A Deep Learning Model with Self-Supervised Learning and Attention Mechanism for COVID-19 Diagnosis Using Chest X-ray Images
Abstract
:1. Introduction
- Firstly, self-supervised learning is introduced to prevent the overfitting problem caused by the limited number of training images in deep learning. Models Genesis [13] was modified by adding convolutional attention module and trained using 112,120 unlabeled CXR dataset. And it was fine-tuned on a COVID-19 data set containing 1821 X-ray images. The accuracy of our model is 98.6%.
- We improved the performance of Models Genesis by adding a convolutional attention module after every convolutional layer. We conducted extensive experiments in which we compared the performance of the modified Models Genesis containing the attention modules with that of the original for the COVID-19 classification.
- For qualitative evaluation of model results, we considered a visually explainable AI approach, Score-CAM [14]. By using it, we investigated how the proposed model makes correct/incorrect classifications to identify critical factors related to COVID-19 cases. The Score-CAM used in this paper is an improved method which resolve issues of the Grad-CAM [15].
2. Materials and Methods
2.1. Datasets
2.2. Existing Models
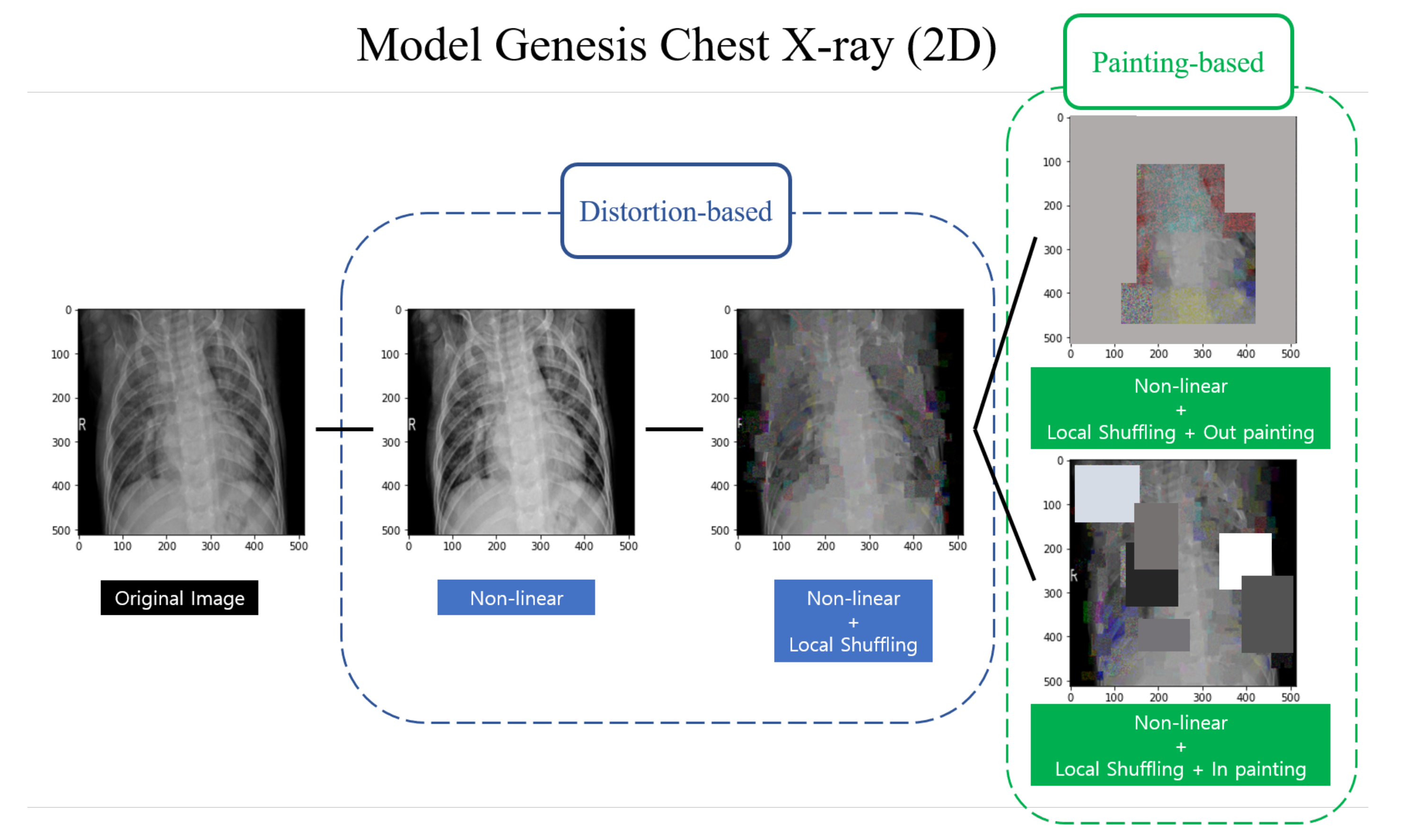
2.3. Self-Supervised Learning
2.4. Convolutional Attention Module
2.5. Our Proposed System
2.5.1. Self-Supervised Learning with Convolutional Attention Module
2.5.2. Fine-Tuing the Encoder
3. Experimental Study
3.1. Experimental Details
3.2. Experimental Results
4. Discussion
4.1. AI over RT-PCR Using CXR
4.2. Interpretation of Classification Results Using Score-Cam
4.3. Comparison with Other Methods for COVID-19 Classification
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.; Zhang, H.; Xie, J.; Lin, M.; Ying, L.; Pang, P.; Ji, W. Sensitivity of chest CT for COVID-19: Comparison to RT-PCR. Radiology 2020, 296, 115–117. [Google Scholar] [CrossRef]
- Wikramaratna, P.; Paton, R.S.; Ghafari, M.; Lourenco, J. Estimating false-negative detection rate of SARS-CoV-2 by RT-PCR. Euro Surveill. 2020, 25, 2000568. [Google Scholar] [CrossRef] [PubMed]
- Pham, D.T.; Pham, P.T.N. Artificial intelligence in engineering. Int. J. Mach. Tools Manuf. 1999, 39, 937–949. [Google Scholar] [CrossRef]
- Dirican, C. The impacts of robotics, artificial intelligence on business and economics. Procedia-Soc. Behav. Sci. 2015, 195, 564–573. [Google Scholar] [CrossRef] [Green Version]
- Parveen, N.; Sathik, M.M. Detection of pneumonia in chest X-ray images. J. Ray Sci. Technol. 2011, 19, 423–428. [Google Scholar] [CrossRef]
- Farooq, M.; Hafeez, A. Covid-resnet: A deep learning framework for screening of covid19 from radiographs. arXiv 2020, arXiv:2003.14395. [Google Scholar]
- Narin, A.; Kaya, C.; Pamuk, Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. arXiv 2020, arXiv:2003.10849. [Google Scholar]
- Oh, Y.; Park, S.; Ye, J.C. Deep learning covid-19 features on cxr using limited training data sets. IEEE Trans. Med. Imaging 2020, 39, 2688–2700. [Google Scholar] [CrossRef]
- Minaee, S.; Kafieh, R.; Sonka, M.; Yazdani, S.; Soufi, G.J. Deep-covid: Predicting covid-19 from chest x-ray images using deep transfer learning. Med. Image Anal. 2020, 65, 101794. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.S.; Kim, J.Y.; Jeon, E.T.; Choi, W.S.; Kim, N.H.; Lee, K.Y. Evaluation of Scalability and Degree of Fine-Tuning of Deep Convolutional Neural Networks for COVID-19 Screening on Chest X-ray Images Using Explainable Deep-Learning Algorithm. J. Pers. Med. 2020, 10, 213. [Google Scholar] [CrossRef]
- Mikołajczyk, A.; Grochowski, M. Data augmentation for improving deep learning in image classification problem. In Proceedings of the 2018 International Interdisciplinary PhD Workshop (IIPhDW), Swinoujscie, Poland, 9–12 May 2018; pp. 117–122. [Google Scholar]
- Zhou, Z.; Sodha, V.; Siddiquee, M.M.R.; Feng, R.; Tajbakhsh, N.; Gotway, M.B.; Liang, J. Models genesis: Generic autodidactic models for 3d medical image analysis. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; pp. 384–393. [Google Scholar]
- Wang, H.; Wang, Z.; Du, M.; Yang, F.; Zhang, Z.; Ding, S.; Mardziel, P.; Hu, X. Score-CAM: Score-weighted visual explanations for convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, 14–19 June 2020; pp. 24–25. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; So, K.I. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Jaeger, S.; Candemir, S.; Antani, S.; Wáng, Y.X.J.; Lu, P.X.; Thoma, G. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg. 2014, 4, 475. [Google Scholar] [PubMed]
- Cohen, J.P.; Morrison, P.; Dao, L.; Roth, K.; Duong, T.Q.; Ghassemi, M. Covid-19 image data collection: Prospective predictions are the future. arXiv 2020, arXiv:2006.11988. [Google Scholar]
- Chung, A. Figure 1 COVID-19 Chest X-ray Data Initiative. 2020. Available online: https://github.com/agchung/Figure1-COVID-chestxray-dataset (accessed on 4 May 2020).
- Chung, A. Actualmed COVID-19 Chest X-ray Data Initiative. 2020. Available online: https://github.com/agchung/Actualmed-COVID-chestxray-dataset (accessed on 6 May 2020).
- Rahman, T.; Chowdhury, M.; Khandakar, A. COVID-19 Radiography Database; Kaggle: San Francisco, CA, USA, 2020. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Das, S.; Roy, S.D.; Malakar, S.; Velásquez, J.D.; Sarkar, R. Bi-Level Prediction Model for Screening COVID-19 Patients Using Chest X-Ray Images. Big Data Res. 2021, 25, 100233. [Google Scholar] [CrossRef]
- Rahimzadeh, M.; Attar, A. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2. Inform. Med. Unlocked 2020, 19, 100360. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in deep residual networks. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 8–16 October 2016; pp. 630–645. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Rahaman, M.M.; Li, C.; Yao, Y.; Kulwa, F.; Rahman, M.A.; Wang, Q.; Qi, S.; Kong, F.; Zhu, X.; Zhao, X. Identification of COVID-19 samples from chest X-ray images using deep learning: A comparison of transfer learning approaches. J. Ray Sci. Technol. 2020, 1–19, in preprint. [Google Scholar] [CrossRef]
- Rehman, A.; Naz, S.; Khan, A.; Zaib, A.; Razzak, I. Improving coronavirus (COVID-19) diagnosis using deep transfer learning. medRxiv 2020. Available online: https://www.medrxiv.org/content/early/2020/04/17/2020.04.11.20054643.full.pdf (accessed on 17 August 2021).
- Wong, H.Y.F.; Lam, H.Y.S.; Fong, A.H.T.; Leung, S.T.; Chin, T.W.Y.; Lo, C.S.Y.; Lui, M.M.S.; Lee, J.C.Y.; Chiu, K.W.H.; Chung, T.W.H.; et al. Frequency and distribution of chest radiographic findings in patients positive for COVID-19. Radiology 2020, 296, E72–E78. [Google Scholar] [CrossRef] [Green Version]
- Gidaris, S.; Singh, P.; Komodakis, N. Unsupervised representation learning by predicting image rotations. arXiv 2018, arXiv:1803.07728. [Google Scholar]
- Zhai, X.; Oliver, A.; Kolesnikov, A.; Beyer, L. S4l: Self-supervised semi-supervised learning. In Proceedings of the IEEE International Conference on Computer Vision, Seoul, Korea, 27 October–3 November 2019; pp. 1476–1485. [Google Scholar]
- Larsson, G.; Maire, M.; Shakhnarovich, G. Learning representations for automatic colorization. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 8–16 October 2016; pp. 577–593. [Google Scholar]
- Zhang, R.; Isola, P.; Efros, A.A. Colorful image colorization. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 8–16 October 2016; pp. 649–666. [Google Scholar]
- Hendrycks, D.; Mazeika, M.; Kadavath, S.; Song, D. Using self-supervised learning can improve model robustness and uncertainty. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 2712–2721. [Google Scholar]
- Itti, L.; Koch, C.; Niebur, E. A model of saliency-based visual attention for rapid scene analysis. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20, 1254–1259. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 2921–2929. [Google Scholar]
- Channappayya, S.S.; Bovik, A.C.; Heath, R.W. Rate bounds on SSIM index of quantized images. IEEE Trans. Image Process. 2008, 17, 1624–1639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azulay, A.; Weiss, Y. Why do deep convolutional networks generalize so poorly to small image transformations? J. Mach. Learn. Res. 2018, 20, 1–25. [Google Scholar]
- Zhang, Z. Improved adam optimizer for deep neural networks. In Proceedings of the IEEE/ACM 26th International Symposium on Quality of Service (IWQoS), Banff, AB, Canada, 4–6 June 2018; pp. 1–2. [Google Scholar]
- Perez, L.; Wang, J. The effectiveness of data augmentation in image classification using deep learning. Convolutional Neural Netw. Vis. Recognit. 2017, 11, 1–8. [Google Scholar]
- Girosi, F.; Jones, M.; Poggio, T. Regularization theory and neural networks architectures. Neural Comput. 1995, 7, 219–269. [Google Scholar] [CrossRef]
- Han, X.; Dai, Q. Batch-normalized Mlpconv-wise supervised pre-training network in network. Appl. Intell. 2018, 48, 142–155. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Gulli, A.; Pal, S. Deep Learning with Keras; Packt Publishing Ltd.: Birmingham, UK, 2017. [Google Scholar]
- Ng, M.Y.; Lee, E.Y.; Yang, J.; Yang, F.; Li, X.; Wang, H.; Lui, M.M.; Lo, C.S.; Leung, B.; Khong, P.L.; et al. Imaging profile of the COVID-19 infection: Radiologic findings and literature review. Radiol. Cardiothorac. Imaging 2020, 2, e200034. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Liu, F.; Li, J.; Zhang, T.; Wang, D.; Lan, W. Clinical and CT imaging features of the COVID-19 pneumonia: Focus on pregnant women and children. J. Infect. 2020, 80, 7–13. [Google Scholar] [CrossRef]
- Fiszman, M.; Chapman, W.W.; Aronsky, D.; Evans, R.S.; Haug, P.J. Automatic detection of acute bacterial pneumonia from chest X-ray reports. J. Am. Med. Inform. Assoc. 2000, 7, 593–604. [Google Scholar] [CrossRef]
- Zhao, D.; Yao, F.; Wang, L.; Zheng, L.; Gao, Y.; Ye, J.; Guo, F.; Zhao, H.; Gao, R. A comparative study on the clinical features of COVID-19 pneumonia to other pneumonias. Clin. Infect. Dis. 2020, 71, 756–761. [Google Scholar] [CrossRef] [Green Version]
- Ouchicha, C.; Ammor, O.; Meknassi, M. CVDNet: A novel deep learning architecture for detection of coronavirus (Covid-19) from chest X-ray images. Chaos Solitons Fractals 2020, 140, 110245. [Google Scholar] [CrossRef] [PubMed]
- Marques, G.; Agarwal, D.; de la Torre Díez, I. Automated medical diagnosis of COVID-19 through EfficientNet convolutional neural network. Appl. Soft Comput. 2020, 96, 106691. [Google Scholar] [CrossRef] [PubMed]
- Hassantabar, S.; Ahmadi, M.; Sharifi, A. Diagnosis and detection of infected tissue of COVID-19 patients based on lung X-ray image using convolutional neural network approaches. Chaos Solitons Fractals 2020, 140, 110170. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.I.; Shah, J.L.; Bhat, M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest X-ray images. Comput. Methods Prog. Biomed. 2020, 196, 105581. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Afshar, P.; Heidarian, S.; Naderkhani, F.; Oikonomou, A.; Plataniotis, K.N.; Mohammadi, A. Covid-caps: A capsule network-based framework for identification of covid-19 cases from X-ray images. Pattern Recognit. Lett. 2020, 138, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Sriram, A.; Muckley, M.; Sinha, K.; Shamout, F.; Pineau, J.; Geras, K.J.; Azour, L.; Aphinyanaphongs, Y.; Yakubova, N.; Moore, W. COVID-19 Prognosis via Self-Supervised Representation Learning and Multi-Image Prediction. arXiv 2021, arXiv:2101.04909. [Google Scholar]
- Goel, T.; Murugan, R.; Mirjalili, S.; Chakrabartty, D.K. OptCoNet: An optimized convolutional neural network for an automatic diagnosis of COVID-19. Appl. Intell. 2021, 51, 1351–1366. [Google Scholar] [CrossRef]









| Class | Sources | Number of Data |
|---|---|---|
| Normal | NIH CXRs dataset | 607 |
| Pneumonia | NIH CXRs dataset | 607 |
| COVID-19 | COVID-19 image data collection [19] | 468 |
| Figure 1 COVID-19 CXRs [20] | 35 | |
| Actualmed COVID-19 CXRs [21] | 58 | |
| COVID-19 Radiography Database [22] | 46 |
| Model Name | Class | Accuracy | Specificity | Sensitivity | AUC | F1 Score | Averaged Accuracy |
|---|---|---|---|---|---|---|---|
| Lee et al. [11] | N | 0.980 | 0.992 | 0.982 | 0.981 | 0.968 | 0.978 |
| P | 0.981 | 0.992 | 0.982 | 0.970 | 0.964 | ||
| C | 0.975 | 0.996 | 0.992 | 0.934 | 0.982 | ||
| Das et al. [24] | N | 0.964 | 0.959 | 0.962 | 0.964 | 0.956 | 0.954 |
| P | 0.954 | 0.949 | 0.952 | 0.954 | 0.955 | ||
| C | 0.954 | 0.959 | 0.957 | 0.954 | 0.953 | ||
| Rahimzadeh et al. [25] | N | 0.982 | 0.985 | 0.987 | 0.979 | 0.980 | 0.983 |
| P | 0.983 | 0.987 | 0.976 | 0.977 | 0.971 | ||
| C | 0.995 | 0.992 | 0.981 | 0.982 | 0.980 | ||
| ResNet50 [29] | N | 0.983 | 0.984 | 0.964 | 0.917 | 0.964 | 0.975 |
| P | 0.978 | 0.975 | 0.953 | 0.950 | 0.968 | ||
| C | 0.994 | 1.0 | 1.0 | 0.976 | 0.982 | ||
| ResNet101 [29] | N | 0.972 | 0.984 | 0.964 | 0.970 | 0.955 | 0.975 |
| P | 0.972 | 0.974 | 0.952 | 0.967 | 0.960 | ||
| C | 0.994 | 0.995 | 0.992 | 0.974 | 0.980 | ||
| MobileNet [30] | N | 0.975 | 0.988 | 0.973 | 0.979 | 0.960 | 0.972 |
| P | 0.974 | 0.987 | 0.973 | 0.976 | 0.971 | ||
| C | 0.994 | 1.0 | 1.0 | 0.956 | 0.982 | ||
| MobileNetV2 [31] | N | 0.945 | 0.936 | 0.847 | 0.917 | 0.904 | 0.961 |
| P | 0.945 | 0.982 | 0.969 | 0.950 | 0.924 | ||
| C | 0.994 | 1.0 | 1.0 | 0.986 | 0.982 | ||
| Gen + CBAM (ours) | N | 0.984 | 0.996 | 0.991 | 0.980 | 0.981 | 0.986 |
| P | 0.978 | 0.975 | 0.953 | 0.988 | 0.984 | ||
| C | 0.995 | 0.996 | 0.992 | 0.994 | 0.992 |
| Authors [Reference Number] | Used Method | Base Architecture | Classes | Metrics | % |
|---|---|---|---|---|---|
| Das et al. [24] | Deep transfer learning with machine learning | VGG19 | Normal, Pneumonia, COVID-19 | Accuracy | 99.26% |
| Rahimzadeh et al. [25] | Deep learning | Xception & ResNet50V2 | Normal, Pneumonia, COVID-19 | Accuracy | 91.4% |
| Hassantabar et al. [56] | Deep learning | MLP, CNN | Normal, Pneumonia, COVID-19 | Accuracy | 93.2% |
| Khan et al. [57] | Deep transfer learning | Xception | Normal, Pneumonia bacterial & viral, COVID-19 | Accuracy | 95% |
| Ozturk et al. [58] | Deep learning | Darknet | Normal, COVID-19 | Accuracy | 98.08% |
| Normal, Pneumonia, COVID-19 | Accuracy | 87.2% | |||
| Afshar et al. [60] | Deep transfer learning | Capsul network | Normal, Pneumonia bacterial & viral, COVID-19 | Accuracy | 95.7% |
| Sriram et al. [61] | Self-supervised learning | DenseNet | ICU transfer, intubation, mortality | AUC | 74.2% |
| Goel et al. [62] | GWO | CNN | Normal, Pneumonia, COVID-19 | Accuracy | 97.78% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.; Kwak, I.-Y.; Lim, C. A Deep Learning Model with Self-Supervised Learning and Attention Mechanism for COVID-19 Diagnosis Using Chest X-ray Images. Electronics 2021, 10, 1996. https://doi.org/10.3390/electronics10161996
Park J, Kwak I-Y, Lim C. A Deep Learning Model with Self-Supervised Learning and Attention Mechanism for COVID-19 Diagnosis Using Chest X-ray Images. Electronics. 2021; 10(16):1996. https://doi.org/10.3390/electronics10161996
Chicago/Turabian StylePark, Junghoon, Il-Youp Kwak, and Changwon Lim. 2021. "A Deep Learning Model with Self-Supervised Learning and Attention Mechanism for COVID-19 Diagnosis Using Chest X-ray Images" Electronics 10, no. 16: 1996. https://doi.org/10.3390/electronics10161996
APA StylePark, J., Kwak, I.-Y., & Lim, C. (2021). A Deep Learning Model with Self-Supervised Learning and Attention Mechanism for COVID-19 Diagnosis Using Chest X-ray Images. Electronics, 10(16), 1996. https://doi.org/10.3390/electronics10161996






