; McDaniel, P. Ensemble Adversarial Training: Attacks and Defenses. arXiv 2020, arXiv:1705.07204. Available online: http://arxiv.org/abs/1705.07204 (accessed on 4 June 2021).
- Xie, C.; Wang, J.; Zhang, Z.; Ren, Z.; Yuille, A. Mitigating Adversarial Effects Through Randomization. arXiv 2018, arXiv:1711.01991. Available online: http://arxiv.org/abs/1711.01991 (accessed on 4 June 2021).
- Guo, Y.; Zhang, C.; Zhang, C.; Chen, Y. Sparse DNNs with Improved Adversarial Robustness. arXiv 2019, arXiv:1810.09619. Available online: http://arxiv.org/abs/1810.09619 (accessed on 4 June 2021).
- Wang, Y.; Jha, S.; Chaudhuri, K. Analyzing the Robustness of Nearest Neighbors to Adversarial Examples. In Proceedings of the 35th International Conference on Machine Learning, Stockholm, Sweden, 10–15 July 2018; Volume 80, pp. 5133–5142. [Google Scholar]
- Liu, X.; Li, Y.; Wu, C.; Hsieh, C.-J. Adv-BNN: Improved Adversarial Defense through Robust Bayesian Neural Network. arXiv 2019, arXiv:1810.01279. Available online: http://arxiv.org/abs/1810.01279 (accessed on 4 June 2021).
- Xiao, C.; Deng, R.; Li, B.; Yu, F.; Liu, M.; Song, D. Characterizing Adversarial Examples Based on Spatial Consistency Information for Semantic Segmentation. In Computer Vision—ECCV 2018; Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 11214, pp. 220–237. [Google Scholar] [CrossRef]
- Ma, X.; Li, B.; Wang, Y.; Erfani, S.M.; Wijewickrema, S.; Schoenebeck, G.; Song, D.; Houle, M.E.; Bailey, J. Characterizing Adversarial Subspaces Using Local Intrinsic Dimensionality. arXiv 2018, arXiv:1801.02613. Available online: http://arxiv.org/abs/1801.02613 (accessed on 4 June 2021).
- Metzen, J.H.; Genewein, T.; Fischer, V.; Bischoff, B. On Detecting Adversarial Perturbations. arXiv 2017, arXiv:1702.04267. Available online: http://arxiv.org/abs/1702.04267 (accessed on 4 June 2021).
- Finlayson, S.G.; Chung, H.W.; Kohane, I.S.; Beam, A.L. Adversarial Attacks Against Medical Deep Learning Systems. arXiv 2019, arXiv:1804.05296. Available online: http://arxiv.org/abs/1804.05296 (accessed on 4 June 2021).
- FDA Permits Marketing of Artificial Intelligence-Based Device to Detect Certain Diabetes-Related Eye. Available online: https://www.healthcare.digital/single-post/2018/04/20/fda-permits-marketing-of-artificial-intelligence-based-device-to-detect-certain-diabetes (accessed on 4 June 2021).
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. Available online: http://arxiv.org/abs/1704.04861 (accessed on 4 June 2021).
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Jegou, S.; Drozdzal, M.; Vazquez, D.; Romero, A.; Bengio, Y. The One Hundred Layers Tiramisu: Fully Convolutional DenseNets for Semantic Segmentation. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Honolulu, HI, USA, 21–26 July 2017; pp. 1175–1183. [Google Scholar] [CrossRef]
- Wetstein, S.C.; González-Gonzalo, C.; Bortsova, G.; Liefers, B.; Dubost, F.; Katramados, I.; Hogeweg, L.; van Ginneken, B.; Pluim, J.P.W.; de Bruijne, M.; et al. Adversarial Attack Vulnerability of Medical Image Analysis Systems: Unexplored Factors. arXiv 2020, arXiv:2006.06356. Available online: http://arxiv.org/abs/2006.06356 (accessed on 4 June 2021).
- Cheng, G.; Ji, H. Adversarial Perturbation on MRI Modalities in Brain Tumor Segmentation. IEEE Access 2020, 8, 206009–206015. [Google Scholar] [CrossRef]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Bermudez, C.; Chen, Y.; Landman, B.A.; Vorobeychik, Y. Anatomical context protects deep learning from adversarial perturbations in medical imaging. Neurocomputing 2020, 379, 370–378. [Google Scholar] [CrossRef]
- Huq, A.; Pervin, M.T. Analysis of Adversarial Attacks on Skin Cancer Recognition. In Proceedings of the 2020 International Conference on Data Science and Its Applications (ICoDSA), Bandung, Indonesia, 5–6 August 2020; pp. 1–4. [Google Scholar] [CrossRef]
- On the Assessment of Robustness of Telemedicine Applications against Adversarial Machine Learning Attacks | SpringerLink. Available online: https://link.springer.com/chapter/10.1007/978-3-030-79457-6_44?error=cookies_not_supported&code=3acd5697-d1ba-4ca5-8077-3d1b5d9bae9a (accessed on 10 August 2021).
- Pal, B.; Gupta, D.; Rashed-Al-Mahfuz, M.; Alyami, S.A.; Moni, M.A. Vulnerability in Deep Transfer Learning Models to Adversarial Fast Gradient Sign Attack for COVID-19 Prediction from Chest Radiography Images. Appl. Sci. 2021, 11, 4233. [Google Scholar] [CrossRef]
- Bortsova, G.; Dubost, F.; Hogeweg, L.; Katramados, I.; de Bruijne, M. Adversarial Heart Attack: Neural Networks Fooled to Segment Heart Symbols in Chest X-Ray Images. arXiv 2021, arXiv:2104.00139. Available online: http://arxiv.org/abs/2104.00139 (accessed on 10 August 2021).
- Anand, D.; Tank, D.; Tibrewal, H.; Sethi, A. Self-Supervision vs. Transfer Learning: Robust Biomedical Image Analysis Against Adversarial Attacks. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1159–1163. [Google Scholar] [CrossRef]
- Mendeley Data—Labeled Optical Coherence Tomography (OCT) and Chest X-Ray Images for Classification. Available online: https://data.mendeley.com/datasets/rscbjbr9sj/2 (accessed on 5 June 2021).
- Petitjean, C.; Zuluaga, M.A.; Bai, W.; Dacher, J.-N.; Grosgeorge, D.; Caudron, J.; Ruan, S.; Ayed, I.B.; Cardoso, M.J.; Chen, H.-C.; et al. Right ventricle segmentation from cardiac MRI: A collation study. Med. Image Anal. 2015, 19, 187–202. [Google Scholar] [CrossRef]
- Noroozi, M.; Favaro, P. Unsupervised Learning of Visual Representations by Solving Jigsaw Puzzles. arXiv 2017, arXiv:1603.09246. Available online: http://arxiv.org/abs/1603.09246 (accessed on 4 June 2021).
- Paul, R.; Schabath, M.; Gillies, R.; Hall, L.; Goldgof, D. Mitigating Adversarial Attacks on Medical Image Understanding Systems. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1517–1521. [Google Scholar] [CrossRef]
- Paul, R.; Hawkins, S.H.; Schabath, M.B.; Gillies, R.J.; Hall, L.O.; Goldgof, D.B. Predicting malignant nodules by fusing deep features with classical radiomics features. J. Med. Imaging 2018, 5, 1. [Google Scholar] [CrossRef]
- Risk Susceptibility of Brain Tumor Classification to Adversarial Attacks|SpringerLink. Available online: https://link.springer.com/chapter/10.1007/978-3-030-31964-9_17 (accessed on 4 June 2021).
- Miyato, T.; Maeda, S.; Koyama, M.; Nakae, K.; Ishii, S. Distributional Smoothing with Virtual Adversarial Training. arXiv 2016, arXiv:1507.00677. Available online: http://arxiv.org/abs/1507.00677 (accessed on 4 June 2021).
- Brain Tumor Dataset. Available online: https://figshare.com/articles/dataset/brain_tumor_dataset/1512427 (accessed on 4 June 2021).
- Shah, A.; Lynch, S.; Niemeijer, M.; Amelon, R.; Clarida, W.; Folk, J.; Russell, S.; Wu, X.; Abramoff, M.D. Susceptibility to misdiagnosis of adversarial images by deep learning based retinal image analysis algorithms. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 1454–1457. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Paszke, A.; Chaurasia, A.; Kim, S.; Culurciello, E. ENet: A Deep Neural Network Architecture for Real-Time Semantic Segmentation. arXiv 2016, arXiv:1606.02147. Available online: http://arxiv.org/abs/1606.02147 (accessed on 4 June 2021).
- Abràmoff, M.D.; Lou, Y.; Erginay, A.; Clarida, W.; Amelon, R.; Folk, J.C.; Niemeijer, M. Improved Automated Detection of Diabetic Retinopathy on a Publicly Available Dataset Through Integration of Deep Learning. Invest. Ophthalmol. Vis. Sci. 2016, 57, 5200. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, V.; Voynov, D. Influence of Control Parameters and the Size of Biomedical Image Datasets on the Success of Adversarial Attacks. In Pattern Recognition and Information Processing; Ablameyko, S.V., Krasnoproshin, V.V., Lukashevich, M.M., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 1055, pp. 301–311. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, Z.; Zhou, Y.; Xia, Y.; Shen, W.; Fishman, E.K.; Yuille, A.L. Volumetric Medical Image Segmentation: A 3D Deep Coarse-to-fine Framework and Its Adversarial Examples. arXiv 2019, arXiv:2010.16074. Available online: http://arxiv.org/abs/2010.16074 (accessed on 4 June 2021). [CrossRef]
- Roth, H.R.; Lu, L.; Farag, A.; Shin, H.-C.; Liu, J.; Turkbey, E.; Summers, R.M. DeepOrgan: Multi-level Deep Convolutional Networks for Automated Pancreas Segmentation. arXiv 2015, arXiv:1506.06448. Available online: http://arxiv.org/abs/1506.06448 (accessed on 4 June 2021).
- Zhou, Y.; Xie, L.; Fishman, E.K.; Yuille, A.L. Deep Supervision for Pancreatic Cyst Segmentation in Abdominal CT Scans. arXiv 2017, arXiv:1706.07346. Available online: http://arxiv.org/abs/1706.07346 (accessed on 4 June 2021).
- Allyn, J.; Allou, N.; Vidal, C.; Renou, A.; Ferdynus, C. Adversarial attack on deep learning-based dermatoscopic image recognition systems: Risk of misdiagnosis due to undetectable image perturbations. Medicine 2020, 99, e23568. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef]
- Hirano, H.; Minagi, A.; Takemoto, K. Universal adversarial attacks on deep neural networks for medical image classification. BMC Med. Imaging 2021, 21, 9. [Google Scholar] [CrossRef] [PubMed]
- Hirano, H.; Koga, K.; Takemoto, K. Vulnerability of deep neural networks for detecting COVID-19 cases from chest X-ray images to universal adversarial attacks. PLoS ONE 2020, 15, e0243963. [Google Scholar] [CrossRef]
- Wang, L.; Lin, Z.Q.; Wong, A. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-ray8: Hospital-Scale Chest X-Ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Honolulu, HI, USA, 21–26 July 2017; pp. 2097–2106. [Google Scholar] [CrossRef]
- ISIC Archive. Available online: https://www.isic-archive.com/ (accessed on 5 June 2021).
- Diabetic Retinopathy Detection|Kaggle. Available online: https://www.kaggle.com/c/diabetic-retinopathy-detection (accessed on 5 June 2021).
- Feinman, R.; Curtin, R.R.; Shintre, S.; Gardner, A.B. Detecting Adversarial Samples from Artifacts. arXiv 2017, arXiv:1703.00410. Available online: http://arxiv.org/abs/1703.00410 (accessed on 4 June 2021).
- Byra, M.; Styczynski, G.; Szmigielski, C.; Kalinowski, P.; Michalowski, L.; Paluszkiewicz, R.; Ziarkiewicz-Wroblewska, B.; Zieniewicz, K.; Nowicki, A. Adversarial Attacks on Deep Learning Models for Fatty Liver Disease Classification by Modification of Ultrasound Image Reconstruction Method. arXiv 2020, arXiv:2009.03364. Available online: http://arxiv.org/abs/2009.03364 (accessed on 4 June 2021).
- Chen, P.-Y.; Zhang, H.; Sharma, Y.; Yi, J.; Hsieh, C.-J. ZOO: Zeroth Order Optimization Based Black-box Attacks to Deep Neural Networks without Training Substitute Models. In Proceedings of the 10th ACM Workshop on Artificial Intelligence and Security, Dallas, TX, USA, 3 November 2017; pp. 15–26. [Google Scholar] [CrossRef]
- Ozbulak, U.; Van Messem, A.; De Neve, W. Impact of Adversarial Examples on Deep Learning Models for Biomedical Image Segmentation. arXiv 2019, arXiv:1907.13124. Available online: http://arxiv.org/abs/1907.13124 (accessed on 4 June 2021).
- Pena-Betancor, C.; Gonzalez-Hernandez, M.; Fumero-Batista, F.; Sigut, J.; Medina-Mesa, E.; Alayon, S.; Gonzalez de la Rosa, M. Estimation of the Relative Amount of Hemoglobin in the Cup and Neuroretinal Rim Using Stereoscopic Color Fundus Images. Investig. Ophthalmol. Vis. Sci. 2015, 56, 1562–1568. [Google Scholar] [CrossRef] [PubMed]
- Codella, N.C.F.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin Lesion Analysis Toward Melanoma Detection: A Challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), Hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2018, arXiv:1710.05006. Available online: http://arxiv.org/abs/1710.05006 (accessed on 4 June 2021).
- Chen, L.; Bentley, P.; Mori, K.; Misawa, K.; Fujiwara, M.; Rueckert, D. Intelligent Image Synthesis to Attack a Segmentation CNN Using Adversarial Learning. arXiv 2019, arXiv:1909.11167. Available online: http://arxiv.org/abs/1909.11167 (accessed on 4 June 2021).
- Kugler, D. Physical Attacks in Dermoscopy: An Evaluation of Robustness for clinical Deep-Learning. J. Mach. Learn. Biomed. Imaging 2021, 7, 1–32. [Google Scholar]
- Yao, Q.; He, Z.; Lin, Y.; Ma, K.; Zheng, Y.; Zhou, S.K. A Hierarchical Feature Constraint to Camouflage Medical Adversarial Attacks. arXiv 2021, arXiv:2012.09501. Available online: http://arxiv.org/abs/2012.09501 (accessed on 4 June 2021).
- Shao, M.; Zhang, G.; Zuo, W.; Meng, D. Target attack on biomedical image segmentation model based on multi-scale gradients. Inf. Sci. 2021, 554, 33–46. [Google Scholar] [CrossRef]
- REFUGE Challenge: A unified framework for evaluating automated methods for glaucoma assessment from fundus photographs—ScienceDirect. Available online: https://www.sciencedirect.com/science/article/abs/pii/S1361841519301100 (accessed on 10 August 2021).
- Alom, M.Z.; Yakopcic, C.; Hasan, M.; Taha, T.M.; Asari, V.K. Recurrent residual U-Net for medical image segmentation. J. Med. Imag. 2019, 6, 1. [Google Scholar] [CrossRef]
- Attention U-Net: Learning Where to Look for the Pancreas. Available online: https://arxiv.org/abs/1804.03999 (accessed on 10 August 2021).
- Qi, G.; Gong, L.; Song, Y.; Ma, K.; Zheng, Y. Stabilized Medical Image Attacks. arXiv 2021, arXiv:2103.05232. Available online: http://arxiv.org/abs/2103.05232 (accessed on 10 August 2021).
- Semi-Supervised Classification with Graph Convolutional Networks. Available online: https://arxiv.org/abs/1609.02907 (accessed on 10 August 2021).
- Attentive CT Lesion Detection Using Deep Pyramid Inference with Multi-scale Booster|SpringerLink. Available online: https://link.springer.com/chapter/10.1007/978-3-030-32226-7_34?error=cookies_not_supported&code=b33ccaa9-9f15-438c-8f5b-b2aabb1aa1fa (accessed on 10 August 2021).
- Wu, D.; Liu, S.; Ban, J. Classification of Diabetic Retinopathy Using Adversarial Training. IOP Conf. Ser. Mater. Sci. Eng. 2020, 806, 012050. [Google Scholar] [CrossRef]
- He, X.; Yang, S.; Li, G.; Li, H.; Chang, H.; Yu, Y. Non-Local Context Encoder: Robust Biomedical Image Segmentation against Adversarial Attacks. AAAI 2019, 33, 8417–8424. [Google Scholar] [CrossRef]
- Novikov, A.A.; Lenis, D.; Major, D.; Hladůvka, J.; Wimmer, M.; Bühler, K. Fully Convolutional Architectures for Multi-Class Segmentation in Chest Radiographs. arXiv 2018, arXiv:1701.08816. Available online: http://arxiv.org/abs/1701.08816 (accessed on 4 June 2021).
- Sarker, M.M.K.; Rashwan, H.A.; Akram, F.; Banu, S.F.; Saleh, A.; Singh, V.K.; Chowdhury, F.U.H.; Abdulwahab, S.; Romani, S.; Radeva, P.; et al. SLSDeep: Skin Lesion Segmentation Based on Dilated Residual and Pyramid Pooling Networks. arXiv 2018, arXiv:1805.10241. Available online: http://arxiv.org/abs/1805.10241 (accessed on 4 June 2021).
- Hwang, S.; Park, S. Accurate Lung Segmentation via Network-Wise Training of Convolutional Networks. arXiv 2017, arXiv:1708.00710. Available online: http://arxiv.org/abs/1708.00710 (accessed on 4 June 2021).
- Yuan, Y. Automatic skin lesion segmentation with fully convolutional-deconvolutional networks. IEEE J. Biomed. Health Inform. 2019, 23, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Taghanaki, S.A.; Das, A.; Hamarneh, G. Vulnerability Analysis of Chest X-Ray Image Classification Against Adversarial Attacks. arXiv 2018, arXiv:1807.02905. Available online: http://arxiv.org/abs/1807.02905 (accessed on 4 June 2021).
- Ren, X.; Zhang, L.; Wei, D.; Shen, D.; Wang, Q. Brain MR Image Segmentation in Small Dataset with Adversarial Defense and Task Reorganization. In Machine Learning in Medical Imaging; Suk, H.-I., Liu, M., Yan, P., Lian, C., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 11861, pp. 1–8. [Google Scholar] [CrossRef]
- Wang, G.; Li, W.; Ourselin, S.; Vercauteren, T. Automatic Brain Tumor Segmentation using Cascaded Anisotropic Convolutional Neural Networks. arXiv 2018, arXiv:1709.00382. Available online: http://arxiv.org/abs/1709.00382 (accessed on 4 June 2021). [CrossRef]
- Pervin, M.T.; Tao, L.; Huq, A.; He, Z.; Huo, L. Adversarial Attack Driven Data Augmentation for Accurate and Robust Medical Image Segmentation. arXiv 2021, arXiv:2105.12106. Available online: http://arxiv.org/abs/2105.12106 (accessed on 10 August 2021).
- Liu, S.; Setio, A.A.A.; Ghesu, F.C.; Gibson, E.; Grbic, S.; Georgescu, B.; Comaniciu, D. No Surprises: Training Robust Lung Nodule Detection for Low-Dose CT Scans by Augmenting with Adversarial Attacks. arXiv 2020, arXiv:2003.03824. Available online: http://arxiv.org/abs/2003.03824 (accessed on 4 June 2021). [CrossRef] [PubMed]
- National Lung Screening Trial Research Team the National Lung Screening Trial: Overview and Study Design. Radiology 2011, 258, 243–253. [CrossRef] [PubMed]
- Vatian, A.; Gusarova, N.; Dobrenko, N.; Dudorov, S.; Nigmatullin, N.; Shalyto, A.; Lobantsev, A. Impact of Adversarial Examples on the Efficiency of Interpretation and Use of Information from High-Tech Medical Images. In Proceedings of the 2019 24th Conference of Open Innovations Association (FRUCT), Moscow, Russia, 8–12 April 2019; pp. 472–478. [Google Scholar] [CrossRef]
- The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A Completed Reference Database of Lung Nodules on CT Scans. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3041807/ (accessed on 5 June 2021).
- Dvořák, P.; Menze, B. Local Structure Prediction with Convolutional Neural Networks for Multimodal Brain Tumor Segmentation. In Medical Computer Vision: Algorithms for Big Data; Menze, B., Langs, G., Montillo, A., Kelm, M., Müller, H., Zhang, S., Cai, W., Metaxas, D., Eds.; Springer International Publishing: Cham, Switzerland, 2016; Volume 9601, pp. 59–71. [Google Scholar] [CrossRef]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.-A.; Cetin, I.; Lekadir, K.; Camara, O.; Gonzalez Ballester, M.A.; et al. Deep Learning Techniques for Automatic MRI Cardiac Multi-Structures Segmentation and Diagnosis: Is the Problem Solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef]
- Cheng, K.; Caliva, F.; Shah, R.; Han, M.; Majumdar, S.; Pedoia, V. Addressing the False Negative Problem of Deep Learning MRI Reconstruction Models by Adversarial Attacks and Robust Training. Proc. Mach. Learn. Res. 2020, 121, 121–135. [Google Scholar]
- Putzky, P.; Welling, M. Invert to Learn to Invert. arXiv 2019, arXiv:1911.10914. Available online: http://arxiv.org/abs/1911.10914 (accessed on 4 June 2021).
- Park, H.; Bayat, A.; Sabokrou, M.; Kirschke, J.S.; Menze, B.H. Robustification of Segmentation Models Against Adversarial Perturbations in Medical Imaging. arXiv 2020, arXiv:2009.11090. Available online: http://arxiv.org/abs/2009.11090 (accessed on 4 June 2021).
- Marcus, D.S.; Wang, T.H.; Parker, J.; Csernansky, J.G.; Morris, J.C.; Buckner, R.L. Open Access Series of Imaging Studies (OASIS): Cross-sectional MRI Data in Young, Middle Aged, Nondemented, and Demented Older Adults. J. Cogn. Neurosci. 2007, 19, 1498–1507. [Google Scholar] [CrossRef]
- Taghanaki, S.A.; Abhishek, K.; Azizi, S.; Hamarneh, G. A Kernelized Manifold Mapping to Diminish the Effect of Adversarial Perturbations. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 16–20 June 2019; pp. 11332–11341. [Google Scholar] [CrossRef][Green Version]
- Uwimana1, A.; Senanayake, R. Out of Distribution Detection and Adversarial Attacks on Deep Neural Networks for Robust Medical Image Analysis. arXiv 2021, arXiv:2107.04882. Available online: http://arxiv.org/abs/2107.04882 (accessed on 10 August 2021).
- Daza, L.; Pérez, J.C.; Arbeláez, P. Towards Robust General Medical Image Segmentation. arXiv 2021, arXiv:2107.04263. Available online: http://arxiv.org/abs/2107.04263 (accessed on 10 August 2021).
- Reliable Evaluation of Adversarial Robustness with an Ensemble of Diverse Parameter-Free Attacks. Available online: http://proceedings.mlr.press/v119/croce20b.html (accessed on 10 August 2021).
- Huang, Y.; Würfl, T.; Breininger, K.; Liu, L.; Lauritsch, G.; Maier, A. Some Investigations on Robustness of Deep Learning in Limited Angle Tomography. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 11070, pp. 145–153. [Google Scholar] [CrossRef]
- Xue, F.-F.; Peng, J.; Wang, R.; Zhang, Q.; Zheng, W.-S. Improving Robustness of Medical Image Diagnosis with Denoising Convolutional Neural Networks. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019; Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.-T., Khan, A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 11769, pp. 846–854. [Google Scholar] [CrossRef]
- Tripathi, A.M.; Mishra, A. Fuzzy Unique Image Transformation: Defense Against Adversarial Attacks on Deep COVID-19 Models. arXiv 2020, arXiv:2009.04004. Available online: http://arxiv.org/abs/2009.04004 (accessed on 4 June 2021).
- Defending Deep Learning-Based Biomedical Image Segmentation from Adversarial Attacks: A Low-Cost Frequency Refinement Approach|SpringerLink. Available online: https://link.springer.com/chapter/10.1007/978-3-030-59719-1_34 (accessed on 4 June 2021).
- Xu, M.; Zhang, T.; Li, Z.; Liu, M.; Zhang, D. Towards evaluating the robustness of deep diagnostic models by adversarial attack. Med. Image Anal. 2021, 69, 101977. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zou, D.; Yi, J.; Bailey, J.; Ma, X.; Gu, Q. Improving adversarial robustness requires revisiting misclassified examples. In Proceedings of the International Conference on Learning Representations, Virtual, 27–30 April 2020. [Google Scholar]
- Li, X.; Zhu, D. Robust Detection of Adversarial Attacks on Medical Images. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1154–1158. [Google Scholar] [CrossRef]
- Li, X.; Pan, D.; Zhu, D. Defending against Adversarial Attacks on Medical Imaging AI System, Classification or Detection? arXiv 2020, arXiv:2006.13555. Available online: http://arxiv.org/abs/2006.13555 (accessed on 4 June 2021).
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131.e9. [Google Scholar] [CrossRef]
- Lee, S.; Lee, H.; Yoon, S. Adversarial Vertex Mixup: Toward Better Adversarially Robust Generalization. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 14–19 June 2020; pp. 269–278. [Google Scholar] [CrossRef]
- Liu, X.; Xiao, T.; Si, S.; Cao, Q.; Kumar, S.; Hsieh, C.-J. How Does Noise Help Robustness? Explanation and Exploration under the Neural SDE Framework. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 14–19 June 2020; pp. 279–287. [Google Scholar] [CrossRef]
- Qin, Y.; Wang, X.; Beutel, A.; Chi, E.H. Improving Uncertainty Estimates through the Relationship with Adversarial Robustness. arXiv 2020, arXiv:2006.16375. Available online: http://arxiv.org/abs/2006.16375 (accessed on 10 August 2021).
- Yi, M.; Hou, L.; Sun, J.; Shang, L.; Jiang, X.; Liu, Q.; Ma, Z.-M. Improved OOD Generalization via Adversarial Training and Pre-training. arXiv 2021, arXiv:2105.11144. Available online: http://arxiv.org/abs/2105.11144 (accessed on 10 August 2021).
- Papernot, N.; Faghri, F.; Carlini, N.; Goodfellow, I.; Feinman, R.; Kurakin, A.; Xie, C.; Sharma, Y.; Brown, T.; Roy, A.; et al. Technical Report on the CleverHans v2.1.0 Adversarial Examples Library. arXiv 2018, arXiv:1610.00768. Available online: http://arxiv.org/abs/1610.00768 (accessed on 4 June 2021).
- Rauber, J.; Zimmermann, R.; Bethge, M.; Brendel, W. Foolbox Native: Fast adversarial attacks to benchmark the robustness of machine learning models in PyTorch, TensorFlow, and JAX. JOSS 2020, 5, 2607. [Google Scholar] [CrossRef]
- Nicolae, M.-I.; Sinn, M.; Tran, M.N.; Buesser, B.; Rawat, A.; Wistuba, M.; Zantedeschi, V.; Baracaldo, N.; Chen, B.; Ludwig, H.; et al. Adversarial Robustness Toolbox v1.0.0. arXiv 2019, arXiv:1807.01069. Available online: http://arxiv.org/abs/1807.01069 (accessed on 4 June 2021).
- Goodman, D.; Xin, H.; Yang, W.; Yuesheng, W.; Junfeng, X.; Huan, Z. Advbox: A Toolbox to Generate Adversarial Examples that Fool Neural Networks. arXiv 2020, arXiv:2001.05574. Available online: http://arxiv.org/abs/2001.05574 (accessed on 4 June 2021).
- Ding, G.W.; Wang, L.; Jin, X. advertorch v0.1: An Adversarial Robustness Toolbox based on PyTorch. arXiv 2019, arXiv:1902.07623. Available online: http://arxiv.org/abs/1902.07623 (accessed on 4 June 2021).
- Ling, X.; Ji, S.; Zou, J.; Wang, J.; Wu, C.; Li, B.; Wang, T. DEEPSEC: A Uniform Platform for Security Analysis of Deep Learning Model. In Proceedings of the 2019 IEEE Symposium on Security and Privacy (SP), San Francisco, CA, USA, 19–23 May 2019; pp. 673–690. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).