Metabolic Responses of Carotenoid and Cordycepin Biosynthetic Pathways in Cordyceps militaris under Light-Programming Exposure through Genome-Wide Transcriptional Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strain and Cultivation
2.2. Determination of Cordycepin and Carotenoid Contents of C. militaris Cultures
2.3. RNA Sequencing, Reads Mapping, and Functional Annotation
2.4. Differentially Expressed Genes Analysis
2.5. Improvement of the Genome-Scale Metabolic Network of C. militaris for Reporter Metabolites Analysis
3. Results and Discussion
3.1. Growth Characteristics and Production of Carotenoid and Cordycepin of C. militaris on Different Carbon Sources
3.2. Genome-Wide Transcriptome of C. militaris
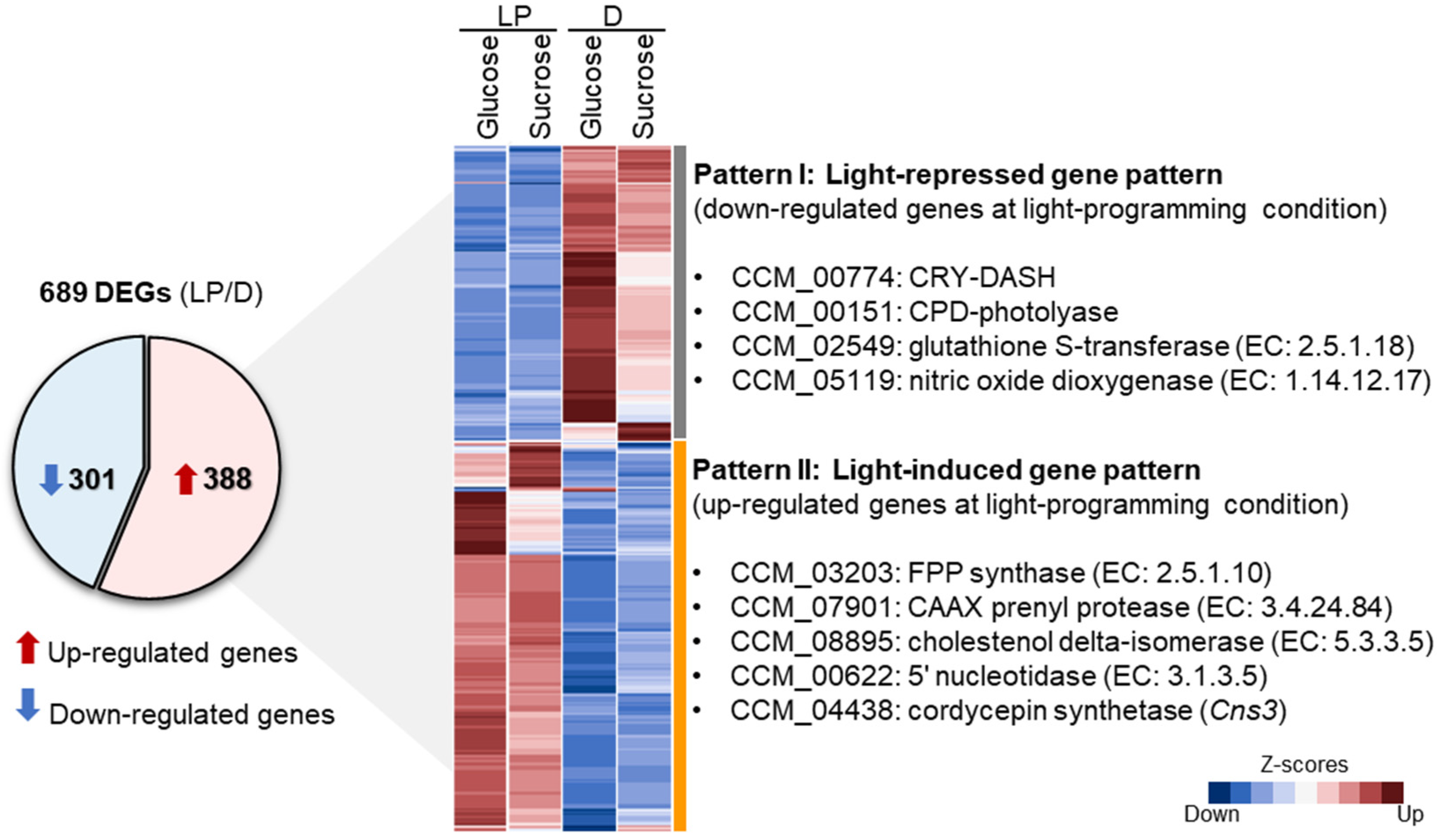
3.3. Differentially Expressed Genes of C. militaris Cultures at Light-Programming and Dark Conditions
3.4. Metabolic Responses of C. militaris in Carotenoid and Cordycepin Biosynthetic Pathways to Light-Programming Condition
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shrestha, B.; Tanaka, E.; Hyun, M.W.; Han, J.G.; Kim, C.S.; Jo, J.W.; Han, S.K.; Oh, J.; Sung, G.H. Coleopteran and lepidopteran hosts of the entomopathogenic genus Cordyceps sensu lato. J. Mycol. 2016, 2016, 7648219. [Google Scholar]
- Dong, J.Z.; Lei, C.; Ai, X.R.; Wang, Y. Selenium enrichment on Cordyceps militaris link and analysis on its main active components. Appl. Biochem. Biotechnol. 2012, 166, 1215–1224. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.Z.; Lei, C.; Zheng, X.J.; Ai, X.R.; Wang, Y.; Wang, Q. Light wavelengths regulate growth and active components of Cordyceps militaris fruit bodies. J. Food Biochem. 2013, 37, 578–584. [Google Scholar]
- Dong, J.Z.; Wang, S.H.; Ai, X.R.; Yao, L.; Sun, Z.W.; Lei, C.; Wang, Y.; Wang, Q. Composition and characterization of cordyxanthins from Cordyceps militaris fruit bodies. J. Funct. Foods 2013, 5, 1450–1455. [Google Scholar] [CrossRef]
- Tuli, H.S.; Sandhu, S.S.; Sharma, A.K. Pharmacological and therapeutic potential of Cordyceps with special reference to cordycepin. 3 Biotech 2014, 4, 1–12. [Google Scholar] [CrossRef]
- Yang, T.; Sun, J.; Lian, T.; Wang, W.; Dong, C.H. Process optimization for extraction of carotenoids from medicinal caterpillar fungus, Cordyceps militaris (ascomycetes). Int. J. Med. Mushrooms 2014, 16, 125–135. [Google Scholar] [CrossRef]
- Lou, H.W.; Zhao, Y.; Tang, H.B.; Ye, Z.W.; Wei, T.; Lin, J.F.; Guo, L.Q. Transcriptome analysis of Cordyceps militaris reveals genes associated with carotenoid synthesis and identification of the function of the Cmtns gene. Front. Microbiol. 2019, 10, 2105. [Google Scholar] [CrossRef]
- Lou, H.W.; Zhao, Y.; Chen, B.X.; Yu, Y.H.; Tang, H.B.; Ye, Z.W.; Lin, J.F.; Guo, L.Q. Cmfhp gene mediates fruiting body development and carotenoid production in Cordyceps militaris. Biomolecules 2020, 10, 410. [Google Scholar] [CrossRef]
- Avalos, J.; Carmen Limón, M. Biological roles of fungal carotenoids. Curr. Genet. 2015, 61, 309–324. [Google Scholar] [CrossRef]
- Echavarri-Erasun, C.; Johnson, E.A. Fungal carotenoids. In Applied Mycology and Biotechnology; Khachatourians, G.G., Arora, D.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2002; pp. 45–85. [Google Scholar]
- Skibsted, L.H. Carotenoids in antioxidant networks. Colorants or radical scavengers. J. Agric. Food Chem. 2012, 60, 2409–2417. [Google Scholar] [CrossRef]
- Chiang, S.S.; Liang, Z.C.; Wang, Y.C.; Liang, C.H. Effect of light-emitting diodes on the production of cordycepin, mannitol and adenosine in solid-state fermented rice by Cordyceps militaris. J. Food Compost. Anal. 2017, 60, 51–56. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, F.; Liu, K.; Liu, Q.; Yang, Y.; Dong, C. Heat and light stresses affect metabolite production in the fruit body of the medicinal mushroom Cordyceps militaris. Appl. Microbiol. Biotechnol. 2018, 102, 4523–4533. [Google Scholar]
- Wang, F.; Liu, Q.; Zhang, J.; Liu, K.; Li, K.; Liu, G.; Dong, C. Comparative transcriptome analysis between a spontaneous albino mutant and its sibling strain of Cordyceps militaris in response to light stress. Front. Microbiol. 2018, 9, 1237. [Google Scholar] [CrossRef] [PubMed]
- Raethong, N.; Wang, H.; Nielsen, J.; Vongsangnak, W. Optimizing cultivation of Cordyceps militaris for fast growth and cordycepin overproduction using rational design of synthetic media. Comput. Struct. Biotechnol. J. 2020, 18, 1–8. [Google Scholar] [CrossRef]
- Chandi, G.K.; Gill, B.S. Production and characterization of microbial carotenoids as an alternative to synthetic colors: A review. Int. J. Food Prop. 2011, 14, 503–513. [Google Scholar] [CrossRef]
- Raethong, N.; Laoteng, K.; Vongsangnak, W. Uncovering global metabolic response to cordycepin production in Cordyceps militaris through transcriptome and genome-scale network-driven analysis. Sci. Rep. 2018, 8, 9250. [Google Scholar] [CrossRef]
- Lin, S.; Yu, L.; Zhang, H. Transcriptomic responses to thermal stress and varied phosphorus conditions in Fugacium kawagutii. Microorganisms 2019, 7, 96. [Google Scholar] [CrossRef] [PubMed]
- Lane, T.; Best, T.; Zembower, N.; Davitt, J.; Henry, N.; Xu, Y.; Koch, J.; Liang, H.; McGraw, J.; Schuster, S.; et al. The green ash transcriptome and identification of genes responding to abiotic and biotic stresses. BMC Genom. 2016, 17, 702. [Google Scholar] [CrossRef]
- Vongsangnak, W.; Chumnanpuen, P.; Sriboonlert, A. Transcriptome analysis reveals candidate genes involved in luciferin metabolism in Luciola aquatilis (Coleoptera: Lampyridae). PeerJ 2016, 4, e2534. [Google Scholar] [CrossRef][Green Version]
- Zheng, P.; Xia, Y.; Xiao, G.; Xiong, C.; Hu, X.; Zhang, S.; Zheng, H.; Huang, Y.; Zhou, Y.; Wang, S.; et al. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional chinese medicine. Genome Biol. 2011, 12, R116. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2013, 42, D699–D704. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, S.; Furió-Tarí, P.; Turrà, D.; Pietro, A.D.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, S.; García-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef]
- Warnes, G.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Mächler, M.; Magnusson, A.; Möller, S. Gplots: Various R Programming Tools for Plotting Data. R package version 2.6.0. 2005; CRAN.R-project.org/package=gplots. [Google Scholar]
- Sirithep, K.; Xiao, F.; Raethong, N.; Zhang, Y.; Laoteng, K.; Hu, G.; Vongsangnak, W. Probing carbon utilization of Cordyceps militaris by sugar transportome and protein structural analysis. Cells 2020, 9, 401. [Google Scholar] [CrossRef]
- Vongsangnak, W.; Raethong, N.; Mujchariyakul, W.; Nguyen, N.N.; Leong, H.W.; Laoteng, K. Genome-scale metabolic network of Cordyceps militaris useful for comparative analysis of entomopathogenic fungi. Gene 2017, 626, 132–139. [Google Scholar] [CrossRef]
- Patil, K.R.; Nielsen, J. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc. Natl. Acad. Sci. USA 2005, 102, 2685–2689. [Google Scholar] [CrossRef]
- Väremo, L.; Nielsen, J.; Nookaew, I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013, 41, 4378–4391. [Google Scholar] [CrossRef] [PubMed]
- Wöstemeyer, J.; Grünler, A.; Schimek, C.; Voigt, K. Genetic regulation of carotenoid biosynthesis in fungi. In Applied Mycology and Biotechnology; Arora, D.K., Berka, R.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2005; Volume 5, pp. 257–274. [Google Scholar]
- Wang, F.; Song, X.; Dong, X.; Zhang, J.; Dong, C. DASH-type cryptochromes regulate fruiting body development and secondary metabolism differently than CmWC-1 in the fungus Cordyceps militaris. Appl. Microbiol. Biotechnol. 2017, 101, 4645–4657. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Guo, M.; Yang, H.; Guo, S.; Dong, C. The blue-light receptor CmWC-1 mediates fruit body development and secondary metabolism in Cordyceps militaris. Appl. Microbiol. Biotechnol. 2016, 100, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Tagua, V.G.; Pausch, M.; Eckel, M.; Gutiérrez, G.; Miralles-Durán, A.; Sanz, C.; Eslava, A.P.; Pokorny, R.; Corrochano, L.M.; Batschauer, A. Fungal cryptochrome with DNA repair activity reveals an early stage in cryptochrome evolution. Proc. Natl. Acad. Sci. USA 2015, 112, 15130–15135. [Google Scholar] [CrossRef] [PubMed]
- Veluchamy, S.; Rollins, J.A. A CRY-DASH-type photolyase/cryptochrome from Sclerotinia sclerotiorum mediates minor UV-A-specific effects on development. Fungal Genet. Biol. 2008, 45, 1265–1276. [Google Scholar] [CrossRef] [PubMed]
- Nimse, S.B.; Pal, D. Free radicals, natural antioxidants, and their reaction mechanisms. RSC Adv. 2015, 5, 27986–28006. [Google Scholar] [CrossRef]
- Gardner, P.R.; Gardner, A.M.; Martin, L.A.; Salzman, A.L. Nitric oxide dioxygenase: An enzymic function for flavohemoglobin. Proc. Natl. Acad. Sci. USA 1998, 95, 10378–10383. [Google Scholar] [CrossRef]
- Ketterer, B. Glutathione S-transferases and prevention of cellular free radical damage. Free Radic. Res. 1998, 28, 647–658. [Google Scholar] [CrossRef]
- Sandmann, G. Antioxidant protection from UV- and light-stress related to carotenoid structures. Antioxidants 2019, 8, 219. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, G.; Li, C.; Ling, J. Cordycepin and pentostatin biosynthesis gene identified through transcriptome and proteomics analysis of Cordyceps kyushuensis kob. Microbiol. Res. 2019, 218, 12–21. [Google Scholar] [CrossRef]
- Wongsa, B.; Raethong, N.; Chumnanpuen, P.; Wong-ekkabut, J.; Laoteng, K.; Vongsangnak, W. Alternative metabolic routes in channeling xylose to cordycepin production of Cordyceps militaris identified by comparative transcriptome analysis. Genomics 2020, 112, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Kimura, A. Methylglyoxal and regulation of its metabolism in microorganisms. In Advances in Microbial Physiology; Poole, R.K., Ed.; Academic Press: Cambridge, MA, USA, 1995; Volume 37, pp. 177–227. [Google Scholar]
- Hoque, T.S.; Hossain, M.A.; Mostofa, M.G.; Burritt, D.J.; Fujita, M.; Tran, L.-S.P. Methylglyoxal: An emerging signaling molecule in plant abiotic stress responses and tolerance. Front. Plant Sci. 2016, 7, 1341. [Google Scholar] [CrossRef] [PubMed]
- Yalcinkaya, T.; Uzilday, B.; Ozgur, R.; Turkan, I.; Mano, J. Lipid peroxidation-derived reactive carbonyl species (RCS): Their interaction with ROS and cellular redox during environmental stresses. Environ. Exp. Bot. 2019, 165, 139–149. [Google Scholar] [CrossRef]




| Phenotypic Characteristics | Light-Programming | Dark | ||
|---|---|---|---|---|
| Glucose | Sucrose | Glucose | Sucrose | |
| Maximum specific growth rate, µmax (h−1) | 0.0116 ± 0.0040 | 0.0122 ± 0.0017 | 0.0079 ± 0.0008 1 | 0.0104 ± 0.0021 1 |
| Biomass productivity (g DCW L−1 h−1) | 0.0122 ± 0.0018 | 0.0112 ± 0.0016 | 0.0096 ± 0.0013 1 | 0.0088 ± 0.0004 1 |
| Extracellular cordycepin titer (mg L−1) | 112.12 ± 0.32 | 95.65 ± 8.56 | 109.14 ± 11.54 1 | 119.32 ± 14.06 1 |
| Carotenoid content (mg g DCW−1) | 1.4692 ± 0.0122 a | 1.4590 ± 0.0052 a | 0.0244 ± 0.0051 b,2 | 0.0234 ± 0.0095 b,2 |
| Summary | Light-Programming | Dark | Average | ||
|---|---|---|---|---|---|
| Glucose | Sucrose | Glucose | Sucrose | ||
| Total clean reads (Mb) | 48.81 | 48.79 | 45.15 * | 44.87 * | 46.91 ± 1.90 |
| Total mapped reads to genome | 39,885,625 (81.72%) | 39,088,706 (80.12%) | 38,097,583 (84.38%) | 37,711,698 (84.05%) | 38,695,903 ± 850,937.56 (82.57 ± 2.02%) |
| Number of expressed genes | 8504 | 8499 | 8570 | 8542 | 8529 ± 33.54 |
| All expressed genes | 8747 | ||||
| Reporter Metabolites | Up-Directional p-Value |
|---|---|
| AMP | 0.0020 * |
| ADP | 0.0020 * |
| ATP | 0.0020 * |
| UDP | 0.0020 * |
| glucose | 0.0020 * |
| UDP-glucose | 0.0020 * |
| thioredoxin dithiol | 0.0200 |
| 2-oxoglutarate | 0.0798 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thananusak, R.; Laoteng, K.; Raethong, N.; Zhang, Y.; Vongsangnak, W. Metabolic Responses of Carotenoid and Cordycepin Biosynthetic Pathways in Cordyceps militaris under Light-Programming Exposure through Genome-Wide Transcriptional Analysis. Biology 2020, 9, 242. https://doi.org/10.3390/biology9090242
Thananusak R, Laoteng K, Raethong N, Zhang Y, Vongsangnak W. Metabolic Responses of Carotenoid and Cordycepin Biosynthetic Pathways in Cordyceps militaris under Light-Programming Exposure through Genome-Wide Transcriptional Analysis. Biology. 2020; 9(9):242. https://doi.org/10.3390/biology9090242
Chicago/Turabian StyleThananusak, Roypim, Kobkul Laoteng, Nachon Raethong, Yu Zhang, and Wanwipa Vongsangnak. 2020. "Metabolic Responses of Carotenoid and Cordycepin Biosynthetic Pathways in Cordyceps militaris under Light-Programming Exposure through Genome-Wide Transcriptional Analysis" Biology 9, no. 9: 242. https://doi.org/10.3390/biology9090242
APA StyleThananusak, R., Laoteng, K., Raethong, N., Zhang, Y., & Vongsangnak, W. (2020). Metabolic Responses of Carotenoid and Cordycepin Biosynthetic Pathways in Cordyceps militaris under Light-Programming Exposure through Genome-Wide Transcriptional Analysis. Biology, 9(9), 242. https://doi.org/10.3390/biology9090242






