Abstract
Abeliophyllum distichum Nakai is known as a monotypic genus endemic to South Korea. Currently, several pharmacological studies have revealed that A. distichum extract exhibits diverse biological functions, including anti-cancer, anti-diabetic, anti-hypertensive, and anti-inflammatory activities. In this study, we present the anti-osteoporotic activity of A. distichum extract by inhibiting osteoclast formation. First, we show that the methanolic extract of the leaves of A. distichum, but not extracts of the branches or fruits, significantly inhibits receptor activator of the NF-κB ligand (RANKL)-induced osteoclast differentiation. Second, our transcriptome analysis revealed that the leaf extract (LE) blocks sets of RANKL-mediated osteoclast-related genes. Third, the LE attenuates the phosphorylation of extracellular signal-related kinase. Finally, treatment with the LE effectively prevents postmenopausal bone loss in ovariectomized mice and glucocorticoid-induced osteoporosis in zebrafish. Our findings show that the extract of A. distichum efficiently suppressed osteoclastogenesis by regulating osteoclast-related genes, thus offering a novel therapeutic strategy for osteoporosis.
1. Introduction
Bone tissue is continuously maintained by osteoclasts and osteoblasts [1]. They are remodeled through life to regenerate damaged bones and maintain mechanical strength. An imbalance between bone resorption and bone formation can cause bone diseases, including osteoporosis and osteopetrosis [2]. Osteoporosis is a metabolic skeletal disorder caused by excessive bone resorption, which is characterized by low bone mass and the microstructural deterioration of bone tissue. These processes decrease the mechanical strength of the bone and increase the risk of fracture [3,4]. Osteoporotic fractures are the leading cause of morbidity and disability in the elderly and can lead to death in the case of hip fracture. Therefore, osteoporosis is becoming a major health problem in the aging population [5]. Osteoporosis has been classified into two main groups: primary and secondary. Primary osteoporosis can be further divided into two subgroups: postmenopausal osteoporosis (type I) and senile osteoporosis (type II) [6]. Postmenopausal osteoporosis, caused by estrogen deficiency, is the most common type of osteoporosis. Glucocorticoid-induced osteoporosis (GIO), the most frequent form of secondary osteoporosis, is triggered by long-term glucocorticoid treatment [7]. These two types of osteoporosis are characterized by an abnormal increase in osteoclast formation and activity. Therefore, regulating osteoclastogenesis has been targeted in current osteoporosis therapies.
Osteoclast differentiation is induced by macrophage colony-stimulating factor (M-CSF) and receptor activator of nuclear factor-kappa Β (NF-κB) ligand (RANKL). M-CSF is bound to its receptor (CSF receptor 1, cFMS) in osteoclast precursors, which induces the expression of RANK, the receptor for RANKL [8,9,10]. The binding of RANKL to RANK leads to RANK trimerization and recruitment of tumor necrosis factor receptor-associated factor 6 (TRAF6), which stimulates downstream signaling pathways, such as the NF-κB, c-JUN N-terminal kinase (JNK), p38 mitogen-activated protein kinase, and extracellular signal-related kinase (ERK) pathways [11,12]. Then, the activated nuclear factor-activated T cells c1 (NFATc1), an essential regulator of osteoclastogenesis, is translocated to the nucleus and cooperates with MITF, c-FOS, and NF-κB to express a series of osteoclast-related genes, such as tartrate-resistant acid phosphatase (TRAP), cathepsin K, osteoclast-associated receptor, and matrix metalloproteinase-9 (MMP-9), in addition to NFATc1 [11,12,13].
Abeliophyllum distichum Nakai (A. distichum), known as white forsythia, is a deciduous shrub and a monotypic genus of the Oleaceae. A. distichum growth is limited to the south and central areas of Korea, and it has been used as a landscape plant. Recent studies reported that A. distichum has diversified pharmacological properties including antioxidant, anti-inflammatory, anti-cancer, anti-diabetic, and antihypertensive activities [14,15,16,17]. However, the pharmacological effects of A. distichum on osteoclastogenesis and bone homeostasis remain unknown.
Here, we report that the leaf extract (LE) of A. distichum significantly inhibited RANKL-induced osteoclastogenesis. Transcriptome profiling showed that LE of A. distichum negatively modulates osteoclast differentiation through the downregulation of NFATc1 expression. Furthermore, our signaling pathway analysis and reporter gene assays revealed that LE of A. distichum not only blocked ERK signaling pathways, but also inhibited the transactivation of NFATc1, p65, and c-FOS as well as the upregulation of NFATc1. Moreover, LE of A. distichum alleviated the osteoporotic phenotype in both a mouse model of ovariectomy-induced bone loss and a zebrafish model of prednisolone-induced osteoporosis.
2. Materials and Methods
2.1. Preparation of Extracts of Abeliophyllum distichum
Fruits, branches, and leaves of A. distichum were obtained from the research forest at Chungbuk National University. The fruit, branches, and leaves were lyophilized and pulverized into a fine powder using a blender. The ground materials (approx. 50 g) were soaked in methanol (1:10 dry weight material to MeOH (mL)) for 24 h, and sonicated (1 h × 3 times) at 40 °C in an ultrasonic bath (Power sonic 420, Hwashin Co., Yeongcheon, Korea). After filtration, the methanol extracts of the fruits (FE), branches (BE), and leaves (LE) were concentrated at 40 °C using a rotary vacuum evaporator under low pressure, and kept in a refrigerator until use.
2.2. Osteoclast Differentiation Assay
Osteoclast precursor (OCP) cells were generated as described previously [18]. Briefly, bone marrow cells were isolated from 6–7-week-old ICR male mouse with α-minimum essential medium (α-MEM) supplemented with 10% FBS and M-CSF (5 ng/mL). Non-adherent cells were seeds on 10-cm culture dish and further cultured with M-CSF (30 ng/mL) for 3 days. After removing floating cells, adherent cells were used as bone marrow-derived macrophage (BMM) cells. To check the effect of extracts of A. distichum on osteoclastogenesis, OCP cells (3 × 104 cells) were seeded in 48-well culture plate and cultured with 30 ng/mL of M-CSF and 100 ng/mL of RANKL, in the absence or presence of an indicated concentration of extracts of A. distichum. After TRAP staining using an acid phosphatase leukocyte kit (Cat. No. 387A, Sigma-Aldrich, St. Louise, MO, USA), TRAP-positive multinucleated cells (MNCs) were counted as osteoclasts.
2.3. Cell Proliferation Assay
OCP cells (5 × 103 cells) were seeded in 96-well culture plate grown with three different concentrations of BE, LE, or FE (0.1, 1, and 10 μg/mL), and cell proliferations were analyzed by MTT assay using Cell Proliferation Kit 1 (Cat. No. 11465007001, Roche, Basel, Switzerland).
2.4. RNA-seq Data Analysis
For RNA-seq, OCP cells (2 × 105 cells) were plated on 6-well culture plate with M-CSF (30 ng/mL). Cells were treated with RANKL in the absence or presence of LE as following: 1) no RANKL (-R); cells treated with M-CSF (30 ng/mL) for 3 days, 2) RANKL (+ R); cells treated with M-CSF (30 ng/mL) and RANKL (100 ng/mL) for 3 days, and 3) RANKL + LE (R + LE); cells treated with M-CSF (30 ng/mL) and RANKL (100 ng/mL) in the presence of LE for 3 days. After libraries construction following total RNA preparation, high-throughput sequencing with 100bp pair-end was performed from E-Biogen Inc. (Seoul, Korea) using HiSeq 2500 (Illumina, CA, USA). mRNA-Seq reads were aligned to reference mouse genome (mm10 assembly) using TopHat. HOMER program was used to analyze RNA-seq data performed as described [18]. The cut-off parameters were set at fold change ≥ 2 and FDR < 0.05 (Benjamin-Hochberg). For K-means clustering and gene ontology (GO) analysis, we used the Morpheus web site (https://software.boradinstitute.org/morpheus/), Metascape tool, and Gene Set Enrichment Analysis (GSEA) of MsigDB gen sets [19,20].
2.5. Real-Time Quantitative PCR (qRT-PCR)
For qRT-PCR analysis, OCP cells were differentiated with M-CSF and RANKL in the absence or presence of LE as described in RNA-seq. Total RNA was prepared from the cultured cells using Tri-RNA reagents (Cat. No. FATRR 001, Favorgen, Ping-Tung, Taiwan). Total RNA (2 µg) was reverse-transcribed to cDNA using the Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase (Cat. No. M170B, Promega, Madison, WI, USA). Quantitative real-time PCR was carried out using IQ SYBR Green SuperMix (Cat. No. 1708882, Bio-Rad, Hercules, CA, USA). The following primers were used for qPCR: Nfatc1 5′-CTCGAAAGACAGCACTGGAGCAT-3′ (forward) and 5′-CGGCTGCCTTCCGTCTCATAG-3′ (reverse); p65 5′-GGAGTTCCAGTACTTGCC-3′ (forward) and 5′-GTCCTTTTGCGCTTCTCT-3′ (reverse); c-Fos 5′-CCAGTCAAGAGCATCAGCAA-3′ (forward) and 5′-AAGTAGTCGCAGCCCCGAGTA-3′ (reverse); Traf6 5′-AAACCACGAAGAGGTCATGG-3′ (forward) and 5′-GCGGGTAGAGACTTCACAGC-3′ (reverse); Actin 5′-GCAAGTGCTTCTAGGCGGAC-3′ (forward) and 5′-AAGAAAGGGTGTAAAACGCAGC-3′ (reverse); Ccr1 5′-ACTCCACTCCATGCCAAAAG-3′ (forward) and 5′-CTAGGACATTGCCCACCACT-3′ (reverse); Mitf 5′-GGAACAGCAACGAGCTAAGG-3′ (forward) and 5′-TGATGATCCGATTCACCAGA-3′ (reverse); Ctsk 5′-ACGGAGGCATTGACTCTGAAGATG-3′ (forward) and 5′-GGAACCACCAACGAGAGGAGAAAT-3′ (reverse). The relative mRNA expressions were assessed by the ΔΔCt method, using β-Actin as internal control.
2.6. Immunoblotting
OCP cells grown with M-CSF (30 ng/mL) were treated with RANKL (100 ng/mL) in the absence or presence of LE as the indicated times. Whole cell lysates were prepared with standard lysis buffer containing protease inhibitor, 1 mM sodium orthovanadate, and 2.5 mM sodium pyrophosphate. The cell lysates were subjected to SDS-PAGE, followed by immunoblotting with the indicated antibodies. The resulting blots were analyzed by enhanced chemiluminescence detection using Amersham ECL reagent (Cat. No. RPN2106, Amersham, Buckinghamshire, UK). The following antibodies were used in this study: p-AKT (1:1000 dilution, cat. No. 9272), AKT (1:1000 dilution, cat. No. 9271), p-ERK (1:500 dilution, cat. No. 9101), and ERK (1:500 dilution, cat. No. 9102) antibodies from Cell Signaling Technology (Massachusetts, MA, United States); p-IκB (1:500 dilution, cat. No. sc-8404), NFATc1 (1:200 dilution, cat. No. sc-7294) and p65 (1:200 dilution, cat. No. sc-8008), Lamin A/C (1:200 dilution, cat. No. sc-376248) antibodies from Santa Cruz Biotechnology (Santa Cruz, CA, USA); IκB (1:500 dilution, cat. No. 12045-R116) and β-Actin (1:2000 dilution, cat. No. 100166-MM10) antibodies from Sino Biological (Beijing, China); p-p38 (1:500 dilution, cat. No. MABS1754), p38 (1:500 dilution, cat. No. 05-1059), p-JNK (1:500 dilution, cat. No. 559304), JNK (1:500 dilution, cat. No. PS1019) antibodies from Millipore (Burlington, MA, USA); and HRP anti-mouse (1:5000 dilution, cat. No. 405306) and HRP anti-rabbit (1:5000 dilution, cat. No. 406401) from Biolegend (San Diego, CA, USA).
2.7. Reporter Gene Assay
Reporter gene assays were performed as described previously [21]. Briefly, 293T cells were transfected with an Nfatc1-Luc reporter plasmid and vectors encoding c-FOS, p65 and NFATc1 with or without LE of A. distichum (10 μg/mL) for 24 h. Cells were lysed and assayed for luciferase activity using SpectraMax i3x (Molecular Devices, San Jose, CA, USA).
2.8. Micro-Computed Tomography (Micro-CT) Analysis
Eight-week-old female C57BL/6 mice (20 g) were randomly divided into three groups (n = 5 mice per group): sham-operated mice, bilateral ovariectomized (OVX) mice treated with vehicle, and OVX mice treated with LE of A. distichum. One week after operation, mice were injected intraperitoneally with vehicle or LE (10 mg/kg) once a week for 8 weeks. For micro-CT, the distal femur was scanned using a Quantum GX Micro-CT imaging system (PerkinElmer, Hopkinton, MA, USA) with the following settings: 90 kV, 88 μA and a 4 min scanning time. The trabecular bone parameters were analyzed by Analyze 12.0 software (AnalyzeDirect, Overland Park, KS, USA). Maintenance, use, and treatment of all animals were approved by the Animal Care and Use committee at the Korea Basic Science Institute (KBSI-AEC 1915).
2.9. Bone Histomorphometry
For hematoxylin and eosin (H&E) and tartrate-resistant acid phosphatase (TRAP) staining, the femurs of mice were excised, fixed in 4% paraformaldehyde solution for two days, washed with PBS for three times, decalcified in 0.5 M ethylenediaminetetraacetic acid (EDTA, pH 7.4) for 3 weeks. After complete decalcification, the femurs were then dehydrated with ethanol and clarified with xylene. The paraffin-embedded femur sections (5 μm thick) were stained with H&E and TRAP. Histological analysis was performed as recently described [22].
2.10. Zebrafish Maintenance
Adult wild-type zebrafish were maintained under standard conditions in a circulating water system at 28 °C with day–night (14 h light/10 h dark) cycles. The male and female zebrafish were chosen for spawning. Embryos were kept at 28 °C. At 10 dpf, the larvae were treated with 25 µM prednisolone ± LE of A. distichum. At 13 dpf, the larvae were collected for whole-mount skeletal staining [23]. All experimental protocols were approved by the Animal Care and Use committee of the Chungbuk National University, Korea (CBNUA-1391-20-01).
2.11. Whole-Mount Skeletal Staining
Whole-mount skeletal staining on zebrafish larvae was performed as described [18]. The larvae at 13 dpf were fixed in 10% neutral buffered formalin, and rinsed with tap water several times. After bleaching pigmentation with 3% H2O2 solution, the larvae were stained with 1 mg/mL alizarin red stain/1% KOH. To quantify bone mineral density, the areas of the first five stained vertebrae (V1–V5) were measured using the ImageJ densitometry program.
2.12. Statistical Analysis
All quantitative data are presented as mean ± SD. Statistical analysis was performed with GraphPad prism 7 software (one-way ANOVA test, followed by Tukey’s multiple comparison test or Dunnett’s multiple comparison test).
2.13. Accession Numbers
RNA-seq data were available at NCBI Gene Expression Omnibus (GSE152479).
3. Results
3.1. Leaf Extract of Abeliophyllum distichum Inhibits Osteoclast Differentiation
To investigate the effect of extracts from various tissues of A. distichum on osteoclast differentiation, we prepared extracts from the branches, fruits, or leaves of A. distichum. Osteoclast precursors (OCPs) were grown in osteoclastogenic medium in the presence or absence of extracts of A. distichum, in doses of 0.1, 1, 10 μg/mL. As shown in Figure 1A, LE significantly suppressed TRAP-positive osteoclast formation in a dose-dependent manner (IC50 = 1 μg/mL). By contrast, branch extract (BE) or fruit extract (FE) decreased the number of osteoclasts at higher doses (IC50 of FE = 10 μg/mL, IC50 of BE = not determined). Considering the fact that osteoclast differentiation involves several distinct steps such as cell proliferation, we next determined the effect of those extracts on OCP cell proliferation. As shown in Figure 1B, we observed that LE extracts had no effect on the proliferation of osteoclast precursors, indicating that the LE of A. distichum directly modulates the differentiation potential of osteoclast precursors.
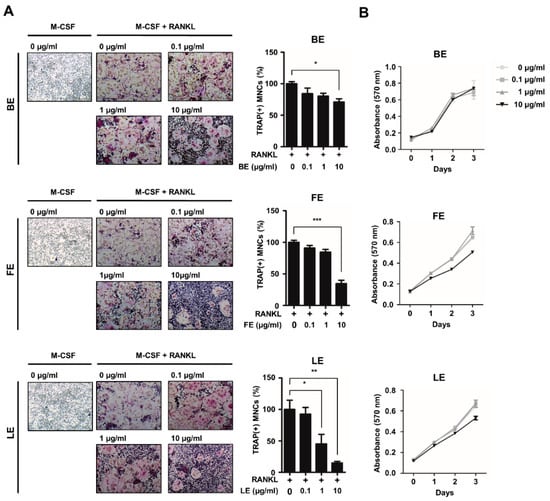
Figure 1.
Abeliophyllum distichum extracts suppress osteoclast differentiation. (A) BMMs were treated with M-CSF (30 ng/mL) and RANKL (100 ng/mL) in the absence or the presence of increasing concentration of three tissue extracts such as branch extract (BE), fruit extract (FE) and leaf extract (LE) of A. distichum for 3 days. Cells were fixed and stained for TRAP. The representative TRAP staining was shown (left panel) and the number of TRAP-positive multinuclear cells was counted (right panel). (B) BMMs were cultured with M-CSF (30 ng/mL) in the presence of extracts of A. distichum and cell proliferation was measured by MTT assays. Error bars represent the mean result ± SD of three independent experiments; * p < 0.05, ** p < 0.01, *** p < 0.001.
3.2. Leaf Extract of Abeliophyllum distichum Regulates the Expression of a Set of Osteoclastogenic Genes during Osteoclastogenesis
To identify differentially expressed genes after treatment with the LE of A. distichum, we performed high-throughput RNA-sequencing of BMMs under three different conditions: no RANKL (−R), RANKL (+ R), RANKL + LE (R + LE). Genome-wide transcriptome analysis revealed that 2573 genes were differentially expressed in any pairwise comparison among the three conditions. K-means clustering classified genes into six gene clusters that were differentially modulated by RANKL and LE of A. distichum, as shown in Figure 2A and Supplementary Table S1. GO analysis revealed that distinct functional terms were enriched in each cluster (Figure 2B). Because RANKL upregulates osteoclast-specific gene expression and LE treatment suppressed RANKL-induced osteoclastogenesis (Figure 1A), we focused on the inhibitory effect of LE on RANKL-induced genes. Among RANKL-induced genes (948 genes, upregulated two-fold), 213 genes (12 upregulated and 201 downregulated genes) were differentially expressed after LE treatment (Figure 2C and Supplementary Table S2). Intriguingly, most of the downregulated genes belonged to cluster IV, which is mainly associated with osteoclast differentiation and tissue remodeling, as shown in Figure 2D. Furthermore, GSEA scoring plots showed significant enrichment in pathways related to bone resorption and the regulation of osteoclast differentiation (Figure 2E,F). Exploration of the leading-edge subset of these genes identified six genes related to bone resorption and nine genes related to the regulation of osteoclast differentiation, respectively. The results from GSEA were further confirmed by qRT-PCR analysis (Figure 2G).
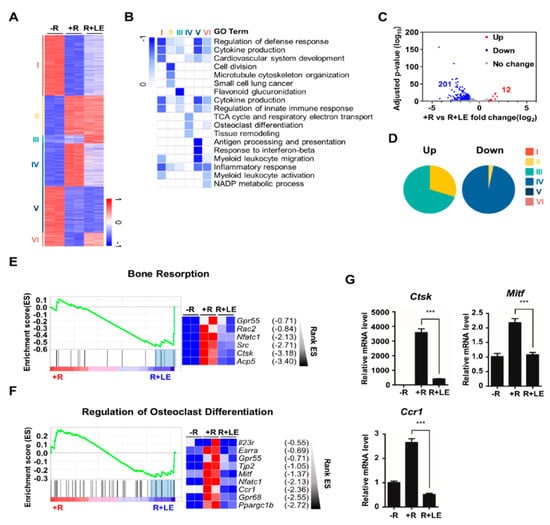
Figure 2.
Leaf extract of Abeliophyllum distichum alters gene expression profiling during osteoclastogenesis. (A) K-means (K = 6) clustering of 2,573 differentially expressed genes (DEGs) in any pairwise comparison among three conditions. Clusters are indicated on the left (-R; no RANKL, +R; RANKL (100 ng/mL), R + LE; RANKL (100 ng/mL) + LE of A. distichum (10 μg/mL) for 3 days. (B) Heatmap showing the P-value significance of GO term enrichment for genes in each cluster. (C) Volcano plot of transcriptomic changes of RANKL-induced genes by LE treatment; colored dots correspond to genes with significant (FDR < 0.05) and greater than two-fold expression changes. (D) Pie chart showing each cluster portion of up or down DEGs. Up: Cluster 1 (33.2%), Cluster 2 (1.7%), Cluster 3 (21.3%), Cluster 4 (0%), Cluster 5 (0%), and Cluster 6 (43.8%). Down: Cluster 1 (0%), Cluster 2 (1.7%), Cluster 3 (0%), Cluster 4 (89.8%), Cluster 5 (7.6%), and Cluster 6 (0%). (E-F) GSEA analysis plot and heatmap showing decreased gene expression of the Bone Resorption and Regulation of Osteoclast Differentiation in LE compared to +R. (G) BMM cells were cultured for 3 days in the presence of M-CSF (30 ng/mL) and RANKL (100 ng/mL) in the absence or the presence of LE. To quantify relative mRNA levels, qRT-PCR was performed using primers specific for Ctsk, Mitf, and Ccr1. The results shown are mean values from three independent experiments; *** p < 0.001.
3.3. Leaf Extract of Abeliophyllum distichum Decreases RANKL-Induced NFATc1 Expression
RANKL/RANK signals to downstream targets such as c-FOS, NF-κB, and NFATc1, which are key transcription factors implicated in NFATc1 transactivation during osteoclastogenesis [12]. To examine the influence of LE of A. distichum on mRNA expression of Nfatc1, c-Fos, and p65, we first checked the reads per kilobase per million of those mRNA from RNA-seq data. As shown in Figure 3A, LE blocked Nfatc1 expression induced by RANKL, but LE had less of an effect on c-Fos and p65 expression. We further confirmed the selective inhibitory effect of LE on Nfatc1 expression by qRT-PCR analysis (Figure 3B). Consistent with qRT-PCR results, LE significantly reduced the expression of RANKL-induced NFATc1 expression, as shown in Figure 3C.
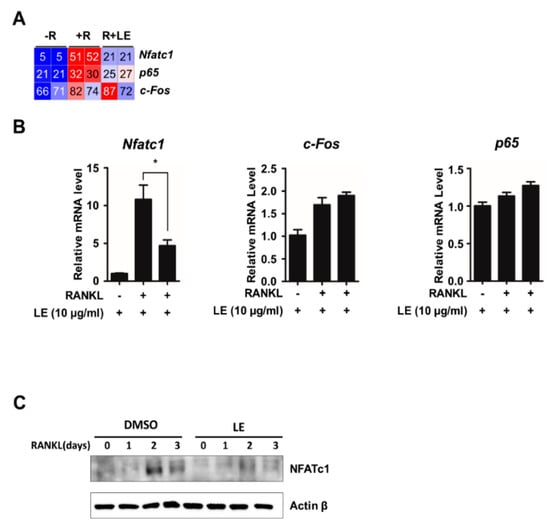
Figure 3.
Leaf extract of Abeliophyllum distichum inhibits RANKL-induced NFATc1 expression. (A) Heatmap showing the reads per kilobase per million (RPKM) of c-Fos, p65 and Nfatc1. Numbers within the figure indicate RPKM values. (B) BMM cells were cultured for 3 days in the presence of M-CSF (30 ng/mL) and RANKL (100 ng/mL) in the absence or the presence of LE. Total RNA was isolated from cell lysates, and qRT-PCR was performed using primers specific for c-Fos, p65 and Nfatc1. The mRNA levels were normalized against an internal β-actin control. The results shown are mean values from three independent experiments; * p < 0.05. (C) Whole cell lysates were prepared from M-CSF/RANKL-treated BMMs with or without LE (10 μg/mL) for 0, 1, 2, and 3 days, and analyzed by immunoblotting with antibodies against NFATc1 or β-Actin.
3.4. Leaf Extract of Abeliophyllum distichum Suppresses NFATc1 Expression by the Inhibition of Phosphorylation of ERK and the Inhibition of Transcriptional Activities of NFATc1, c-FOS, and p65
RANKL induces NFATc1 expression via diverse signaling pathways [24,25]. To elucidate the mechanism through which LE regulates the expression of NFATc1, we first examined the relevant signaling pathways such as the ERK, JNK, and p38, AKT, and NF-κB signaling pathways. BMMs pre-treated with LE for 2 h were stimulated with RANKL for the indicated times (Figure 4A). Our immunoblot analysis showed that LE inhibited RANKL-induced phosphorylation of ERK, whereas LE had less of an effect on the RANKL-induced phosphorylation of AKT, p38, JNK, and degradation of Iκ-B (Figure 4A). These results strongly suggested that LE controls NFATc1 expression induced by RANKL, mainly through the ERK signaling pathway.
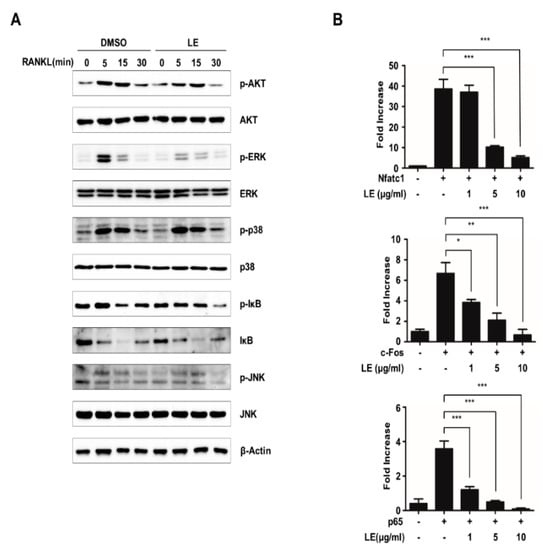
Figure 4.
Leaf extract of Abeliophyllum distichum suppresses NFATc1 expression by blocking NFATc1, p65 and c-FOS-mediated transactivity. (A) BMMs were pre-treated with DMSO and LE (10 μg/mL) for 2 h, and stimulated with RANKL for 0, 5, 15, and 30 min. The cells were lysed and immunoblotted with antibodies against p-AKT, AKT, p-ERK, ERK, p-IκB, IκB, p-p38, p38, p-JNK, and JNK. (B) 293T cells were transiently transfected with the reporter plasmid Nfatc1-Luc along with c-Fos, p65, or Nfatc1 in the presence or absence of LE. Luciferase activity was measured after 24h post-transfection. Each bar represents the mean ± SD of three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001.
It is well known that the ERK signaling pathway plays a critical role in osteoclast differentiation through the regulation of key transcription factors such as c-FOS and NFATc1 [26,27]. It has also been reported that several transcription factors, including c-FOS, p65, and NFATc1, are recruited to the Nfatc1 promoter during osteoclastogenesis [12,28,29]. To explore whether LE affects the transactivities of c-FOS, p65, and NFATc1 to the Nfatc1 promoter, we performed reporter gene assays using a luciferase reporter plasmid driven by the Nfatc1 promoter. The expression of c-FOS, p65, and NFATc1 induced NFATc1 reporter gene transcription (Figure 4B). However, the transactivities of c-FOS, p65, or NFATc1 were inhibited by LE of A. distichum. Collectively, these results suggest that LE of A. distichum suppresses the transcriptional activities of c-FOS, p65, and NFATc1 as well as ERK signaling, leading to the downregulation of NFATc1 expression.
3.5. Leaf Extract of Abeliophyllum distichum Prevents Bone Loss in Both Ovariectomized Mice and Prednisolone-Treated Zebrafish
To investigate the effect of LE on postmenopausal osteoporosis, we used an ovariectomized (OVX) mouse model. OVX mice were injected with the vehicle or LE once a week for eight weeks, and the bone microarchitecture was analyzed by micro-computed tomography (micro-CT) (Figure 5A). Our micro-CT analysis revealed that OVX caused a decrease in bone mineral density and trabecular bone volume (BV/TV), compared to the sham-operated mice, as shown in Figure 5B,C. In contrast, the treatment with LE of A. distichum alleviated bone loss in comparison to the treatment with the vehicle in OVX mice. These results were further confirmed by hematoxylin and eosin staining of decalcified bone sections (Figure 5E). Femoral sections from the OVX mice treated with vehicle showed that the trabeculae are scarce and thin compared to those from sham-operated mice. However, LE treatment in OVX mice markedly increased bone density and significantly increased the trabecular density and thickness, as shown in Figure 5D,E. Furthermore, TRAP staining showed that OVX mice with LE displayed lower osteoclast numbers per unit bone perimeter (N.Oc/B.Pm), osteoclast surface per bone surface (Oc.S/BS) and eroded surface per bone surface (ES/BS) (Figure 5F). Since zebrafish are a useful model system for GIO, we employed zebrafish to examine the in vivo effect of LE on GIO [30,31]. Zebrafish larvae at ten days post-fertilization were treated with prednisolone (25 μM) in the presence or absence of LE of A. distichum for three days, and zebrafish bone mineral density was analyzed. As shown in Figure 5G, prednisolone significantly attenuated bone mineralization in zebrafish larvae, which was counteracted by LE treatment. These results strongly suggest that LE prevents both postmenopausal osteoporosis and GIO by inhibiting osteoclast differentiation.
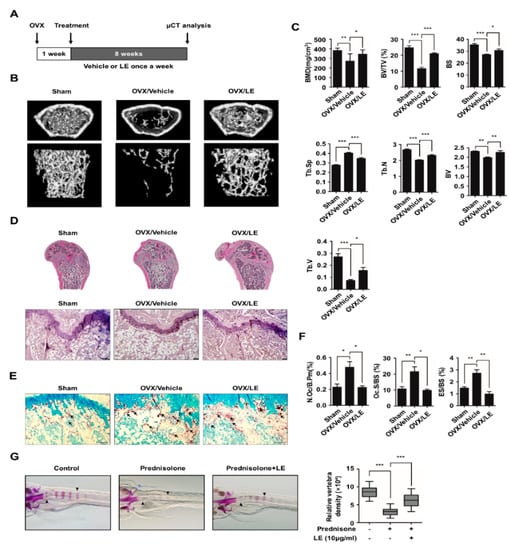
Figure 5.
Leaf extract of Abeliophyllum distichum ameliorates bone loss in osteoporotic models. (A) Schematic representation of the experimental design of the ovariectomized mouse study. (B) Representative μCT images of femurs of 18-week-old sham, vehicle-treated OVX, LE-treated OVX are shown. (C) Bone parameters including bone mineral density (BMD) and bone volume per total volume (BV/TV) were analyzed. (D, E) Histomorphometric analysis of bone. Tibial sections were stained with hematoxylin and eosin staining (D) and TRAP staining (E). Arrows indicate TRAP-positive osteoclast cells. (F) Parameters for osteoclastic bone resorption during bone morphometric analysis. Osteoclast surface per bone surface (Oc.S./B.S.), number of TRAP-positive osteoclasts per bone perimeter (N.Oc./B.Pm.), eroded surface per bone surface (ES/BS) are shown. (G) The larvae at 10 dpf. (days post-fertilization) were treated with 25 μM prednisolone in the presence or absence of LE (10 μg/mL) for 3 days. Whole-mount Alizarin red staining was performed to analyzed the mineralized bone. Relative vertebral bone density was assessed by measuring the areas of the first five stained vertebrae (V1—V5, indicated by arrowhead). * p < 0.05, ** p < 0.01, *** p < 0.001.
4. Discussion
Osteoporosis is characterized by decreased bone mass and changes in the microarchitecture of bone, which increases bone fragility. It becomes a major health problem since the prevalence of this disease is increasing due to an aging population. Variable conventional treatments such as bisphosphonates, hormone therapy, calcitonin treatment, and selective estrogen receptor modulators are available for osteoporosis. However, since these treatments have been shown to cause severe adverse effects, new treatments with fewer adverse effects are needed for osteoporosis [32]. Recently, a series of natural products and herbal medicines have been reported for their anti-osteoporotic activities [33,34,35]. A. distichum has been shown to have several pharmaceutical properties including anti-cancer activity, anti-diabetic activity, and anti-hypertensive activity [15,16,17]. In the present study, we demonstrated that the LE of A. distichum has anti-osteoporotic activity by suppressing osteoclast differentiation. Furthermore, the transcriptome analysis showed that LE decreased the expression of genes associated with osteoclast differentiation. Specifically, LE of A. distichum downregulated NFATc1 expression without affecting c-Fos and p65 expression during osteoclastogenesis. Moreover, LE inhibited RANKL-induced ERK phosphorylation and the transactivities of NFATc1, c-FOS, and p65 to regulate Nfatc1 gene expression. Finally, we observed that LE prevents bone loss in both OVX mice and prednisolone-induced osteoporotic zebrafish model. When we were preparing this manuscript, Kim et al. [36] reported that A.distichum alleviates postmenopausal osteoporosis in ovariectomized rats. They identified that ethyl acetate fraction of A.distichum inhibited the expression of c-FOS/NFATc1 and phosphorylation of MAPK (ERK, JNK and p38). Our mechanistic results are somewhat different from theirs. Such inconsistency may be due to the difference in materials (extract versus fraction), cells (primary cell versus cell line) and so on. Nevertheless, we believe that our data is the first study to report the detailed mechanism of the inhibitory effect of A.distichum on osteoclast formation through systematic approaches: 1) three types of tissues, 2) transcriptome analysis, and 3) two types of osteoporosis models.
NFATc1 is a key regulator of osteoclast differentiation, modulating a series of osteoclast-specific genes including TRAP, cathepsin K, osteoclast-associated receptor, and MMP-9 as well as NFATc1 itself. Recently, the expression and activation of NFATc1 have been reported to be regulated via diverse mechanisms, including transcription, acetylation, and non-coding RNAs [12]. RANKL stimulates signaling pathways including ERK, JNK, AKT, p38, and NF-κB, leading to the binding of several transcription factors such as c-FOS, NF-κB, MITF, and NFATc1 itself to Nfatc1 promoter [12,26]. In this study, we observed that LE of A. distichum induced a decrease of RANKL-stimulated mRNA expression of Nfatc1 and protein level of NFATc1 while it did not affect the expression of c-Fos and p65 that are involved in Nfatc1 expression. Moreover, LE did not affect RANKL-induced p65 translocation into the nucleus, as shown in Supplementary Figure 1. Interestingly, it was observed that the LE of A. distichum significantly inhibited NFATc1/c-FOS/p65 transactivation for Nfatc1 expression. Additionally, LE of A. distichum suppressed ERK phosphorylation. These findings suggest that LE of A. distichum may downregulate RANKL-induced NFATc1 expression by targeting the ERK pathway and Nfatc1 transcription by NFATc1, c-FOS, and p65. Further studies will be needed to elucidate the exact molecular mechanisms underlying the effects of LE of A. distichum on NFATc1 expression. An unsolved question in this study is which compound(s) of LE exhibited anti-osteoclastogenic activity. Recent studies demonstrated that A. distichum methanol extract possesses more than 20 polyphenolic compounds [14,15,37]. Considered that seven compounds (acteoside, chlorogenic acid, caffeic acid, ferulic acid, naringenin, taxifolin, and quercetin) among the polyphenolic compounds are known to inhibit RANKL-mediated osteoclastogenesis [38,39,40,41,42,43,44], we assume that these bioactive compounds might play a role in anti-osteoclstogenesis.
Postmenopausal osteoporosis stems from estrogen deficiency which affects all types of bone cells and results in an increase in bone turnover. Meanwhile, GIO is a common form of secondary osteoporosis [7]. Glucocorticoids have been used as anti-inflammatory and immunosuppressive agents [45] but have been shown to increase the differentiation of osteoclasts as well as decrease the function of osteoblasts during long-term treatment. In this study, we employed two osteoporosis animal models, including OVX mice and a zebrafish osteoporotic model-induced by prednisolone, to evaluate the effects of LE on bone mass in postmenopausal osteoporosis and GIO. LE of A. distichum rescued the OVX- or GIO-induced decrease in bone mass. These results suggest that LE of A. distichum could have therapeutic value in treating postmenopausal osteoporosis and GIO.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-7737/9/8/212/s1, Table S1: K-means clustering of 2574 differentially expressed genes (DEGs), Table S2: List of affected genes by LE treatment.
Author Contributions
K.L., S.-J.Y., and K.K. conceived and design the study; K.L. analyzed transcriptome data; H.L., Y.-J.J., H.L., E.K., Y.K., T.-K.Y., T.K.H., J.-I.P., and S.-J.Y. performed the experiments; E.K. and J.-I.P. provided technical assistance; K.L., S.-J.Y., and K.K. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was carried out with the support of the National Research Foundation of Korea (2020R1A2C1008179 to K.K.; 2019R1I1A1A01061125 to S.-J.Y.), the Korea Institute of Oriental Medicine, Ministry of Education, Science and Technology, Republic of Korea (KSN2012330, W20102 to J.-I.P.) and R&D Program for Forest Science Technology (Project No. 2019139A00-1920-0001 and 2020249A00-2021-0001 to K.L.) provided by Korea Forest Service (Korea Forestry Promotion Institute).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| RANKL | Receptor activator of the NF-κB ligand |
| NFATc1 | Nuclear factor of activated T-cells, cytoplasmic 1 |
| ERK | Extracellular-signal-regulated kinase |
| BMM | Bone marrow-derived macrophage |
| GIO | Glucocorticoid-induced osteoporosis |
| TRAP | Tartrate resistant acid phosphatase |
References
- Yi, S.J.; Lee, H.; Lee, J.; Lee, K.; Kim, J.; Kim, Y.; Park, J.I.; Kim, K. Bone Remodeling: Histone Modifications as Fate Determinants of Bone Cell Differentiation. Int. J. Mol. Sci. 2019, 20, 3147. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; McDonald, J.M. Disorders of bone remodeling. Annu. Rev. Pathol. 2011, 6, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Glaser, D.L.; Kaplan, F.S. Osteoporosis. Definition and clinical presentation. Spine 1997, 22, 12S–16S. [Google Scholar] [CrossRef] [PubMed]
- Compston, J.E.; McClung, M.R.; Leslie, W.D. Osteoporosis. Lancet 2019, 393, 364–376. [Google Scholar] [CrossRef]
- Cauley, J.A. Public health impact of osteoporosis. J. Gerontol. Ser. A Biol. Sci. Med Sci. 2013, 68, 1243–1251. [Google Scholar] [CrossRef] [PubMed]
- Dobbs, M.B.; Buckwalter, J.; Saltzman, C. Osteoporosis: The increasing role of the orthopaedist. Iowa Orthop. J. 1999, 19, 43–52. [Google Scholar]
- Mazziotti, G.; Angeli, A.; Bilezikian, J.P.; Canalis, E.; Giustina, A. Glucocorticoid-induced osteoporosis: An update. Trends Endocrinol. Metab. 2006, 17, 144–149. [Google Scholar] [CrossRef]
- Hsu, H.; Lacey, D.L.; Dunstan, C.R.; Solovyev, I.; Colombero, A.; Timms, E.; Tan, H.L.; Elliott, G.; Kelley, M.J.; Sarosi, I.; et al. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc. Natl. Acad. Sci. USA 1999, 96, 3540–3545. [Google Scholar] [CrossRef]
- Liu, C.; Walter, T.S.; Huang, P.; Zhang, S.; Zhu, X.; Wu, Y.; Wedderburn, L.R.; Tang, P.; Owens, R.J.; Stuart, D.I.; et al. Structural and functional insights of RANKL-RANK interaction and signaling. J. Immunol. 2010, 184, 6910–6919. [Google Scholar] [CrossRef]
- Nakagawa, N.; Kinosaki, M.; Yamaguchi, K.; Shima, N.; Yasuda, H.; Yano, K.; Morinaga, T.; Higashio, K. RANK is the essential signaling receptor for osteoclast differentiation factor in osteoclastogenesis. Biochem. Biophys. Res. Commun. 1998, 253, 395–400. [Google Scholar] [CrossRef]
- Lee, N.K.; Choi, Y.G.; Baik, J.Y.; Han, S.Y.; Jeong, D.W.; Bae, Y.S.; Kim, N.; Lee, S.Y. A crucial role for reactive oxygen species in RANKL-induced osteoclast differentiation. Blood 2005, 106, 852–859. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kim, N. Regulation of NFATc1 in Osteoclast Differentiation. J. Bone Metab. 2014, 21, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Boyce, B.F.; Yamashita, T.; Yao, Z.Q.; Zhang, Q.; Li, F.; Xing, L.P. Roles for NF-kappa B and c-Fos in osteoclasts. J. Bone Miner. Metab. 2005, 23, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Seo, E.-J.; Sung, J.; Choi, K.M.; Kim, H.; Kim, J.-S.; Lee, J.; Efferth, T.; Hyun, T.K. Polyphenolic compounds, antioxidant and anti-inflammatory effects of Abeliophyllum distichum Nakai extract. J. Appl. Bot. Food Qual. 2017, 90. [Google Scholar] [CrossRef]
- Li, H.M.; Kim, J.K.; Jang, J.M.; Cui, C.B.; Lim, S.S. Analysis of the inhibitory activity of Abeliophyllum distichum leaf constituents against aldose reductase by using high-speed counter current chromatography. Arch. Pharmacal. Res. 2013, 36, 1104–1112. [Google Scholar] [CrossRef]
- Oh, H.; Kang, D.G.; Kwon, T.O.; Jang, K.K.; Chai, K.Y.; Yun, Y.G.; Chung, H.T.; Lee, H.S. Four glycosides from the leaves of Abeliophyllum distichum with inhibitory effects on angiotensin converting enzyme. Phytother. Res. 2003, 17, 811–813. [Google Scholar] [CrossRef]
- Park, G.H.; Park, J.H.; Eo, H.J.; Song, H.M.; Woo, S.H.; Kim, M.K.; Lee, J.W.; Lee, M.H.; Lee, J.R.; Koo, J.S.; et al. The induction of activating transcription factor 3 (ATF3) contributes to anti-cancer activity of Abeliophyllum distichum Nakai in human colorectal cancer cells. BMC Complement. Altern. Med. 2014, 14, 487. [Google Scholar] [CrossRef]
- Lee, H.; Lee, K.; Lee, S.; Lee, J.; Jeong, W.T.; Lim, H.B.; Hyun, T.K.; Yi, S.J.; Kim, K. Ethyl Acetate Fraction of Aqueous Extract of Lentinula edodes Inhibits Osteoclastogenesis by Suppressing NFATc1 Expression. Int. J. Med. Sci. 2020, 17, 21–32. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- An, D.; Kim, K.; Lu, W. Defective entry into mitosis 1 (Dim1) negatively regulates osteoclastogenesis by inhibiting the expression of nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (NFATc1). J. Biol. Chem. 2014, 289, 24366–24373. [Google Scholar] [CrossRef] [PubMed]
- Dempster, D.W.; Compston, J.E.; Drezner, M.K.; Glorieux, F.H.; Kanis, J.A.; Malluche, H.; Meunier, P.J.; Ott, S.M.; Recker, R.R.; Parfitt, A.M. Standardized nomenclature, symbols, and units for bone histomorphometry: A 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J. Bone Miner. Res. 2013, 28, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, J.; Lee, H.; Shin, W.R.; Lee, S.; Lee, J.; Park, J.I.; Jhun, B.H.; Kim, Y.H.; Yi, S.J.; et al. Tetracycline Analogs Inhibit Osteoclast Differentiation by Suppressing MMP-9-Mediated Histone H3 Cleavage. Int. J. Mol. Sci. 2019, 20, 4038. [Google Scholar] [CrossRef] [PubMed]
- Abu-Amer, Y. NF-kappaB signaling and bone resorption. Osteoporos. Int. 2013, 24, 2377–2386. [Google Scholar] [CrossRef]
- Asagiri, M.; Takayanagi, H. The molecular understanding of osteoclast differentiation. Bone 2007, 40, 251–264. [Google Scholar] [CrossRef]
- Lee, K.; Seo, I.; Choi, M.H.; Jeong, D. Roles of Mitogen-Activated Protein Kinases in Osteoclast Biology. Int. J. Mol. Sci. 2018, 19, 3004. [Google Scholar] [CrossRef]
- Monje, P.; Hernandez-Losa, J.; Lyons, R.J.; Castellone, M.D.; Gutkind, J.S. Regulation of the transcriptional activity of c-Fos by ERK. A novel role for the prolyl isomerase PIN1. J. Biol. Chem. 2005, 280, 35081–35084. [Google Scholar] [CrossRef]
- Kim, K.; Kim, J.H.; Lee, J.; Jin, H.M.; Kook, H.; Kim, K.K.; Lee, S.Y.; Kim, N. MafB negatively regulates RANKL-mediated osteoclast differentiation. Blood 2007, 109, 3253–3259. [Google Scholar] [CrossRef]
- Zhao, B.; Ivashkiv, L.B. Negative regulation of osteoclastogenesis and bone resorption by cytokines and transcriptional repressors. Arthritis Res. Ther. 2011, 13, 234. [Google Scholar] [CrossRef]
- Barrett, R.; Chappell, C.; Quick, M.; Fleming, A. A rapid, high content, in vivo model of glucocorticoid-induced osteoporosis. Biotechnol. J. 2006, 1, 651–655. [Google Scholar] [CrossRef]
- He, H.; Wang, C.; Tang, Q.; Yang, F.; Xu, Y. Possible mechanisms of prednisolone-induced osteoporosis in zebrafish larva. Biomed. Pharmacother. 2018, 101, 981–987. [Google Scholar] [CrossRef] [PubMed]
- Compston, J.; Cooper, A.; Cooper, C.; Gittoes, N.; Gregson, C.; Harvey, N.; Hope, S.; Kanis, J.A.; McCloskey, E.V.; Poole, K.E.S.; et al. UK clinical guideline for the prevention and treatment of osteoporosis. Arch. Osteoporos. 2017, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Cheon, Y.H.; Baek, J.M.; Park, S.H.; Ahn, S.J.; Lee, M.S.; Oh, J.; Kim, J.Y. Stauntonia hexaphylla (Lardizabalaceae) leaf methanol extract inhibits osteoclastogenesis and bone resorption activity via proteasome-mediated degradation of c-Fos protein and suppression of NFATc1 expression. BMC Complement. Altern. Med. 2015, 15, 280. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.H.; Jang, S.A.; Kim, T.; Ha, H. Forsythia suspensa Protects against Bone Loss in Ovariectomized Mice. Nutrients 2019, 11, 1831. [Google Scholar] [CrossRef] [PubMed]
- Suvarna, V.; Sarkar, M.; Chaubey, P.; Khan, T.; Sherje, A.; Patel, K.; Dravyakar, B. Bone Health and Natural Products—An Insight. Front. Pharmacol. 2018, 9, 981. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Kim, J.H.; Kim, M.; Park, J.H.; Sohn, Y.; Jung, H.S. Abeliophyllum distichum Nakai alleviates postmenopausal osteoporosis in ovariectomized rats and prevents RANKL-induced osteoclastogenesis In Vitro. J. Ethnopharmacol. 2020, 257, 112828. [Google Scholar] [CrossRef] [PubMed]
- Yoo, T.K.; Kim, J.S.; Hyun, T.K. Polyphenolic Composition and Anti-Melanoma Activity of White Forsythia (Abeliophyllum distichum Nakai) Organ Extracts. Plants 2020, 9, 757. [Google Scholar] [CrossRef]
- Lee, S.Y.; Lee, K.S.; Yi, S.H.; Kook, S.H.; Lee, J.C. Acteoside suppresses RANKL-mediated osteoclastogenesis by inhibiting c-Fos induction and NF-kappaB pathway and attenuating ROS production. PLoS ONE 2013, 8, e80873. [Google Scholar] [CrossRef]
- Ang, E.S.; Pavlos, N.J.; Chai, L.Y.; Qi, M.; Cheng, T.S.; Steer, J.H.; Joyce, D.A.; Zheng, M.H.; Xu, J. Caffeic acid phenethyl ester, an active component of honeybee propolis attenuates osteoclastogenesis and bone resorption via the suppression of RANKL-induced NF-kappaB and NFAT activity. J. Cell. Physiol. 2009, 221, 642–649. [Google Scholar] [CrossRef]
- Kwak, S.C.; Lee, C.; Kim, J.Y.; Oh, H.M.; So, H.S.; Lee, M.S.; Rho, M.C.; Oh, J. Chlorogenic acid inhibits osteoclast differentiation and bone resorption by down-regulation of receptor activator of nuclear factor kappa-B ligand-induced nuclear factor of activated T cells c1 expression. Biol. Pharm. Bull. 2013, 36, 1779–1786. [Google Scholar] [CrossRef]
- Doss, H.M.; Samarpita, S.; Ganesan, R.; Rasool, M. Ferulic acid, a dietary polyphenol suppresses osteoclast differentiation and bone erosion via the inhibition of RANKL dependent NF-kappaB signalling pathway. Life Sci. 2018, 207, 284–295. [Google Scholar] [CrossRef] [PubMed]
- La, V.D.; Tanabe, S.; Grenier, D. Naringenin inhibits human osteoclastogenesis and osteoclastic bone resorption. J. Periodontal Res. 2009, 44, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Masuhara, M.; Tsukahara, T.; Tomita, K.; Furukawa, M.; Miyawaki, S.; Sato, T. A relation between osteoclastogenesis inhibition and membrane-type estrogen receptor GPR30. Biochem. Biophys. Rep. 2016, 8, 389–394. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kyung, T.W.; Lee, J.E.; Shin, H.H.; Choi, H.S. Rutin inhibits osteoclast formation by decreasing reactive oxygen species and TNF-alpha by inhibiting activation of NF-kappaB. Exp. Mol. Med. 2008, 40, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Fraser, L.A.; Adachi, J.D. Glucocorticoid-induced osteoporosis: Treatment update and review. Ther. Adv. Musculoskelet. Dis. 2009, 1, 71–85. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).