Phylogenetic Diversity of Trichoderma Strains and Their Antagonistic Potential against Soil-Borne Pathogens under Stress Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Trichoderma Strains
2.2. Pathogenic Fungi
2.3. TEF1 Sequencing for Identification of Trichoderma Strains
2.4. Evaluation of the Genetic Diversity of Trichoderma Strains
2.4.1. RAPD Analysis of Trichoderma Strains
2.4.2. SSR Analysis of Trichoderma Strains
2.5. Specific Trichoderma Genes
2.5.1. Calmodulin (cal) Gene
2.5.2. β-1,3-endoglucanase
2.6. Activities of Trichoderma Strains under Stress Conditions
2.6.1. Growth under Thermal Stress and Different Substrates
2.6.2. Ability to Solubilize Insoluble Phosphate
2.6.3. The Antifungal Ability of Trichoderma Strains against Soil-Borne Pathogenic Fungi
2.6.4. Rate and Speed of Growth
2.7. T. harzianum (Th6) against F. graminearum
2.7.1. Mycoparasitic Activity at Different Temperatures
2.7.2. Slide Culture Method
2.8. Statistical Analysis
3. Results and Discussion
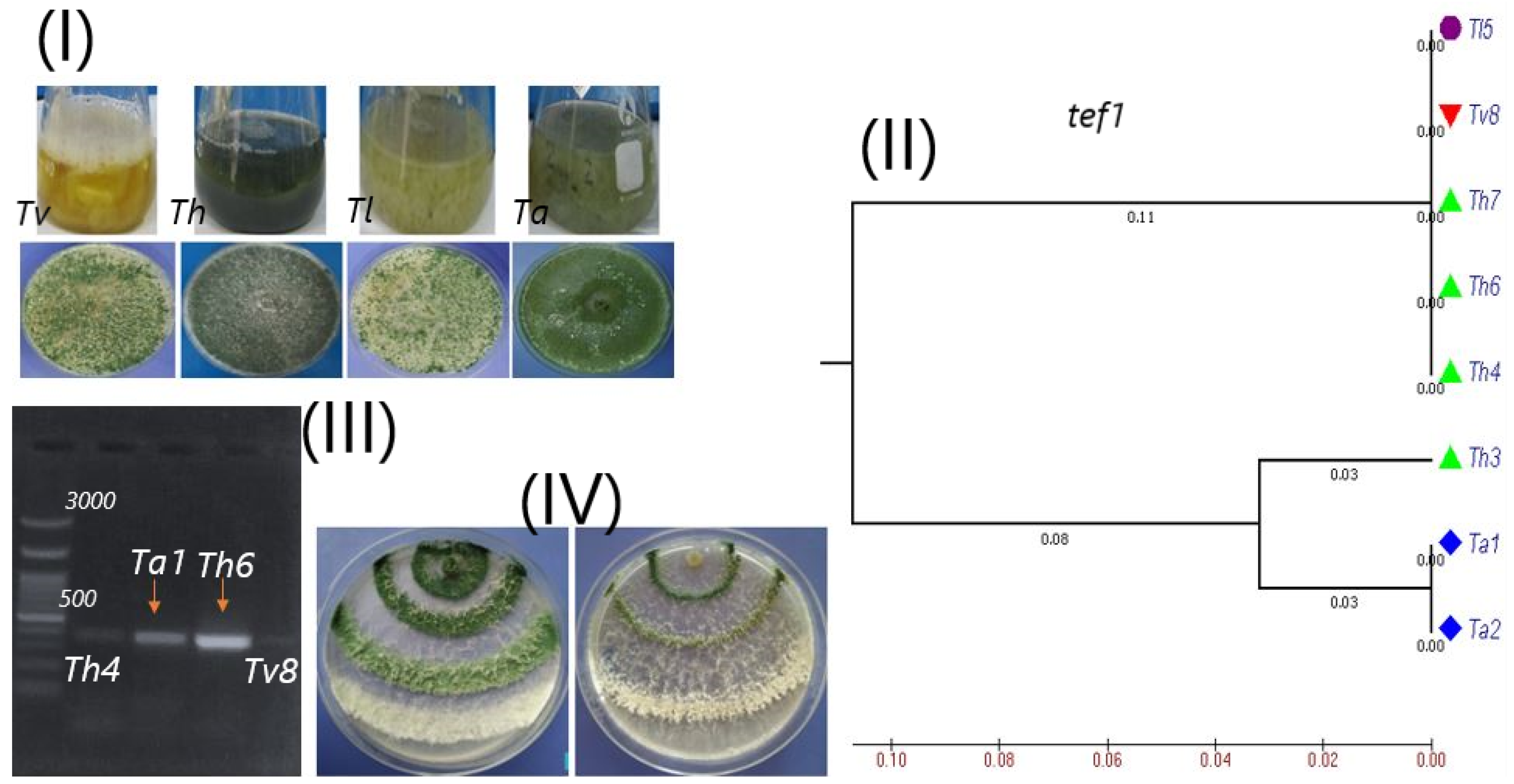
3.1. Molecular Experiments
3.1.1. TEF1 Gene for Confirming the Identification of Trichoderma Strains
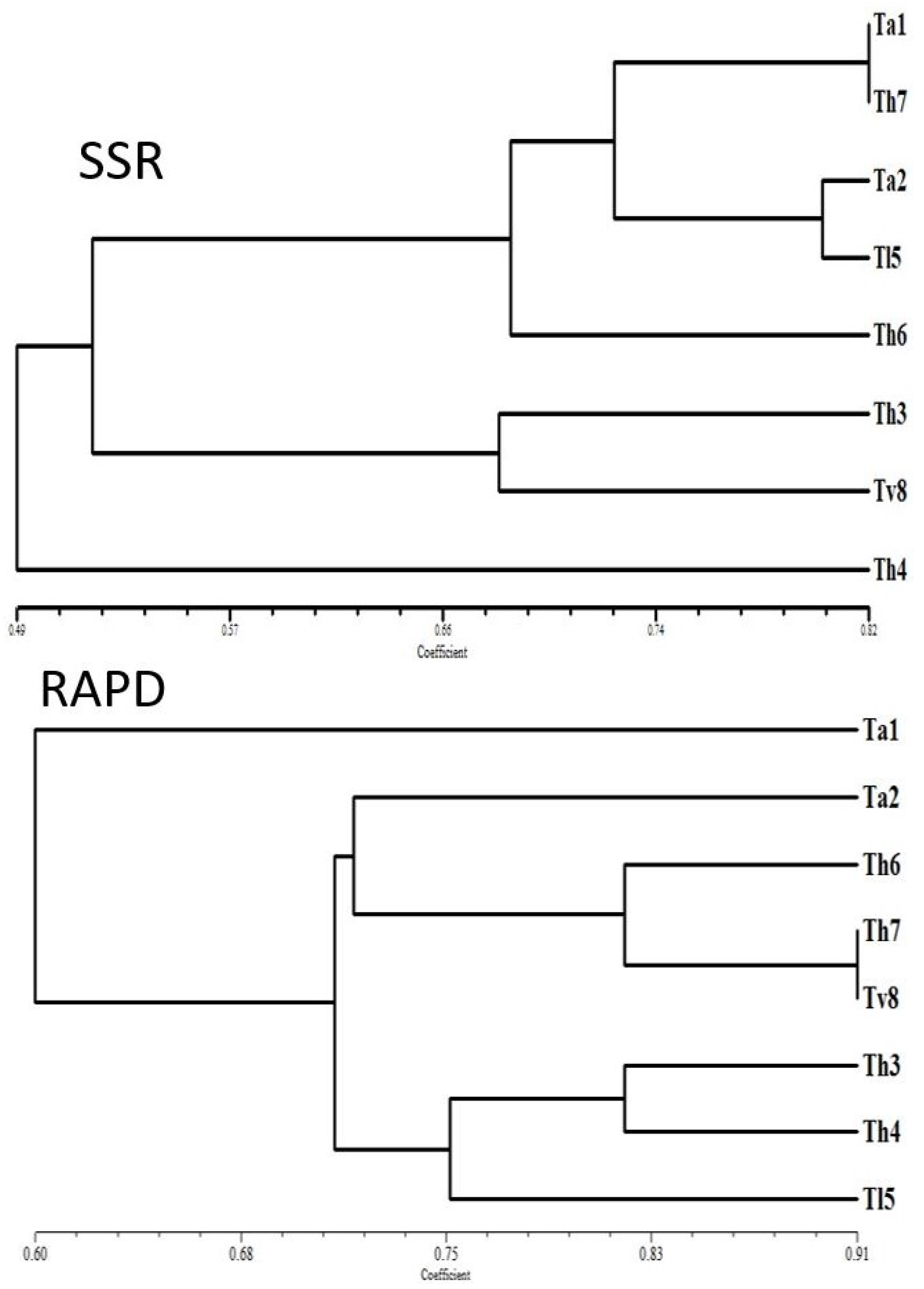
3.1.2. RAPD Analysis of Trichoderma Strains
3.1.3. SSR Analysis of Trichoderma Strains
3.2. Activities of Trichoderma Strains under Stress Conditions
3.2.1. Growth under Thermal Stress and Different Substrates
3.2.2. Ability to Solubilize Insoluble Phosphate
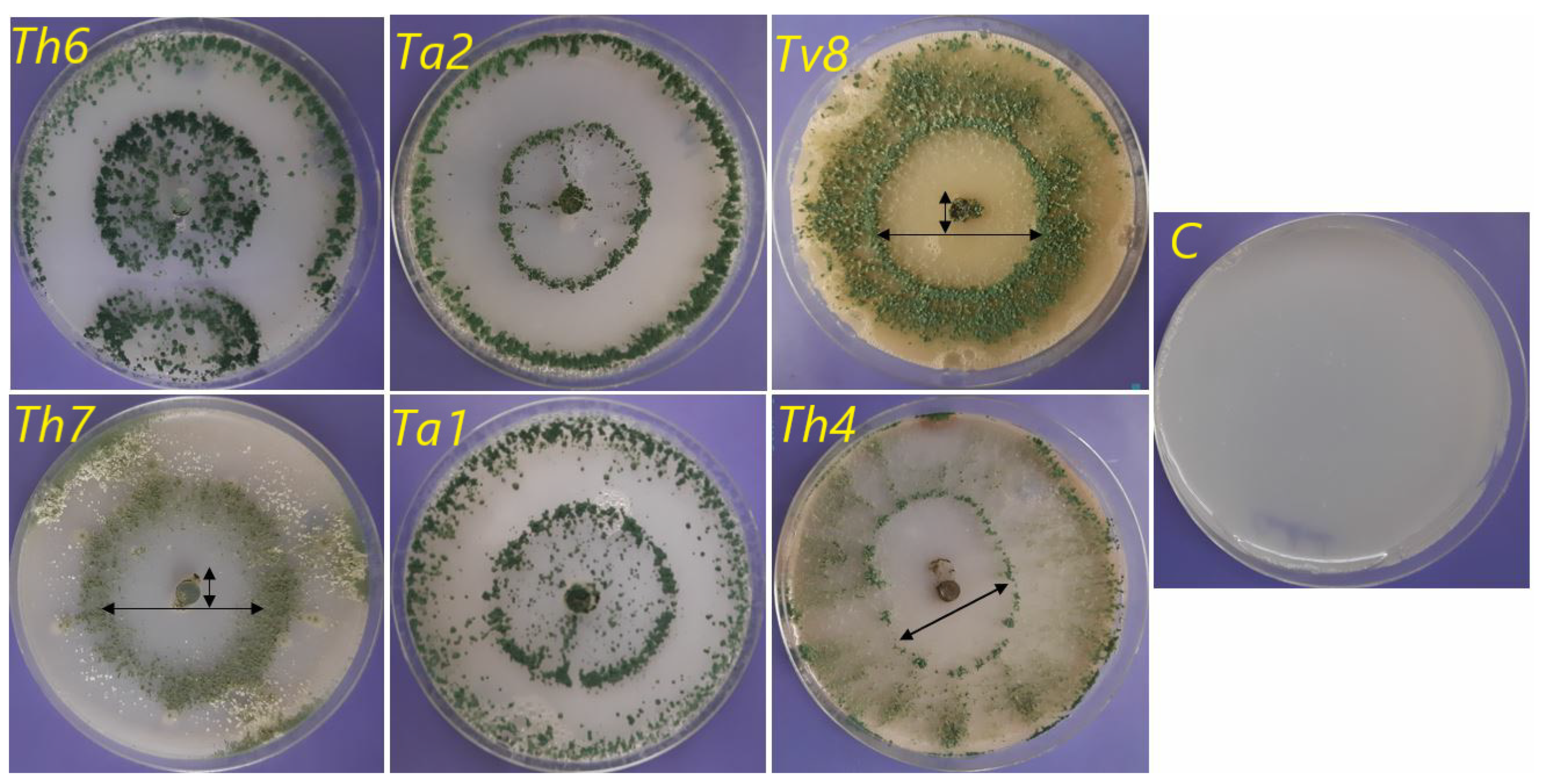
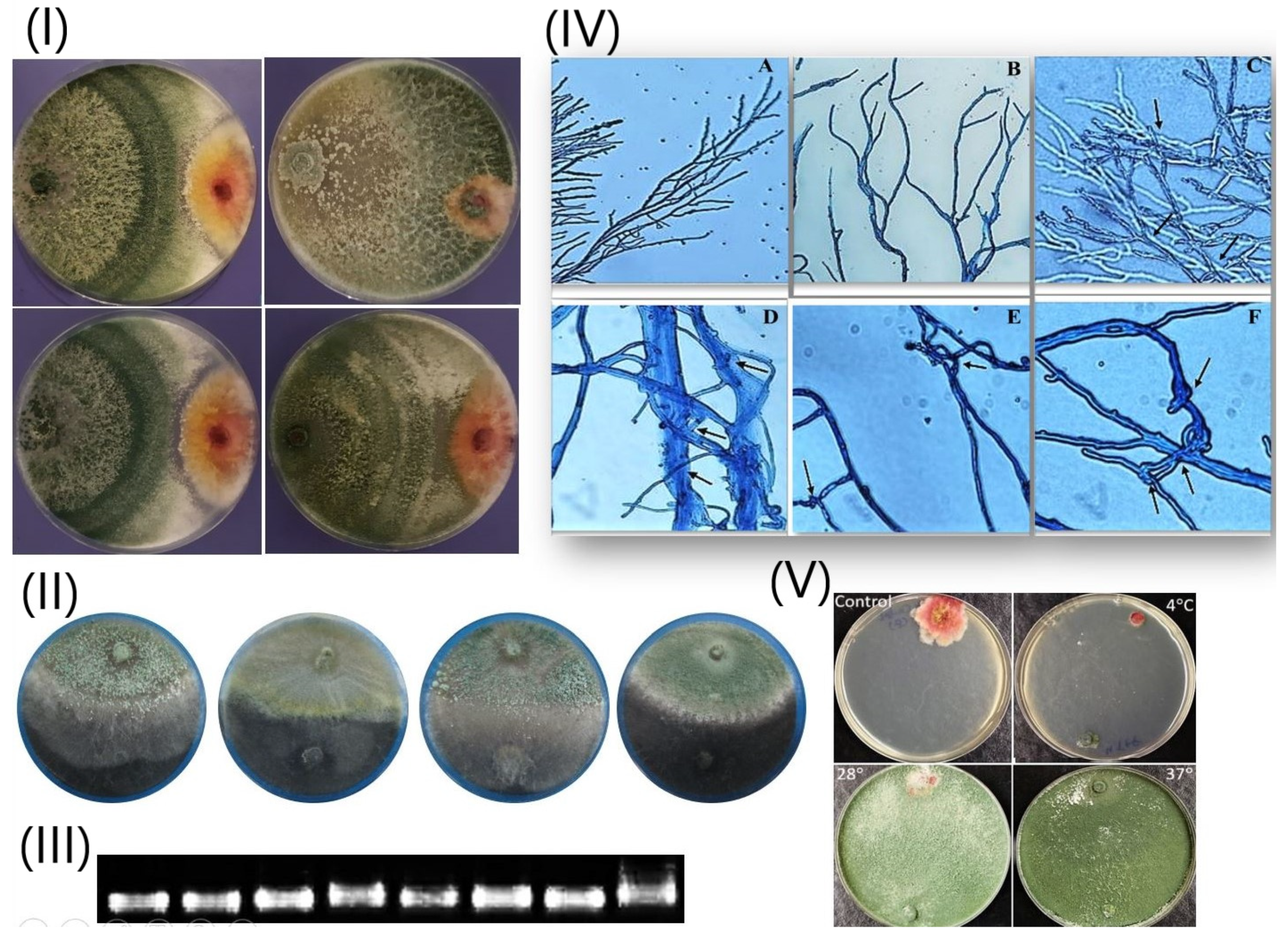
3.2.3. Ability to Antagonize Soil-Borne Pathogenic Fungi
3.2.4. Rate and Speed of Growth
3.3. T. harzianum (Th6) against F. graminearum
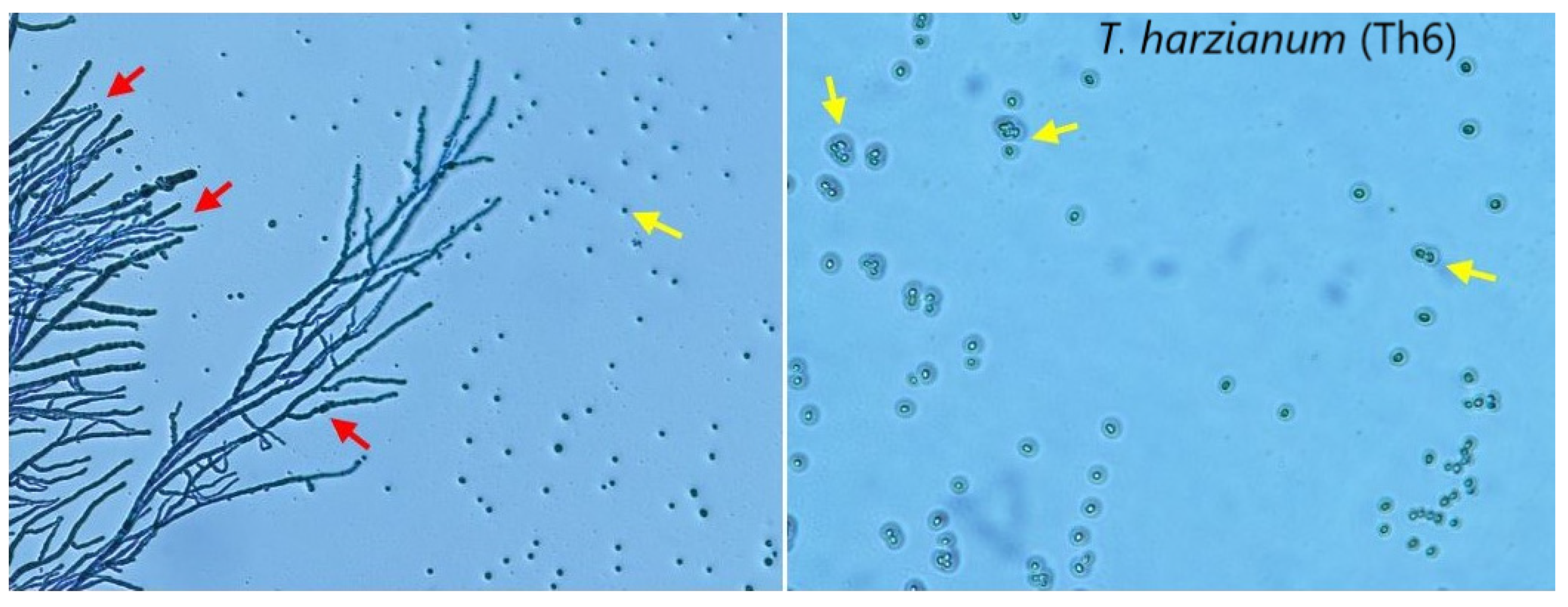
3.3.1. Mycoparasitic Activity at Different Temperatures
3.3.2. Light Microscopic Examination
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tandon, A.; Fatima, T.; Shukla, D.; Tripathi, P.; Srivastava, S.; Singh, P.C. Phosphate solubilization by Trichoderma koningiopsis (NBRI-PR5) under abiotic stress conditions. J. King Saud Univ. Sci. 2020, 32, 791–798. [Google Scholar] [CrossRef]
- Caon, L.; Vargas, R. Threats to Soils: Global trends and Perspectives; Working Paper; United Nations: New York, NY, USA, 2017. [Google Scholar]
- Yedidia, I.; Srivastva, A.K.; Kapulnik, Y.; Chet, I. Effect of Trichoderma harzianum on microelement concentrations and increased growth of cucumber plants. Plant Soil 2001, 235, 235–242. [Google Scholar] [CrossRef]
- Druzhinina, I.S.; Seidl-Seiboth, V.; Herrera-Estrella, A.; Horwitz, B.A.; Kenerley, C.M.; Monte, E.; Mukherjee, P.K.; Zeilinger, S.; Grigoriev, I.V.; Kubicek, C.P. Trichoderma: The genomics of opportunistic success. Nat. Rev. Microbiol. 2011, 9, 749–759. [Google Scholar] [CrossRef]
- Harman, G.E.; Howell, C.R.; Viterbo, A.; Chet, I.; Lorito, M. Trichoderma species—Opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2004, 2, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Sinha, A.; Harshita, D.; Singh, R.; Rao, S.G.; Verma, A. Comprehensive evaluation of Trichoderma harzianum and Trichoderma viride on different culture media & at different temperature and pH. Pharma Innov. J. 2018, 7, 193–195. [Google Scholar]
- Zehra, A.; Dubey, M.; Meena, M.; Upadhyay, R. Effect of different environmental conditions on growth and sporulation of some Trichoderma species. J. Environ. Biol. 2017, 38, 197. [Google Scholar] [CrossRef]
- Borges Chagas, L.; Chagas Junior, A.; Rodrigues de Carvalho, M.; de Oliveira Miller, L.; Colonia, O. Evaluation of the phosphate solubilization potential of Trichoderma strains (Trichoplus JCO) and effects on rice biomass. J. Soil Sci. Plant Nutr. 2015, 15, 794–804. [Google Scholar] [CrossRef]
- Rawat, R.; Tewari, L. Effect of abiotic stress on phosphate solubilization by biocontrol fungus Trichoderma sp. Curr. Microbiol. 2011, 62, 1521–1526. [Google Scholar] [CrossRef]
- Hewedy, O.A.; Abdel-Lateif, K.S.; Bakr, R.A. Genetic diversity and biocontrol efficacy of indigenous Trichoderma isolates against Fusarium wilt of pepper. J. Basic Microbiol. 2020, 60, 126–135. [Google Scholar] [CrossRef]
- López-Mondéjar, R.; Ros, M.; Pascual, J.A. Mycoparasitism-related genes expression of Trichoderma harzianum isolates to evaluate their efficacy as biological control agent. Biol. Control 2011, 56, 59–66. [Google Scholar] [CrossRef]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef] [PubMed]
- Rao, G.S.; Reddy, N.N.R.; Surekha, C. Induction of plant systemic resistance in Legumes cajanus cajan, Vigna radiata, Vigna mungo against plant pathogens Fusarium oxysporum and alternaria alternata–a Trichoderma viride mediated reprogramming of plant defense mechanism. Int. J. Recent Sci. Res 2015, 6, 4270–4280. [Google Scholar]
- Maurya, M.K.; Srivastava, M.; Singh, A.; Pandey, S.; Ratan, V. Effect of different temperature and culture media on the mycelia growth of Trichoderma viride isolates. Int. J. Curr. Microbiol. Appl. Sci 2017, 60, 266–269. [Google Scholar] [CrossRef][Green Version]
- Pastirčáková, K. In vitro interactions between Trichoderma harzianum and pathogenic fungi damaging horse-chestnut (Aesculus hippocastanum) leaves and fruits. Biol. Lett. 2019, 54, 21–35. [Google Scholar] [CrossRef][Green Version]
- Dłużniewska, J. Reaction of Fungi of Trichoderma Genus to Selected Abiotic Factors. Available online: http://www.ejpau.media.pl/volume6/issue2/agronomy/art-04.Html (accessed on 21 July 2020).
- Narsian, V.; Patel, H. Aspergillus aculeatus as a rock phosphate solubilizer. Soil Biol. Biochem. 2000, 32, 559–565. [Google Scholar] [CrossRef]
- Singh, A.; Shahid, M.; Srivastava, M.; Pandey, S.; Sharma, A.; Kumar, V. Optimal physical parameters for growth of Trichoderma species at varying pH, temperature and agitation. Virol. Mycol. 2014, 3, 127–134. [Google Scholar]
- Domingues, M.V.P.F.; Moura, K.E.d.; Salomão, D.; Elias, L.M.; Patricio, F.R.A. Effect of temperature on mycelial growth of Trichoderma, Sclerotinia minor and S. sclerotiorum, as well as on mycoparasitism. Summa Phytopathol. 2016, 42, 222–227. [Google Scholar] [CrossRef]
- Qiu, Z.; Wu, X.; Zhang, J.; Huang, C. High temperature enhances the ability of Trichoderma asperellum to infect Pleurotus ostreatus mycelia. PLoS ONE 2017, 12, e0187055. [Google Scholar] [CrossRef]
- Rojo, F.G.; Reynoso, M.M.; Ferez, M.; Chulze, S.N.; Torres, A.M. Biological control by Trichoderma species of Fusarium solani causing peanut brown root rot under field conditions. Crop Prot. 2007, 26, 549–555. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Li, Y.; Yu, C.; Wang, Q.Q.; Wang, M.; Sun, J.; Gao, J.X.; Chen, J. Effect of Trichoderma harzianum on maize rhizosphere microbiome and biocontrol of Fusarium Stalk rot. Sci. Rep. 2017, 7, 1–13. [Google Scholar]
- Aly, A.A.; Abdel-Sattar, M.A.; Omar, M.R.; Abd-Elsalam, K.A. Differential antagonism of Trichoderma sp. against Macrophomina phaseolina. J. Plant Prot. Res. 2007, 47, 91–102. [Google Scholar]
- Samuels, G.J.; Dodd, S.L.; Gams, W.; Castlebury, L.A.; Petrini, O. Trichoderma species associated with the green mold epidemic of commercially grown Agaricus bisporus. Mycologia 2002, 94, 146–170. [Google Scholar] [CrossRef] [PubMed]
- Kamala, T.; Devi, S.I.; Sharma, K.C.; Kennedy, K. Phylogeny and taxonomical investigation of Trichoderma spp. from Indian region of Indo-Burma biodiversity hot spot region with special reference to Manipur. BioMed Res. Int. 2015, 2015, 285261. [Google Scholar] [CrossRef] [PubMed]
- Lind, A.L.; Wisecaver, J.H.; Lameiras, C.; Wiemann, P.; Palmer, J.M.; Keller, N.P.; Rodrigues, F.; Goldman, G.H.; Rokas, A. Drivers of genetic diversity in secondary metabolic gene clusters in a fungal population. bioRxiv 2017, 149856. [Google Scholar] [CrossRef]
- Chaverri, P.; Branco-Rocha, F.; Jaklitsch, W.; Gazis, R.; Degenkolb, T.; Samuels, G.J. Systematics of the Trichoderma harzianum species complex and the re-identification of commercial biocontrol strains. Mycologia 2015, 107, 558–590. [Google Scholar] [CrossRef]
- Komoń-Zelazowska, M.; Bissett, J.; Zafari, D.; Hatvani, L.; Manczinger, L.; Woo, S.; Lorito, M.; Kredics, L.; Kubicek, C.P.; Druzhinina, I.S. Genetically closely related but phenotypically divergent Trichoderma species cause green mold disease in oyster mushroom farms worldwide. Appl. Environ. Microbiol. 2007, 73, 7415–7426. [Google Scholar] [CrossRef]
- Atanasova, L.; Druzhinina, I.S.; Jaklitsch, W.M.; Mukherjee, P.; Horwitz, B.; Singh, U. Two hundred Trichoderma species recognized on the basis of molecular phylogeny. In Trichoderma: Biology and Applications; Cabi: Wallingford, UK, 2013; pp. 10–42. [Google Scholar]
- Geiser, D.M.; del Mar Jiménez-Gasco, M.; Kang, S.; Makalowska, I.; Veeraraghavan, N.; Ward, T.J.; Zhang, N.; Kuldau, G.A.; O’donnell, K. FUSARIUM-ID v. 1.0: A DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 2004, 110, 473–479. [Google Scholar] [CrossRef]
- Oskiera, M.; Szczech, M.; Bartoszewski, G. Molecular identification of Trichoderma strains collected to develop plant growth-promoting and biocontrol agents. J. Hortic. Res. 2015, 23, 75–86. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Yu, C.; Dou, K.; Wang, M.; Li, Y.; Chen, J. Biodiversity of Trichoderma community in the tidal flats and wetland of southeastern China. PLoS ONE 2016, 11, e0168020. [Google Scholar] [CrossRef]
- Haddad, P.E.; Leite, L.G.; Lucon, C.M.M.; Harakava, R. Selection of Trichoderma spp. strains for the control of Sclerotinia sclerotiorum in soybean. Pesqui. Agropecuária Bras. 2017, 52, 1140–1148. [Google Scholar] [CrossRef]
- Hussein, M.H.; Abdel-Hamid, A.; Hussein, B.A.; Nasseef, M.; El-Morshedyand, J. The Suitability of RAPD Markers in Identifying Some Hexaploid Wheat Crosses. World Appl. Sci. J. 2013, 21, 732–738. [Google Scholar]
- Shahid, M.; Srivastava, M.; Kumar, V.; Singh, A.; Sharma, A.; Pandey, S.; Rastogi, S.; Pathak, N.; Srivastava, A. Phylogenetic diversity analysis of Trichoderma species based on internal transcribed spacer (ITS) marker. Afr. J. Biotechnol. 2014, 13. [Google Scholar] [CrossRef]
- Hadrys, H.; Balick, M.; Schierwater, B. Applications of random amplified polymorphic DNA (RAPD) in molecular ecology. Mol. Ecol. 1992, 1, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Misra, A.; Gupta, A.; Pandey, B.; Gaur, R. Rapd-Pcr of Trichoderma Isolates and In Vitro Antagonism Against Fusarium Wilt Pathogens of Psidium Guajaval. J. Plant Prot. Res. 2010, 50, 256. [Google Scholar] [CrossRef]
- Chakraborty, B.; Chakraborty, U.; Sunar, K.; Dey, P. RAPD profile and rDNA sequence analysis of Talaromyces flavus and Trichoderma species. NISCAIR-CSIR 2011, 11, 487–495. [Google Scholar]
- El_Komy, M.H.; Saleh, A.A.; Eranthodi, A.; Molan, Y.Y. Characterization of novel Trichoderma asperellum isolates to select effective biocontrol agents against tomato Fusarium wilt. Plant Pathol. J. 2015, 31, 50. [Google Scholar] [CrossRef]
- Pandya, J.; Sabalpara, A.; Mahatma, M. Randomly amplified polymorphic DNA analysis of native Trichoderma isolates. Asian J. Appl. Sci. Technol. 2017, 1, 147–150. [Google Scholar]
- Khattak, B.; Saifullah, S.H.; Ahmad, M.; Ali, A.; Junaid, M.; Khan, I.A.; Khan, T.A.; Hussain, M. Genetic Relatedness among the Indigenous Isolates of Trichoderma harzianum, using RAPD and their Nematocidal Capabilities against Meloidogyne javanica. J. Agric. 2018, 34, 486–493. [Google Scholar] [CrossRef]
- Shahid, M.; Srivastava, M.; Sharma, A.; Kumar, V.; Pandey, S.; Singh, A. Morphological, molecular identification and SSR marker analysis of a potential strain of Trichoderma/Hypocrea for production of a bioformulation. J. Plant Pathol. Microbiol. 2013, 4, 1. [Google Scholar] [CrossRef]
- Mahfooz, S.; Singh, S.P.; Rakh, R.; Bhattacharya, A.; Mishra, N.; Singh, P.C.; Chauhan, P.S.; Nautiyal, C.S.; Mishra, A. A comprehensive characterization of simple sequence repeats in the sequenced Trichoderma genomes provides valuable resources for marker development. Front. Microbiol. 2016, 7, 575. [Google Scholar] [CrossRef]
- Geistlinger, J.; Zwanzig, J.; Heckendorff, S.; Schellenberg, I. SSR markers for Trichoderma virens: Their evaluation and application to identify and quantify root-endophytic strains. Diversity 2015, 7, 360–384. [Google Scholar] [CrossRef]
- Kretsinger, R.H.; Wasserman, R.H. Structure and evolution of calcium-modulated protein. Crit. Rev. Biochem. 1980, 8, 119–174. [Google Scholar] [CrossRef] [PubMed]
- Gadd, G. Signal transduction in fungi. In The Growing Fungus; Springer: Berlin/Heidelberg, Germany, 1995; pp. 183–210. [Google Scholar]
- Muthukumar, G.; Nickerson, K.W. Ca (II)-calmodulin regulation of fungal dimorphism in Ceratocystis ulmi. J. Bacteriol. 1984, 159, 390–392. [Google Scholar] [CrossRef] [PubMed]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Montoya, Q.V.; Meirelles, L.A.; Chaverri, P.; Rodrigues, A. Unraveling Trichoderma species in the attine ant environment: Description of three new taxa. Antonie van Leeuwenhoek 2016, 109, 633–651. [Google Scholar] [CrossRef]
- Druzhinina, I.S.; Kopchinskiy, A.G.; Komoń, M.; Bissett, J.; Szakacs, G.; Kubicek, C.P. An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet. Biol. 2005, 42, 813–828. [Google Scholar] [CrossRef]
- Dodd, S.L.; Lieckfeldt, E.; Chaverri, P.; Overton, B.E.; Samuels, G.J. Taxonomy and phylogenetic relationships of two species of Hypocrea with Trichoderma anamorphs. Mycol. Prog. 2002, 1, 409–428. [Google Scholar] [CrossRef]
- Williams, J.G.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef]
- Samuels, G.J.; Suarez, C.; Solis, K.; Holmes, K.A.; Thomas, S.E.; Ismaiel, A.; Evans, H.C. Trichoderma theobromicola and T. paucisporum: Two new species isolated from cacao in South America. Mycol. Res. 2006, 110, 381–392. [Google Scholar] [CrossRef]
- Vieira, P.M.; Coelho, A.S.G.; Steindorff, A.S.; de Siqueira, S.J.L.; do Nascimento Silva, R.; Ulhoa, C.J. Identification of differentially expressed genes from Trichoderma harzianum during growth on cell wall of Fusarium solanias a tool for biotechnological application. BMC Genom. 2013, 14, 177. [Google Scholar] [CrossRef]
- Jaklitsch, W.M.; Komon, M.; Kubicek, C.P.; Druzhinina, I.S. Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 2005, 97, 1365–1378. [Google Scholar] [CrossRef] [PubMed]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Afzal, A.; Bano, A. Rhizobium and phosphate solubilizing bacteria improve the yield and phosphorus uptake in wheat (Triticum aestivum). Int. J. Agric. Biol. 2008, 10, 85–88. [Google Scholar]
- Morton, D.; Stroube, W. Antagonistic and stimulatory effects of soil microorganisms upon Sclerotium rolfsii. Phytopathology 1955, 45, 417–420. [Google Scholar]
- Edgington, L.; Khew, K.; Barron, G. Fungitoxic spectrum of benzimidazole compounds. Phytopathology 1971, 61, 42–44. [Google Scholar] [CrossRef]
- Hajieghrari, B.; Torabi-Giglou, M.; Mohammadi, M.R.; Davari, M. Biological potantial of some Iranian Trichoderma isolates in the control of soil borne plant pathogenic fungi. Afr. J. Biotechnol. 2008, 967–972. [Google Scholar]
- Mousa, W.K.; Shearer, C.R.; Limay-Rios, V.; Zhou, T.; Raizada, M.N. Bacterial endophytes from wild maize suppress Fusarium graminearum in modern maize and inhibit mycotoxin accumulation. Front. Plant Sci. 2015, 6, 805. [Google Scholar] [CrossRef]
- Jamshidi, S.; Jamshidi, S. NTSYSpc 2.02 e implementation in molecular biodata analysis (clustering, screening, and individual selection). In Proceedings of the 4th International Conference on Environmental and Computer Science, Singapore, 28–29 October 2017; pp. 16–18. [Google Scholar]
- Alhawatema, M.; Al Tawaha, A.A.A.R. Application of Using DNA Barcoding Genes in Identification of Fungi Species, a Review. Biosci. Res. 2019, 16, 1763–1775. [Google Scholar]
- De La Cruz, J.; Pintor-Toro, J.A.; Benitez, T.; Llobell, A.; Romero, L.C. A novel endo-beta-1, 3-glucanase, BGN13. 1, involved in the mycoparasitism of Trichoderma harzianum. J. Bacteriol. 1995, 177, 6937–6945. [Google Scholar] [CrossRef]
- Harman, G.E. Myths and dogmas of biocontrol changes in perceptions derived from research on Trichoderma harzinum T-22. Plant Dis. 2000, 84, 377–393. [Google Scholar] [CrossRef]
- Sharma, P.; Kumar, V.; Ramesh, R.; Saravanan, K.; Deep, S.; Sharma, M.; Mahesh, S.; Dinesh, S. Biocontrol genes from Trichoderma species: A review. Afr. J. Biotechnol. 2011, 10, 19898–19907. [Google Scholar]
- Chen, L.; Zou, G.; Wang, J.; Wang, J.; Liu, R.; Jiang, Y.; Zhao, G.; Zhou, Z. Characterization of the Ca2+-responsive signaling pathway in regulating the expression and secretion of cellulases in Trichoderma reesei Rut-C30. Mol. Microbiol. 2016, 100, 560–575. [Google Scholar] [CrossRef] [PubMed]
- Xiong, F.; Zhong, R.; Han, Z.; Jiang, J.; He, L.; Zhuang, W.; Tang, R. Start codon targeted polymorphism for evaluation of functional genetic variation and relationships in cultivated peanut (Arachishypogaea, L.) genotypes. Mol. Biol. Rep. 2011, 38, 3487–3494. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.; Joshi, D.; Zaidi, N. Biodiversity in biocontrol agent, Trichoderma spp.: Characterization and util. ization. J. Mycol. Plant Pathol. 2006, 36, 461–463. [Google Scholar]
- Choudary, K.; Reddy, K.; Reddy, M. Antifungal activity and genetic variability of Trichoderma harzianum isolates. J. Mycol. Plant Pathol. 2007, 37, 1–6. [Google Scholar]
- Limón, M.; Chacón, M.; Mejías, R.; Delgado-Jarana, J.; Rincón, A.; Codón, A.; Benítez, T. Increased antifungal and chitinase specific activities of Trichoderma harzianum CECT 2413 by addition of a cellulose binding domain. Appl. Microbiol. Biotechnol. 2004, 64, 675–685. [Google Scholar] [CrossRef]
- Azher, M.; Khan, M.A.; Inam-ul-Haq, M.; Khan, S.; Pervez, M.A. Mass multiplication of Trichoderma spp. on organic substrate and their effect in management of seed borne fungi. Pak. J. Phytopathol. 2009, 21, 108–114. [Google Scholar]
- Jahan, N.; Sultana, S.; Adhikary, S.; Rahman, S.; Yasmin, S. Evaluation of the growth performance of Trichoderma harzianum (Rifai.) on different culture media. J. Agri. Vet. Sci. 2013, 3, 44–50. [Google Scholar] [CrossRef]
- Mustafa, A.; Khan, M.A.; Inam-ul-Haq, M.; Pervez, M.A.; Umar, U. Usefulness of different culture media for in vitro evaluation of Trichoderma spp. against seed borne fungi of economic importance. Pak. J. Phytopathol. 2009, 21, 83–88. [Google Scholar]
- Santamarina, M.P.; Rosello, J. Influence of temperature and water activity on the antagonism of Trichoderma harzianum to Verticillium and Rhizoctonia. Crop Prot. 2006, 25, 1130–1134. [Google Scholar] [CrossRef]
- Jackson, A.; Whipps, J.; Lynch, J. Effects of temperature, pH and water potential on growth of four fungi with disease biocontrol potential. World J. Microbiol. Biotechnol. 1991, 7, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Yasser, M.; Mousa, A.; Khalek, M.A. Optimization of factors affecting proliferation and flourishment of Trichoderma harzianum in Egyptian soil. J. Basic Appl. Mycol. 2012, 3, 41–48. [Google Scholar]
- Lei, Z.; Qun, L.; Zhang, Y.Q.; Cui, Q.Y.; Liang, Y.C. Effect of acid phosphatase produced by Trichoderma asperellum Q1 on growth of Arabidopsis under salt stress. J. Integr. Agric. 2017, 16, 1341–1346. [Google Scholar]
- Saxena, J.; Chandra, S.; Nain, L. Synergistic effect of phosphate solubilizing rhizobacteria and arbuscular mycorrhiza on growth and yield of wheat plants. J. Soil Sci. Plant Nutr. 2013, 13, 511–525. [Google Scholar]
- Promwee, A.; Issarakraisila, M.; Intana, W.; Chamswarng, C.; Yenjit, P. Phosphate solubilization and growth promotion of rubber tree (Hevea brasiliensis Muell. Arg.) by Trichoderma strains. J. Agric. Sci. 2014, 6, 8. [Google Scholar] [CrossRef]
- Gaind, S. Phosphate dissolving fungi: Mechanism and application in alleviation of salt stress in wheat. Microbiol. Res. 2016, 193, 94–102. [Google Scholar] [CrossRef]
- Altomare, C.; Norvell, W.; Björkman, T.; Harman, G. Solubilization of phosphates and micronutrients by the plant-growth-promoting and biocontrol fungus Trichoderma harzianum Rifai 1295-22. Appl. Environ. Microbiol. 1999, 65, 2926–2933. [Google Scholar] [CrossRef]
- Li, R.-X.; Cai, F.; Pang, G.; Shen, Q.-R.; Li, R.; Chen, W. Solubilisation of phosphate and micronutrients by Trichoderma harzianum and its relationship with the promotion of tomato plant growth. PLoS ONE 2015, 10, e0130081. [Google Scholar] [CrossRef]
- Illmer, P.; Schinner, F. Solubilization of inorganic calcium phosphates—Solubilization mechanisms. Soil Biol. Biochem. 1995, 27, 257–263. [Google Scholar] [CrossRef]
- Kredics, L.; Manczinger, L.; Antal, Z.; Pénzes, Z.; Szekeres, A.; Kevei, F.; Nagy, E. In vitro water activity and pH dependence of mycelial growth and extracellular enzyme activities of Trichoderma strains with biocontrol potential. J. Appl. Microbiol. 2004, 96, 491–498. [Google Scholar] [CrossRef]
- Kuzmanovska, B.; Rusevski, R.; Jankulovska, M.; Oreshkovikj, K.B. Antagonistic activity of Trichoderma asperellum and Trichoderma harzianum against genetically diverse Botrytis cinerea isolates. Chil. J. Agric. Res. 2018, 78, 391–399. [Google Scholar] [CrossRef]
- Bonaterra, A.; Camps, J.; Montesinos, E. Osmotically induced trehalose and glycine betaine accumulation improves tolerance to desiccation, survival and efficacy of the postharvest biocontrol agent Pantoea agglomerans EPS125. FEMS Microbiol. Lett. 2005, 250, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Živković, S.; Stojanović, S.; Ivanović, Ž.; Gavrilović, V.; Popović, T.; Balaž, J. Screening of antagonistic activity of microorganisms against Colletotrichum acutatum and Colletotrichum gloeosporioides. Arch. Biol. Sci. 2010, 62, 611–623. [Google Scholar] [CrossRef]
- Jegathambigai, V.; Wijeratnam, R.W.; Wijesundera, R. Effect of Trichoderma sp. on Sclerotium rolfsii, the causative agent of collar rot on Zamioculcas zamiifolia and an on farm method to mass produce Trichoderma species. Plant Pathol. J. 2010, 9, 47–55. [Google Scholar]
- Figueirêdo, G.S.d.; Figueirêdo, L.C.d.; Cavalcanti, F.C.N.; Santos, A.C.d.; Costa, A.F.d.; Oliveira, N.T.D. Biological and chemical control of Sclerotinia sclerotiorum using Trichoderma spp. and Ulocladium atrum and pathogenicity to bean plants. Braz. Arch. Biol. Technol. 2010, 53, 1–9. [Google Scholar] [CrossRef]

| Primer Name | Primer Sequence (5′–3′) |
|---|---|
| RAPD | |
| OPA02 | TGCCGAGCTG |
| OPA04 | AATCGGGCTG |
| OPA05 | AGGGGTCTTG |
| OP-A3 | AGTCAGCCAC |
| OP-B3 | CATCCCCCTG |
| OP-B9 | TGGGGGACTC |
| OP-C3 | GGGGGTCTTT |
| SSR [42,44] | |
| SSR1 | F: GAAACAACACCGAAATACAC, R: CAAGTCAGATGAAGTTTG |
| SSR2 | F: GACTCATACTTTGTTCTTAGCAG, R: GAACGGAGCGGTCACATTAG |
| SSR3 | F: CAAGCTGACGCCTATGAAGA, R: CTTTCACTCACTCAACTCTC |
| SSR6 | F: CCATGCATACGTGACTGC, R: GTTGACTGTTGGTGTAAGTG |
| SSR8 | F: GGGAATTTGTGGAGGGAAG, R: CCTCAGAATGTCCCTGTC |
| TvCTTT29 | F: GGAAGATAGCACGATGAAGTCG, R: AACCGTGGAAGTAGGTGTCG |
| TvCAT32 | F: GTGTAGCAGCCCAACAGTCC, R: CAGGTGTCGTGACAGATTCG |
| β-1,3-endoglucanase | F:TCAACATCGCCAACGTCAACGAC, R: TGCCAATACGGGAACCAGTGATC |
| Primers | Band Size (bp) | Total Number of Bands | Number of Polymorphic Bands | Polymorphic Bands Percentage (%) |
|---|---|---|---|---|
| RAPD | ||||
| OPA02 | 200–3000 | 9 | 6 | 66.6 |
| OPA04 | 100–2500 | 6 | 4 | 66.6 |
| OPA05 | 250–3000 | 9 | 8 | 88.88 |
| OP-A3 | 400–1500 | 5 | 4 | 80 |
| OP-B3 | 100–1300 | 5 | 5 | 100 |
| OP-B9 | 200–1000 | 4 | 4 | 100 |
| OP-C3 | 100–1300 | 7 | 5 | 71.4 |
| Total | ------------- | 45 | 36 | -------------- |
| SSR | ||||
| SSR1 | 100–500 | 6 | 5 | 83.3 |
| SSR2 | 100–500 | 5 | 4 | 80 |
| SSR3 | 100–500 | 5 | 4 | 80 |
| SSR6 | 100–500 | 5 | 4 | 80 |
| SSR8 | 100–500 | 4 | 3 | 75 |
| Tvc-29 | 100–400 | 5 | 5 | 100 |
| Tvc-32 | 100–400 | 6 | 6 | 100 |
| Total | ------------- | 36 | 31 | --------------- |
| Trichoderma Strains | 25 °C | 35 °C | ||
|---|---|---|---|---|
| PDA | CMD | PDA | CMD | |
| Ta1 | 6.20 ± 0.01 | 4.75 ± 0.10 | 4.22 ± 0.02 | 2.60 ± 0.41 |
| Ta2 | 5.84 ± 0.06 | 4.20 ± 0.08 | 3.20 ± 0.47 | 2.90 ± 0.18 |
| Th3 | 5.10 ± 0.01 | 5.10 ± 0.29 | 2.75 ± 0.38 | 3.50 ± 0.38 |
| Th4 | 5.33 ± 0.09 | 4.10 ± 0.13 | 2.50 ± 0.31 | 3.20 ± 0.09 |
| Tl5 | 6.10 ± 0.44 | 4.90 ± 0.50 | 3.90 ± 0.44 | 2.75 ± 0.46 |
| Th6 | 6.00 ± 0.47 | 5.30 ± 0.19 | 4.30 ± 0.13 | 3.80 ± 0.56 |
| Th7 | 5.95 ± 0.14 | 5.25 ± 0.14 | 3.50 ± 0.89 | 3.00 ± 0.67 |
| Tv8 | 5.00 ± 0.45 | 4.88 ± 0.72 | 2.70 ± 0.57 | 2.30 ± 0.15 |
| Trichoderma Strains | F. solani | M. phaseolina | F. graminearum | |||
|---|---|---|---|---|---|---|
| RMG (cm) | IMG (%) | RMG (cm) | IMG (%) | RMG (cm) | IMG (%) | |
| Control | 3.50 ± 0.06 | 0.00 | 3.70 ± 0.30 | 0.00 | 6.50 ± 0.16 | 0.00 |
| Ta1 | 1.40 ± 0.09 | 60.00 | 1.20 ± 0.31 | 67.56 | 1.50 ± 0.08 | 76.92 |
| Ta2 | 1.65 ± 0.06 | 52.85 | 1.10 ± 0.10 | 70.27 | 1.20 ± 0.21 | 81.53 |
| Th3 | 1.55 ± 0.15 | 55.71 | 1.30 ± 0.22 | 64.86 | 1.50 ± 0.17 | 76.92 |
| Th4 | 1.75 ± 0.38 | 50.01 | 1.14 ± 0.07 | 69.18 | 1.80 ± 0.49 | 72.30 |
| Tl5 | 1.70 ± 0.32 | 51.42 | 1.01 ± 0.11 | 72.96 | 1.20 ± 0.01 | 81.53 |
| Th6 | 1.00 ± 0.01 | 71.42 | 1.00 ± 0.13 | 72.97 | 1.00 ± 0.15 | 84.61 |
| Th7 | 1.55 ± 0.40 | 55.71 | 1.20 ± 0.20 | 67.56 | 1.01 ± 0.01 | 84.60 |
| Tv8 | 1.75 ± 0.30 | 50.00 | 1.33 ± 0.08 | 64.05 | 2.00 ± 0.32 | 69.23 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hewedy, O.A.; Abdel Lateif, K.S.; Seleiman, M.F.; Shami, A.; Albarakaty, F.M.; M. El-Meihy, R. Phylogenetic Diversity of Trichoderma Strains and Their Antagonistic Potential against Soil-Borne Pathogens under Stress Conditions. Biology 2020, 9, 189. https://doi.org/10.3390/biology9080189
Hewedy OA, Abdel Lateif KS, Seleiman MF, Shami A, Albarakaty FM, M. El-Meihy R. Phylogenetic Diversity of Trichoderma Strains and Their Antagonistic Potential against Soil-Borne Pathogens under Stress Conditions. Biology. 2020; 9(8):189. https://doi.org/10.3390/biology9080189
Chicago/Turabian StyleHewedy, Omar A., Khalid S. Abdel Lateif, Mahmoud F. Seleiman, Ashwag Shami, Fawziah M. Albarakaty, and Rasha M. El-Meihy. 2020. "Phylogenetic Diversity of Trichoderma Strains and Their Antagonistic Potential against Soil-Borne Pathogens under Stress Conditions" Biology 9, no. 8: 189. https://doi.org/10.3390/biology9080189
APA StyleHewedy, O. A., Abdel Lateif, K. S., Seleiman, M. F., Shami, A., Albarakaty, F. M., & M. El-Meihy, R. (2020). Phylogenetic Diversity of Trichoderma Strains and Their Antagonistic Potential against Soil-Borne Pathogens under Stress Conditions. Biology, 9(8), 189. https://doi.org/10.3390/biology9080189








