Modification and Targeted Design of N-Terminal Truncates Derived from Brevinin with Improved Therapeutic Efficacy
Abstract
1. Introduction
2. Materials and Methods
2.1. Fmoc Solid-Phase Peptide Synthesis
2.2. Secondary Structure Prediction and Verification
2.3. Antimicrobial Assay
2.4. Haemolytic Assay
2.5. SYTOX Green Uptake Assay
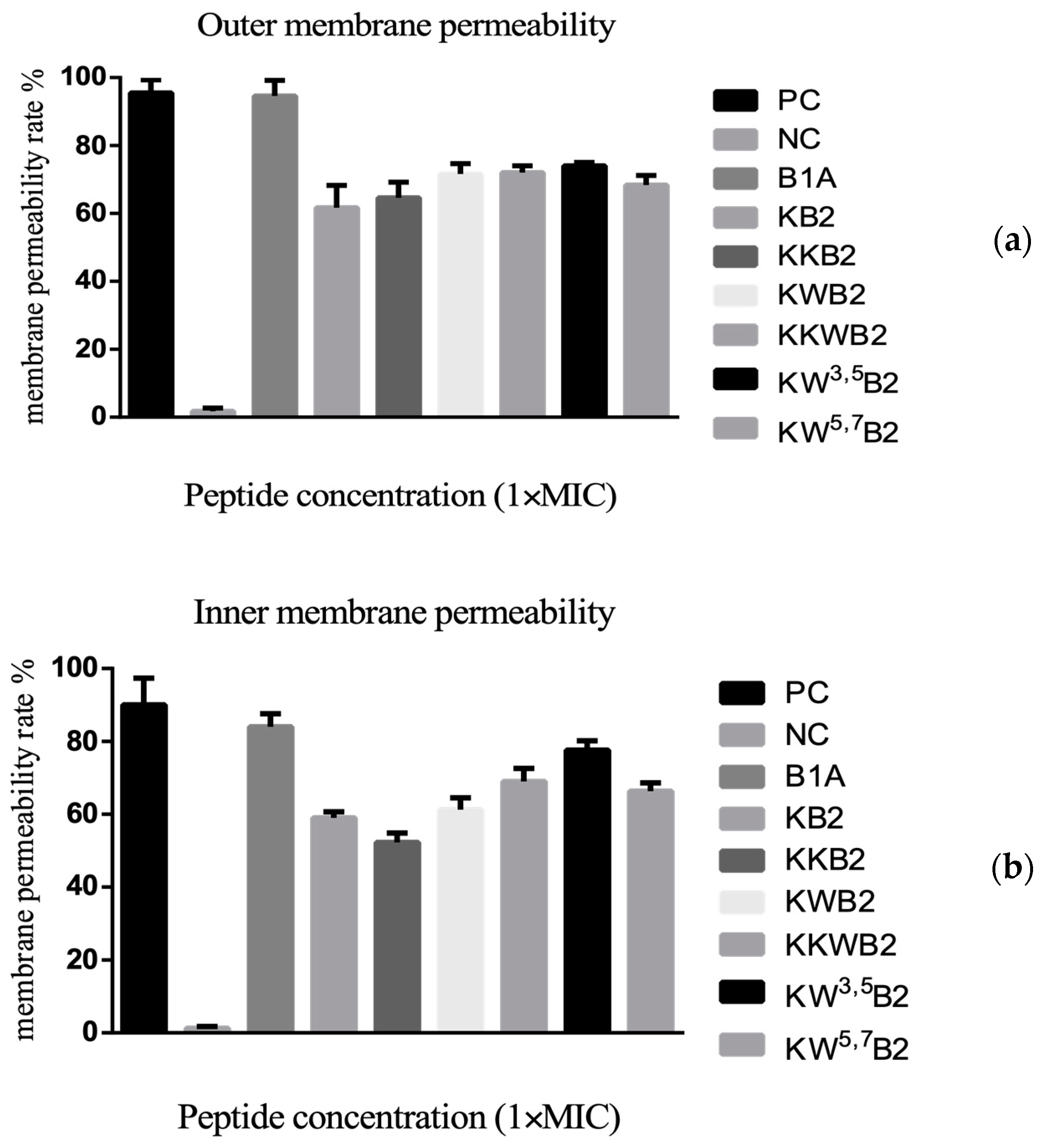
2.6. Outer Membrane Permeability Assay
2.7. Inner Membrane Permeability Assay
2.8. Statistical Analysis
3. Results
3.1. Rational Design of Novel Peptide
3.2. Secondary Structure Analysis
3.3. Cytotoxicity
3.4. Antimicrobial Activities
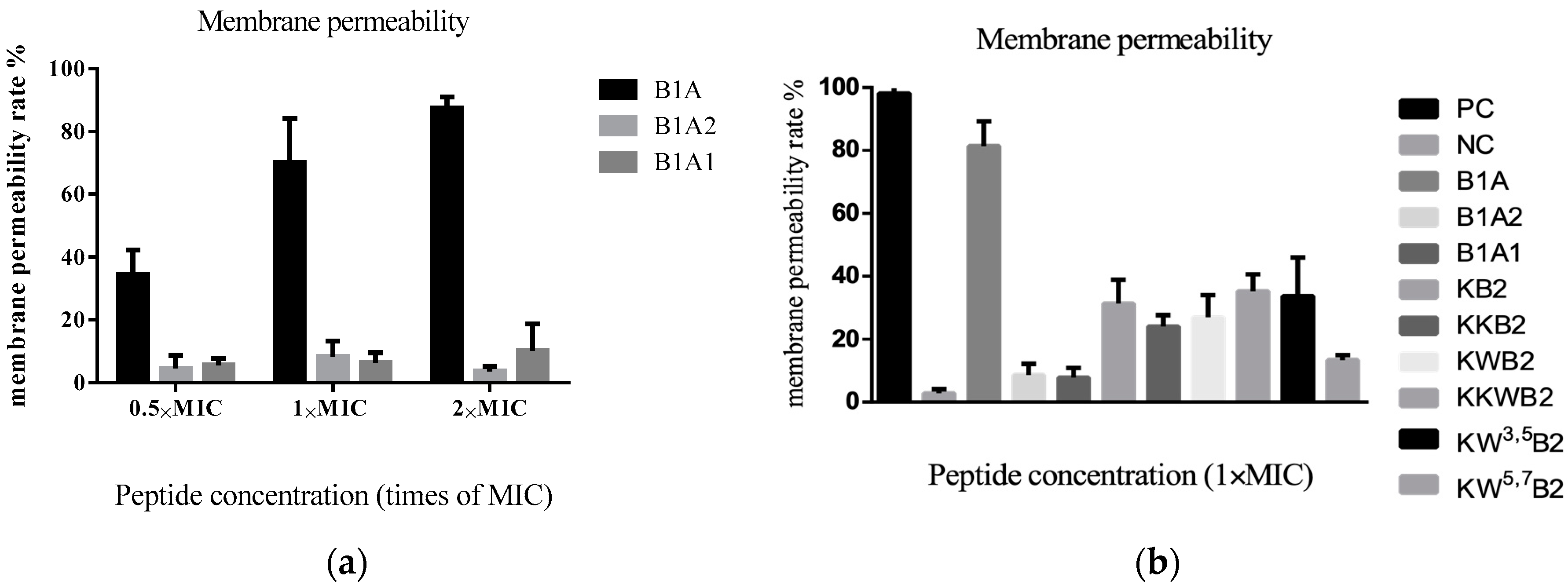
3.5. The Membrane of Gram-Positive Bacteria
3.6. The Membrane of Gram-Negative Bacteria
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Li, B.; Webster, T.J. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. J. Orthop. Res.® 2018, 36, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.; Yedery, R.; Aranha, C. Antimicrobial peptides: Premises and promises. Int. J. Antimicrob. Agents 2004, 24, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Xi, X.; Li, B.; Chen, T.; Kwok, H.F. A review on bradykinin-related peptides isolated from amphibian skin secretion. Toxins 2015, 7, 951–970. [Google Scholar] [CrossRef]
- Lee, J.-K.; Luchian, T.; Park, Y. New antimicrobial peptide kills drug-resistant pathogens without detectable resistance. Oncotarget 2018, 9, 15616. [Google Scholar] [CrossRef]
- Savelyeva, A.; Ghavami, S.; Davoodpour, P.; Asoodeh, A.; Łos, M.J. An Overview of Brevinin Superfamily: Structure, Function and Clinical Perspectives. Anticancer Genes; Springer: London, UK, 2014; pp. 197–212. [Google Scholar]
- Novković, M.; Simunić, J.; Bojović, V.; Tossi, A.; Juretić, D. DADP: The database of anuran defense peptides. Bioinformatics 2012, 28, 1406–1407. [Google Scholar] [CrossRef]
- Clark, D.P.; Durell, S.; Maloy, W.L.; Zasloff, M. Ranalexin. A novel antimicrobial peptide from bullfrog (Rana catesbeiana) skin, structurally related to the bacterial antibiotic, polymyxin. J. Biol. Chem. 1994, 269, 10849–10855. [Google Scholar]
- Kwon, M.-Y.; Hong, S.-Y.; Lee, K.-H. Structure-activity analysis of brevinin 1E amide, an antimicrobial peptide from Rana esculenta. Biochim. et Biophys. Acta (BBA) Protein Struct. Mol. Enzymol. 1998, 1387, 239–248. [Google Scholar] [CrossRef]
- Abraham, P.; Anand Sundaram, A.R.; Reshmy, V.; George, S.; Kumar, K.S. Structure-activity relationship and mode of action of a frog secreted antibacterial peptide B1CTcu5 using synthetically and modularly modified or deleted (SMMD) peptides. PLoS ONE 2015, 10, e0124210. [Google Scholar] [CrossRef]
- Barra, D.; Simmaco, M. Amphibian skin: A promising resource for antimicrobial peptides. Trends Biotechnol. 1995, 13, 205–209. [Google Scholar] [CrossRef]
- Kumari, V.; Nagaraj, R. Structure—Function studies on the amphibian peptide brevinin 1E: Translocating the cationic segment from the C-terminal end to a central position favors selective antibacterial activity. J. Pept. Res. 2001, 58, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Kolodziejek, J.; Nowotny, N. Antimicrobial peptides from ranid frogs: Taxonomic and phylogenetic markers and a potential source of new therapeutic agents. Biochim. et Biophys. Acta 2004, 1696, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Basir, Y.J.; Knoop, F.C.; Dulka, J.; Conlon, J.M. Multiple antimicrobial peptides and peptides related to bradykinin and neuromedin N isolated from skin secretions of the pickerel frog, Rana palustris. Biochim. et Biophys. Acta (BBA) Protein Struct. Mol. Enzymol. 2000, 1543, 95–105. [Google Scholar] [CrossRef]
- Zhou, M.; Wang, L.; Owens, D.E.; Chen, T.; Walker, B.; Shaw, C. Rapid identification of precursor cDNAs encoding five structural classes of antimicrobial peptides from pickerel frog (Rana palustris) skin secretion by single step “shotgun” cloning. Peptides 2007, 28, 1605–1610. [Google Scholar] [CrossRef]
- Lin, Y.; Hu, N.; He, H.; Ma, C.; Zhou, M.; Wang, L.; Chen, T. A hylarana latouchii skin secretion-derived novel bombesin-related pentadecapeptide (ranatensin-HLa) evoke myotropic effects on the in vitro rat smooth muscles. Toxins 2019, 11, 204. [Google Scholar] [CrossRef]
- Sidorova, M.; Molokoedov, A.; Az′muko, A.; Kudryavtseva, E.; Krause, E.; Ovchinnikov, M.; Bespalova, Z.D. The use of hydrogen peroxide for closing disulfide bridges in peptides. Russ. J. Bioorganic Chem. 2004, 30, 101–110. [Google Scholar] [CrossRef]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A web server to screen sequences with specific α-helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef]
- Gao, Y.; Wu, D.; Xi, X.; Wu, Y.; Ma, C.; Zhou, M.; Wang, L.; Yang, M.; Chen, T.; Shaw, C. Identification and characterisation of the antimicrobial peptide, phylloseptin-PT, from the skin secretion of Phyllomedusa tarsius, and comparison of activity with designed, cationicity-enhanced analogues and diastereomers. Molecules 2016, 21, 1667. [Google Scholar] [CrossRef]
- Gao, Y.; Wu, D.; Wang, L.; Lin, C.; Ma, C.; Xi, X.; Zhou, M.; Duan, J.; Bininda-Emonds, O.R.; Chen, T. Targeted modification of a novel amphibian antimicrobial peptide from Phyllomedusa tarsius to enhance its activity against MRSA and microbial biofilm. Front. Microbiol. 2017, 8, 628. [Google Scholar] [CrossRef] [PubMed]
- Kolari, M.; Rautiainen, J.; Hentze, H.-P.; Alakomi, H.-L.; Forssell, P. Biocide Formulation and Method for Treating Water. U.S. Patent 20150351389A1, 10 December 2015. [Google Scholar]
- Loh, B.; Grant, C.; Hancock, R. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1984, 26, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Skerlavaj, B.; Romeo, D.; Gennaro, R. Rapid membrane permeabilization and inhibition of vital functions of gram-negative bacteria by bactenecins. Infect. Immun. 1990, 58, 3724–3730. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, R.; Barton, A.; Daher, K.A.; Harwig, S.; Ganz, T.; Selsted, M.E. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J. Clin. Investig. 1989, 84, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Zohrab, F.; Askarian, S.; Jalili, A.; Oskuee, R.K. Biological properties, current applications and potential therapeautic applications of brevinin peptide superfamily. Int. J. Pept. Res. Ther. 2019, 25, 39–48. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef]
- Won, H.-S.; Kang, S.-J.; Lee, B.-J. Action mechanism and structural requirements of the antimicrobial peptides, gaegurins. Biochim. et Biophys. Acta (BBA) Biomembr. 2009, 1788, 1620–1629. [Google Scholar] [CrossRef]
- Suh, J.-Y.; Lee, K.-H.; Chi, S.-W.; Hong, S.-Y.; Choi, B.-W.; Moon, H.-M.; Choi, B.-S. Unusually stable helical kink in the antimicrobial peptide—A derivative of gaegurin. FEBS Lett. 1996, 392, 309–312. [Google Scholar] [CrossRef]
- Wang, J.; Dou, X.; Song, J.; Lyu, Y.; Zhu, X.; Xu, L.; Li, W.; Shan, A. Antimicrobial peptides: Promising alternatives in the post feeding antibiotic era. Med. Res. Rev. 2019, 39, 831–859. [Google Scholar] [CrossRef]
- Åmand, H.L.; Fant, K.; Nordén, B.; Esbjörner, E.K. Stimulated endocytosis in penetratin uptake: Effect of arginine and lysine. Biochem. Biophys. Res. Commun. 2008, 371, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Situ, A.J.; Kang, S.-M.; Frey, B.B.; An, W.; Kim, C.; Ulmer, T.S. Membrane anchoring of α-helical proteins: Role of tryptophan. J. Phys. Chem. B 2018, 122, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Arias, M.; Nguyen, L.T.; Kuczynski, A.M.; Lejon, T.; Vogel, H.J. Position-dependent influence of the three trp residues on the membrane activity of the antimicrobial peptide, tritrpticin. Antibiotics 2014, 3, 595–616. [Google Scholar] [CrossRef]
- Rekdal, Ø.; Haug, B.E.; Kalaaji, M.; Hunter, H.N.; Lindin, I.; Israelsson, I.; Solstad, T.; Yang, N.; Brandl, M.; Mantzilas, D. Relative spatial positions of tryptophan and cationic residues in helical membrane-active peptides determine their cytotoxicity. J. Biol. Chem. 2012, 287, 233–244. [Google Scholar] [CrossRef] [PubMed]
- de Planque, M.R.; Goormaghtigh, E.; Greathouse, D.V.; Koeppe, R.E.; Kruijtzer, J.A.; Liskamp, R.M.; de Kruijff, B.; Killian, J.A. Sensitivity of single membrane-spanning α-helical peptides to hydrophobic mismatch with a lipid bilayer: Effects on backbone structure, orientation, and extent of membrane incorporation. Biochemistry 2001, 40, 5000–5010. [Google Scholar] [CrossRef]
- Schibli, D.J.; Epand, R.F.; Vogel, H.J.; Epand, R.M. Tryptophan-rich antimicrobial peptides: Comparative properties and membrane interactions. Biochem. Cell Biol. 2002, 80, 667–677. [Google Scholar] [CrossRef]
- Bi, X.; Wang, C.; Ma, L.; Sun, Y.; Shang, D. Investigation of the role of tryptophan residues in cationic antimicrobial peptides to determine the mechanism of antimicrobial action. J. Appl. Microbiol. 2013, 115, 663–672. [Google Scholar] [CrossRef]
- Mishra, A.K.; Choi, J.; Moon, E.; Baek, K.-H. Tryptophan-rich and proline-rich antimicrobial peptides. Molecules 2018, 23, 815. [Google Scholar] [CrossRef]
- Le, C.-F.; Fang, C.-M.; Sekaran, S.D. Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob. Agents Chemother. 2017, 61, e02340-16. [Google Scholar] [CrossRef]
- Ehrenstein, G.; Lecar, H. Electrically gated ionic channels in lipid bilayers. Q. Rev. Biophys. 1977, 10, 1–34. [Google Scholar] [CrossRef]
- Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. et Biophys. Acta (BBA) Biomembr. 1999, 1462, 55–70. [Google Scholar] [CrossRef]
- Tachi, T.; Epand, R.F.; Epand, R.M.; Matsuzaki, K. Position-dependent hydrophobicity of the antimicrobial magainin peptide affects the mode of peptide—Lipid interactions and selective toxicity. Biochemistry 2002, 41, 10723–10731. [Google Scholar] [CrossRef]
- Avrahami, D.; Shai, Y. Conjugation of a magainin analogue with lipophilic acids controls hydrophobicity, solution assembly, and cell selectivity. Biochemistry 2002, 41, 2254–2263. [Google Scholar] [CrossRef] [PubMed]
- Blondelle, S.E.; Houghten, R.A. Hemolytic and antimicrobial activities of the twenty-four individual omission analogs of melittin. Biochemistry 1991, 30, 4671–4678. [Google Scholar] [CrossRef] [PubMed]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Kondejewski, L.H.; Lee, D.L.; Jelokhani-Niaraki, M.; Farmer, S.W.; Hancock, R.E.; Hodges, R.S. Optimization of microbial specificity in cyclic peptides by modulation of hydrophobicity within a defined structural framework. J. Biol. Chem. 2002, 277, 67–74. [Google Scholar] [CrossRef]
- Dathe, M.; Wieprecht, T.; Nikolenko, H.; Handel, L.; Maloy, W.L.; MacDonald, D.L.; Beyermann, M.; Bienert, M. Hydrophobicity, hydrophobic moment and angle subtended by charged residues modulate antibacterial and haemolytic activity of amphipathic helical peptides. FEBS Lett. 1997, 403, 208–212. [Google Scholar] [CrossRef]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of peptide hydrophobicity in the mechanism of action of α-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef]
- Lorenzón, E.; Cespedes, G.; Vicente, E.; Nogueira, L.; Bauab, T.; Castro, M.; Cilli, E.M. Effects of dimerization on the structure and biological activity of antimicrobial peptide Ctx-Ha. Antimicrob. Agents Chemother. 2012, 56, 3004–3010. [Google Scholar] [CrossRef]
- Lorenzon, E.; Sanches, P.; Nogueira, L.; Bauab, T.; Cilli, E.M. Dimerization of aurein 1.2: Effects in structure, antimicrobial activity and aggregation of Candida albicans cells. Amino Acids 2013, 44, 1521–1528. [Google Scholar] [CrossRef] [PubMed]
- Zou, R.; Zhu, X.; Tu, Y.; Wu, J.; Landry, M.P. Activity of antimicrobial peptide aggregates decreases with increased cell membrane embedding free energy cost. Biochemistry 2018, 57, 2606–2610. [Google Scholar] [PubMed]
- Mant, C.T.; Chen, Y.; Hodges, R.S. Temperature profiling of polypeptides in reversed-phase liquid chromatography: I. Monitoring of dimerization and unfolding of amphipathic α-helical peptides. J. Chromatogr. A 2003, 1009, 29–43. [Google Scholar] [CrossRef]
- Mant, C.T.; Tripet, B.; Hodges, R.S. Temperature profiling of polypeptides in reversed-phase liquid chromatography: II. Monitoring of folding and stability of two-stranded α-helical coiled-coils. J. Chromatogr. A 2003, 1009, 45–59. [Google Scholar] [CrossRef]

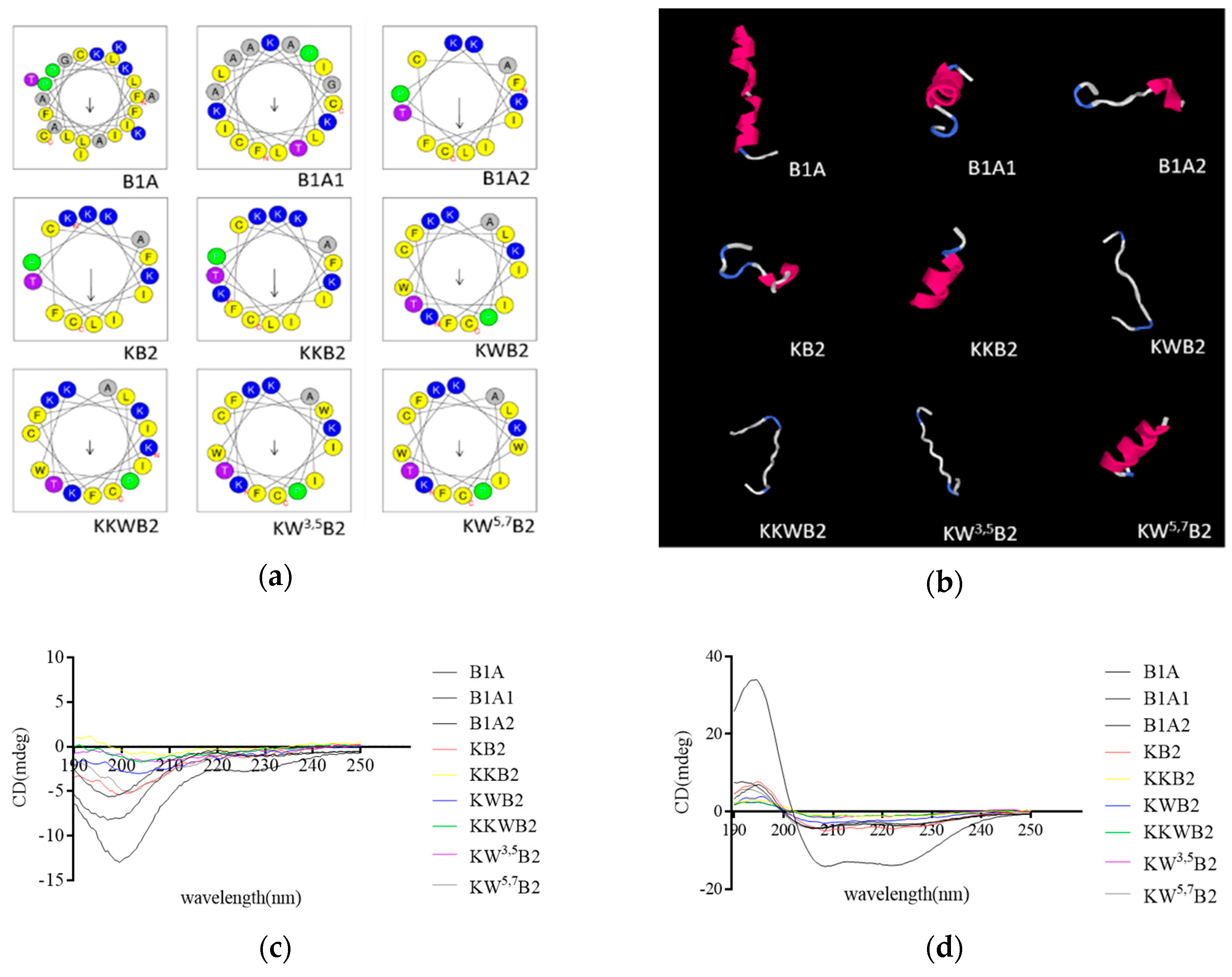
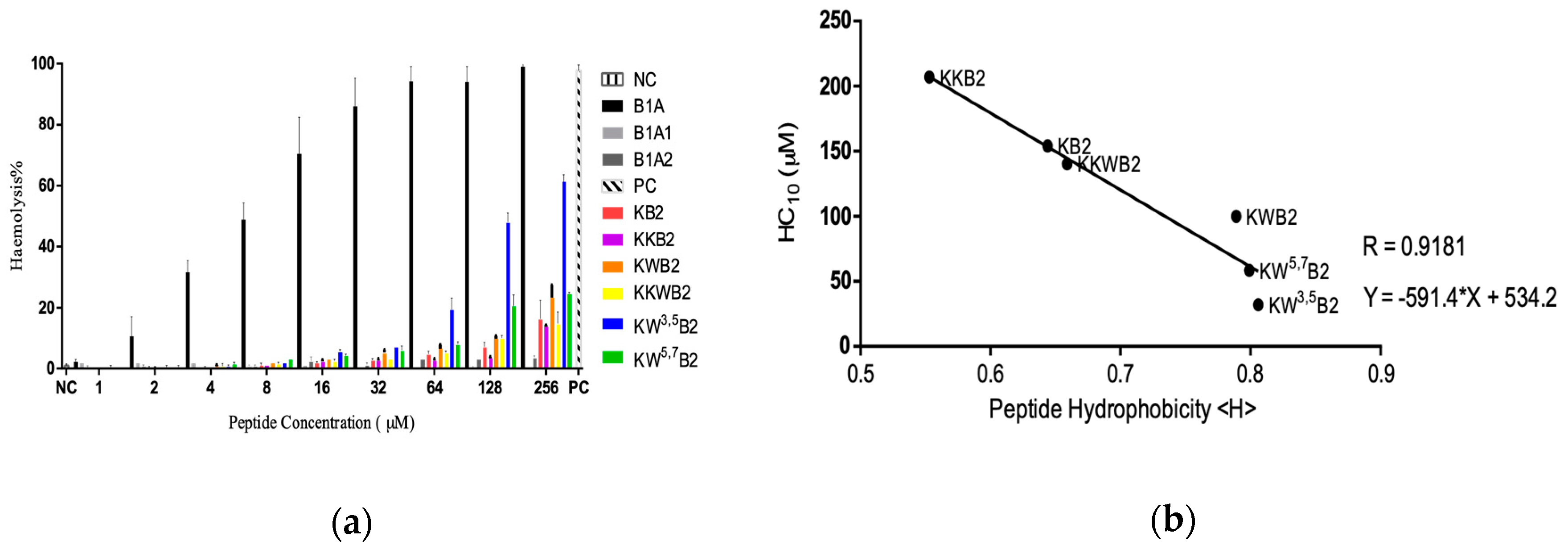
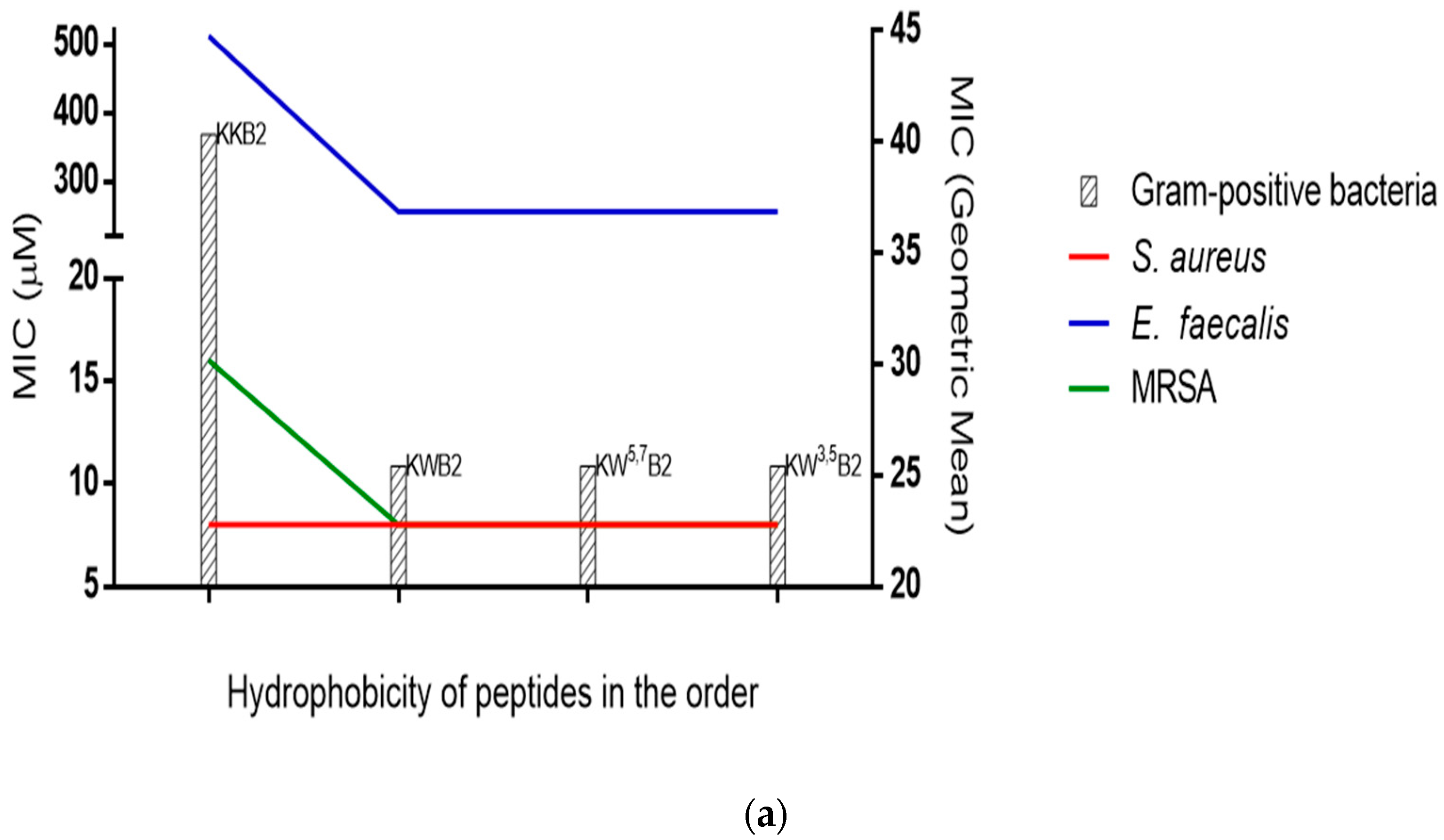
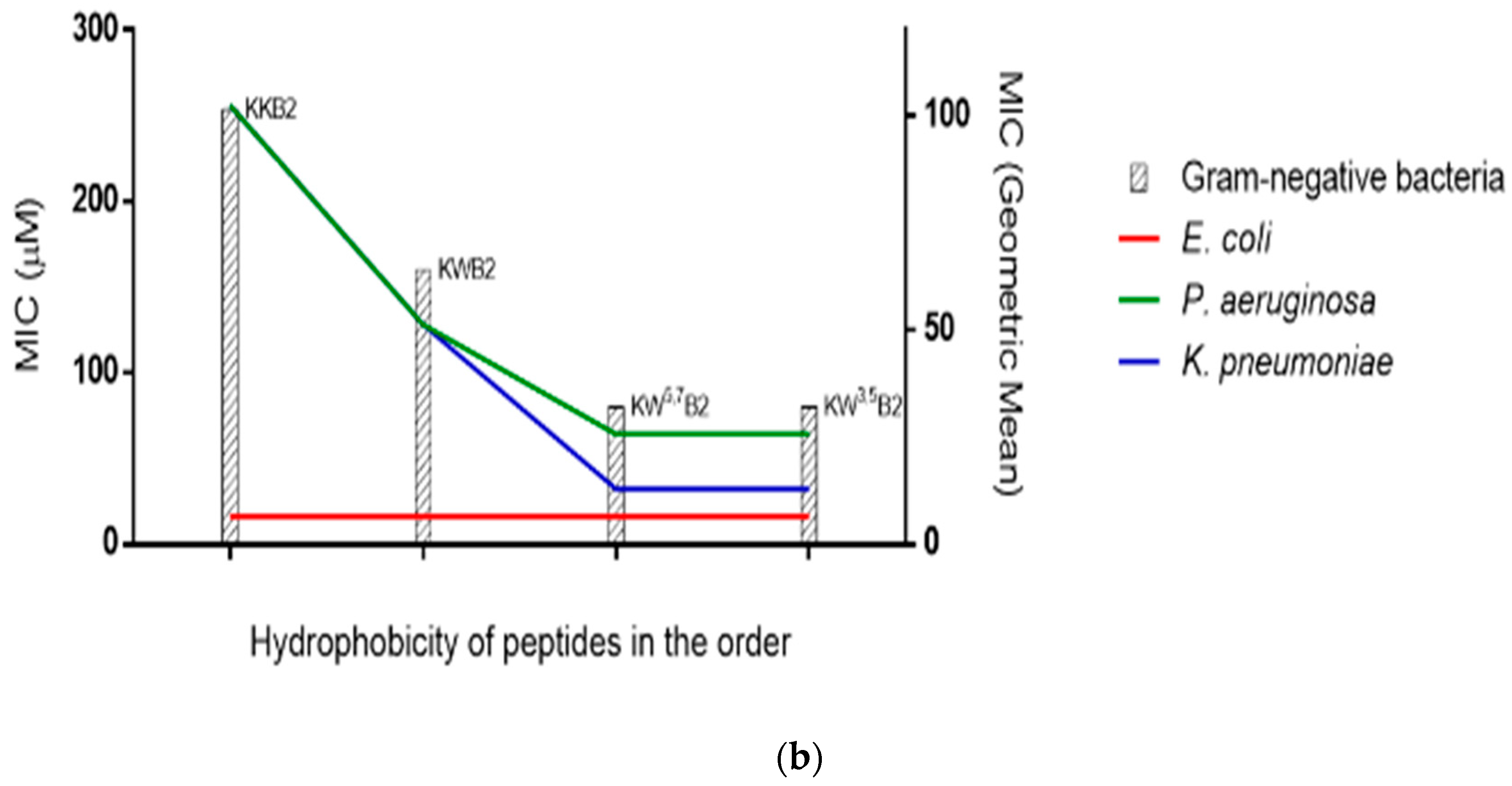
| Peptide | Sequences | Residue No | Charge | Hydrophobicity <H> | Hydrophobic moment <µH> |
|---|---|---|---|---|---|
| B1PLB | FLPLIAGLAANFLPKIFCAITKKC | 24 | 3 | 0.834 | 0.310 |
| B1PLC | FLPVIAGVAAKFLPKIFCAITKKC | 24 | 4 | 0.778 | 0.352 |
| B1A | FLPLIAGLAAKFLPKIFCAITKKC | 24 | 4 | 0.818 | 0.326 |
| B1A1 | FLPLIAGLAAKCAITKKC | 18 | 3 | 0.712 | 0.246 |
| B1A2 | FLPKIFCAITKKC | 13 | 3 | 0.791 | 0.552 |
| KB2 | KFLPKIFCAITKKC | 14 | 4 | 0.644 | 0.577 |
| KKB2 | KKFLPKIFCAITKKC | 15 | 5 | 0.553 | 0.505 |
| KWB2 | KFLPWKIFCAITKKC | 15 | 4 | 0.789 | 0.285 |
| KKWB2 | KKFLPWKIFCAITKKC | 16 | 5 | 0.659 | 0.255 |
| KW3,5B2 | KFWPWKIFCAITKKC | 15 | 4 | 0.806 | 0.263 |
| KW5,7B2 | KFLPWKWFCAITKKC | 15 | 4 | 0.799 | 0.287 |
| Peptide | H2O | 1% SDS | |||||
|---|---|---|---|---|---|---|---|
| No | Name | Helix | Antiparallel | Turn | Helix | Antiparallel | Turn |
| 1 | B1A | 10.1 | 40.3 | 4.1 | 66.7 | 1.4 | 3.8 |
| 2 | B1A1 | 6.1 | 25.7 | 20.9 | 29.3 | 4.9 | 21.4 |
| 3 | B1A2 | 5.1 | 30.7 | 16.1 | 27.5 | 1.6 | 15.3 |
| 4 | KB2 | 10.1 | 21.3 | 17.3 | 20.8 | 9 | 15.2 |
| 5 | KKB2 | 11.4 | 19.0 | 13.8 | 18 | 30.9 | 15.2 |
| 6 | KWB2 | 11.7 | 23.0 | 14.9 | 20.7 | 13.1 | 15.2 |
| 7 | KKWB2 | 10.6 | 28.1 | 15.4 | 18.5 | 13.4 | 14.0 |
| 8 | KW3,5B2 | 6.3 | 33.7 | 14.0 | 10.3 | 38.3 | 11.2 |
| 9 | KW5,7B2 | 4.8 | 31.9 | 14.3 | 20.9 | 9.9 | 16.7 |
| S. aureus | E. coli | C. albicans | MRSA | E. faecalis | P. aeruginosa | K. pneumoniae | HC10 | TI | |
|---|---|---|---|---|---|---|---|---|---|
| B1A | 4/8 | 8/32 | 4/8 | 16/32 | 8/32 | 32/64 | 16/16 | 1.576 | 0.162 |
| B1A1 | 512/>512 | 512/>512 | 512/512 | >512/>512 | >512/>512 | >512/>512 | >512/>512 | >512 | 1 |
| B1A2 | 256/256 | 256/256 | 512/512 | 256/>512 | >512/>512 | >512/>512 | >512/>512 | >512 | 1.682 |
| KB2 | 8/64 | 32/64 | 128/512 | 16/128 | 512/>512 | 512/>512 | 256/256 | 154 | 1.788 |
| KKB2 | 8/64 | 16/32 | 32/64 | 16/64 | 512/512 | 256/>512 | 128/256 | 207 | 3.943 |
| KWB2 | 8/32 | 16/32 | 16/64 | 8/64 | 256/512 | 128/256 | 64/128 | 99.81 | 3.119 |
| KKWB2 | 8/16 | 16/32 | 8/32 | 8/32 | 256/512 | 64/64 | 32/32 | 140.04 | 5.890 |
| KW3,5B2 | 8/16 | 16/32 | 8/32 | 8/32 | 256/256 | 64/128 | 32/64 | 32.07 | 1.349 |
| KW5,7B2 | 8/16 | 16/64 | 16/32 | 8/32 | 256/512 | 64/128 | 32/64 | 58.42 | 2.225 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, H.; Chen, Y.; Ye, Z.; Chen, X.; Ma, C.; Zhou, M.; Xi, X.; Burrows, J.F.; Chen, T.; Wang, L. Modification and Targeted Design of N-Terminal Truncates Derived from Brevinin with Improved Therapeutic Efficacy. Biology 2020, 9, 209. https://doi.org/10.3390/biology9080209
He H, Chen Y, Ye Z, Chen X, Ma C, Zhou M, Xi X, Burrows JF, Chen T, Wang L. Modification and Targeted Design of N-Terminal Truncates Derived from Brevinin with Improved Therapeutic Efficacy. Biology. 2020; 9(8):209. https://doi.org/10.3390/biology9080209
Chicago/Turabian StyleHe, Haoyang, Yuqing Chen, Zhuming Ye, Xiaoling Chen, Chengbang Ma, Mei Zhou, Xinping Xi, James F. Burrows, Tianbao Chen, and Lei Wang. 2020. "Modification and Targeted Design of N-Terminal Truncates Derived from Brevinin with Improved Therapeutic Efficacy" Biology 9, no. 8: 209. https://doi.org/10.3390/biology9080209
APA StyleHe, H., Chen, Y., Ye, Z., Chen, X., Ma, C., Zhou, M., Xi, X., Burrows, J. F., Chen, T., & Wang, L. (2020). Modification and Targeted Design of N-Terminal Truncates Derived from Brevinin with Improved Therapeutic Efficacy. Biology, 9(8), 209. https://doi.org/10.3390/biology9080209






