The Importance of Protein Phosphorylation for Signaling and Metabolism in Response to Diel Light Cycling and Nutrient Availability in a Marine Diatom
Abstract
1. Introduction
2. Materials and Methods
2.1. Growth Conditions and Sampling
2.2. Phosphopeptide Extraction and Analysis
2.3. Phosphopeptide Identification, Quantification and Manual Curation
2.4. Data Analysis
3. Results and Discussion
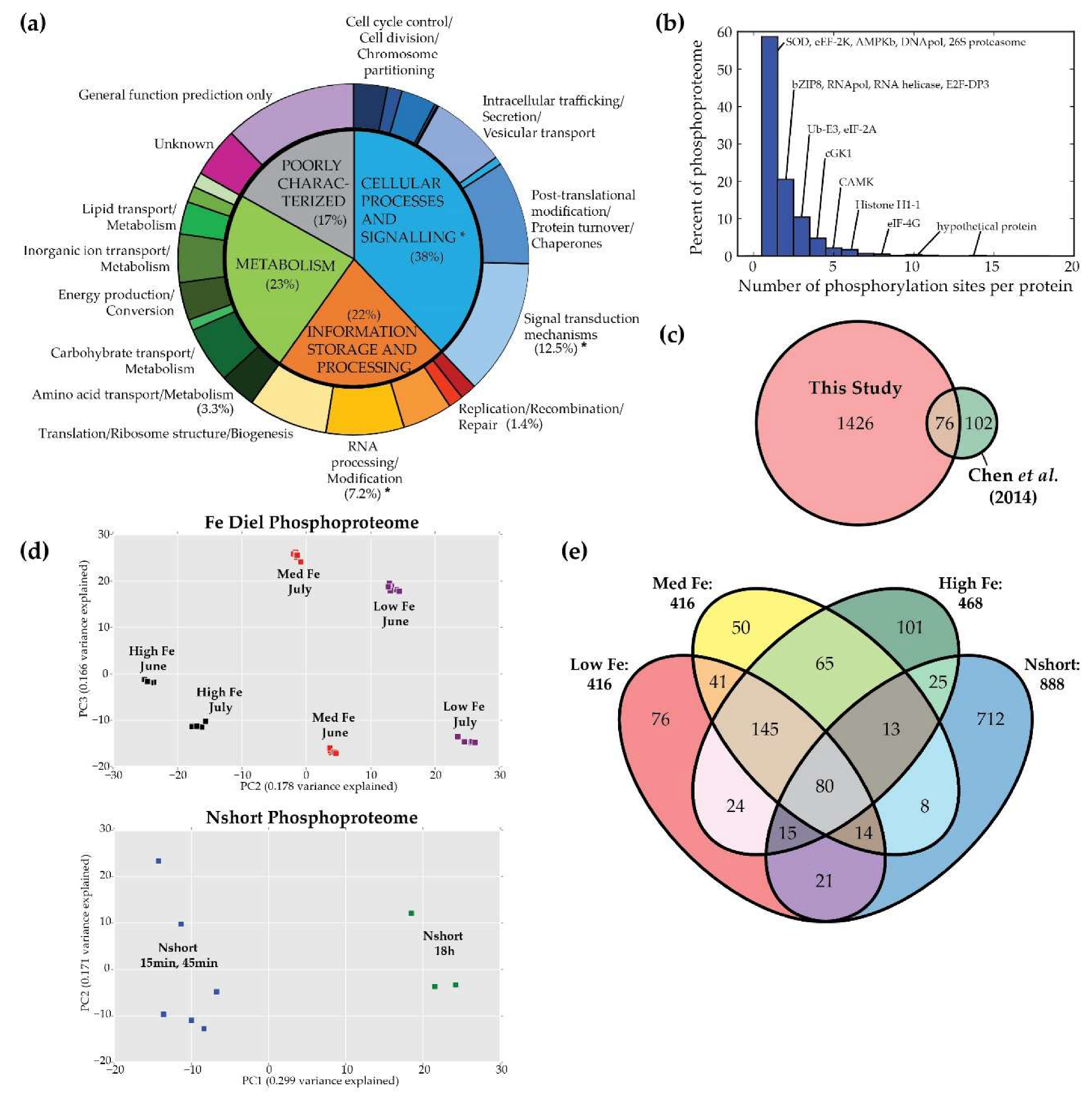
3.1. Overview
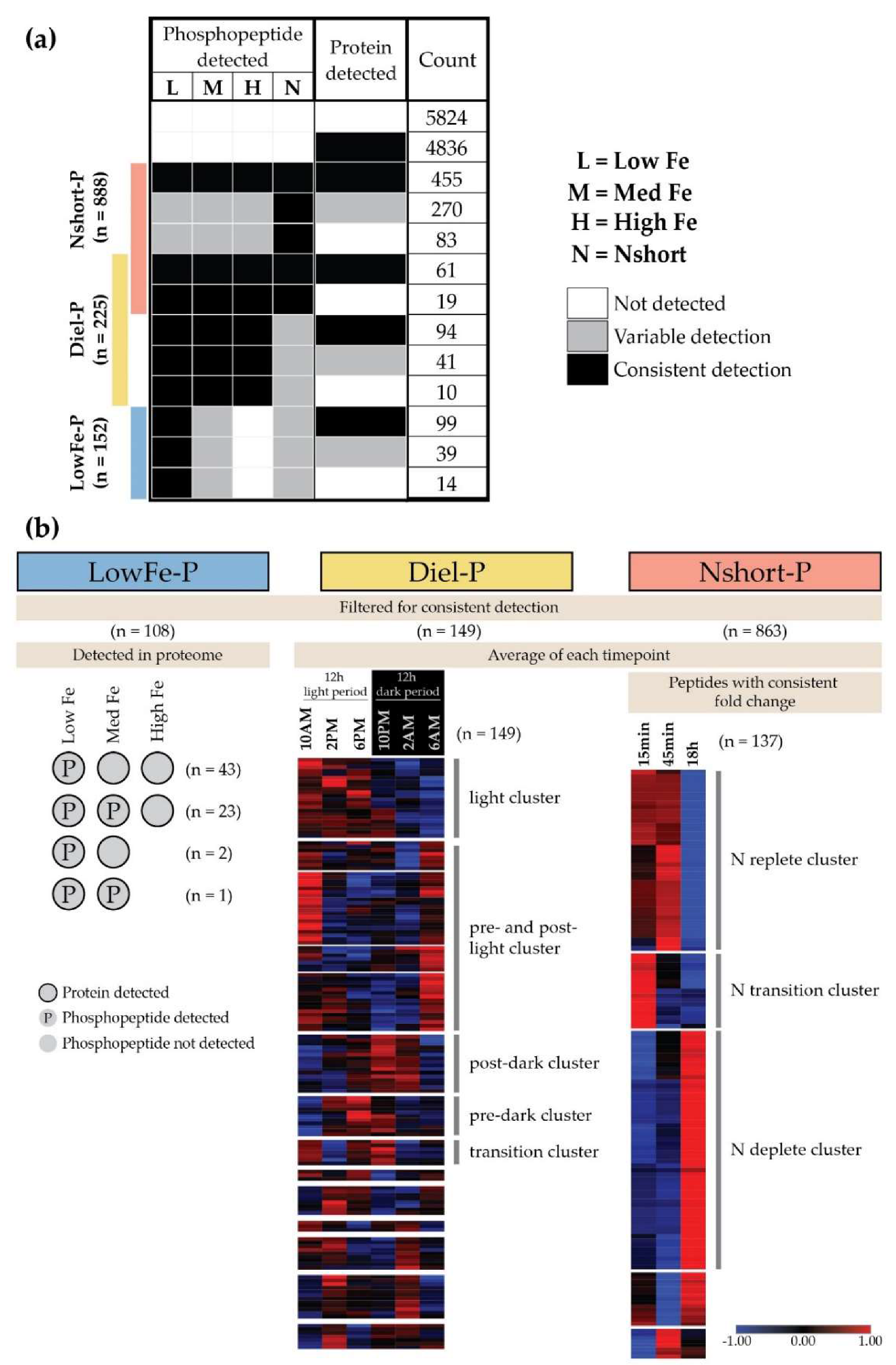
3.2. Conditional Phosphorylation under Fe Limitation, Across Diel Light Cycling, and under N Stress
3.3. Low Fe Phosphorylation
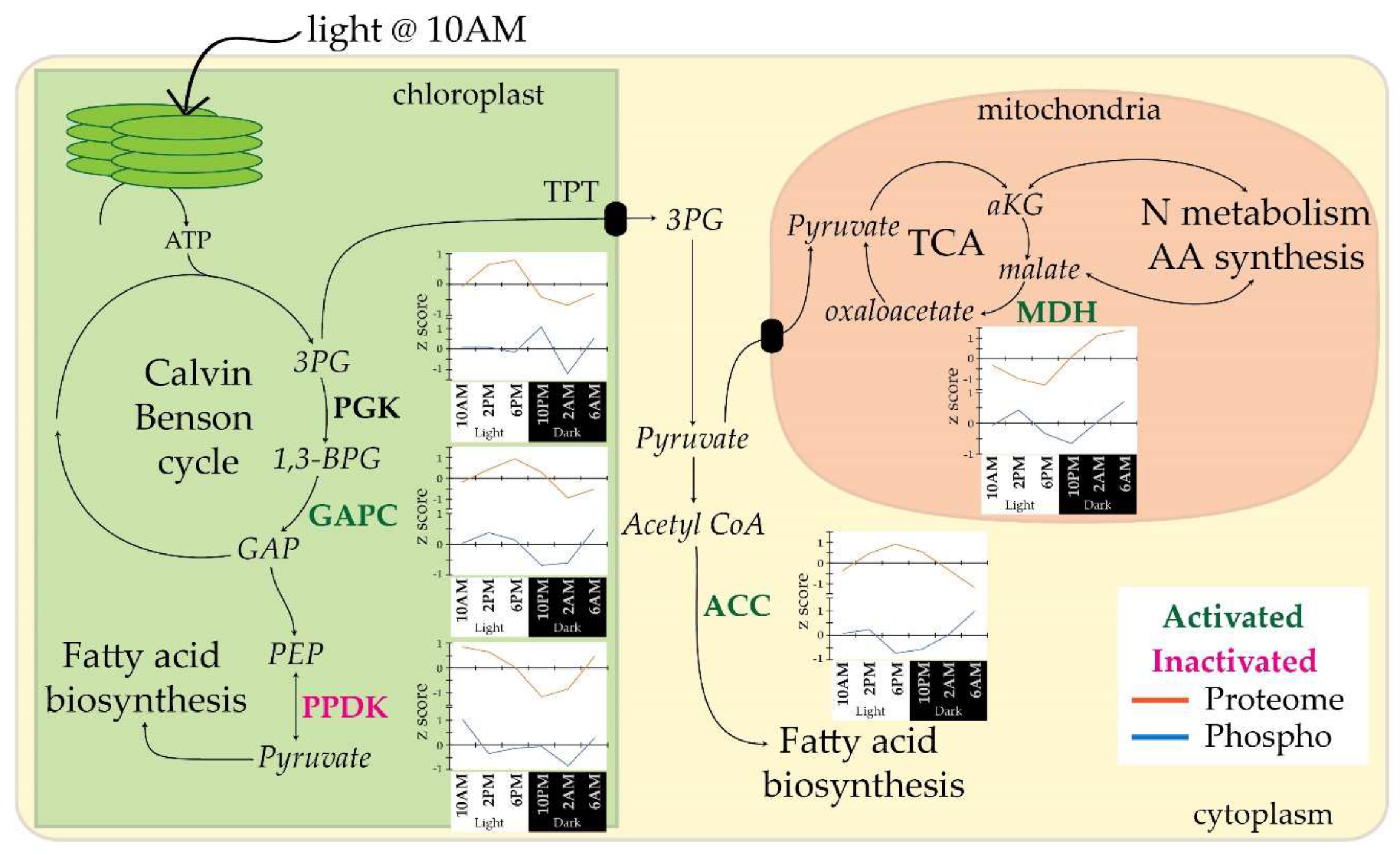
3.4. Diel Profiles of C and N Metabolism Phosphoproteins
3.5. Translation Is Impacted by Coordinated Phosphorylation over the Diel Cycle
3.6. Novel Phosphorylation of Fe Sensitive Proteins
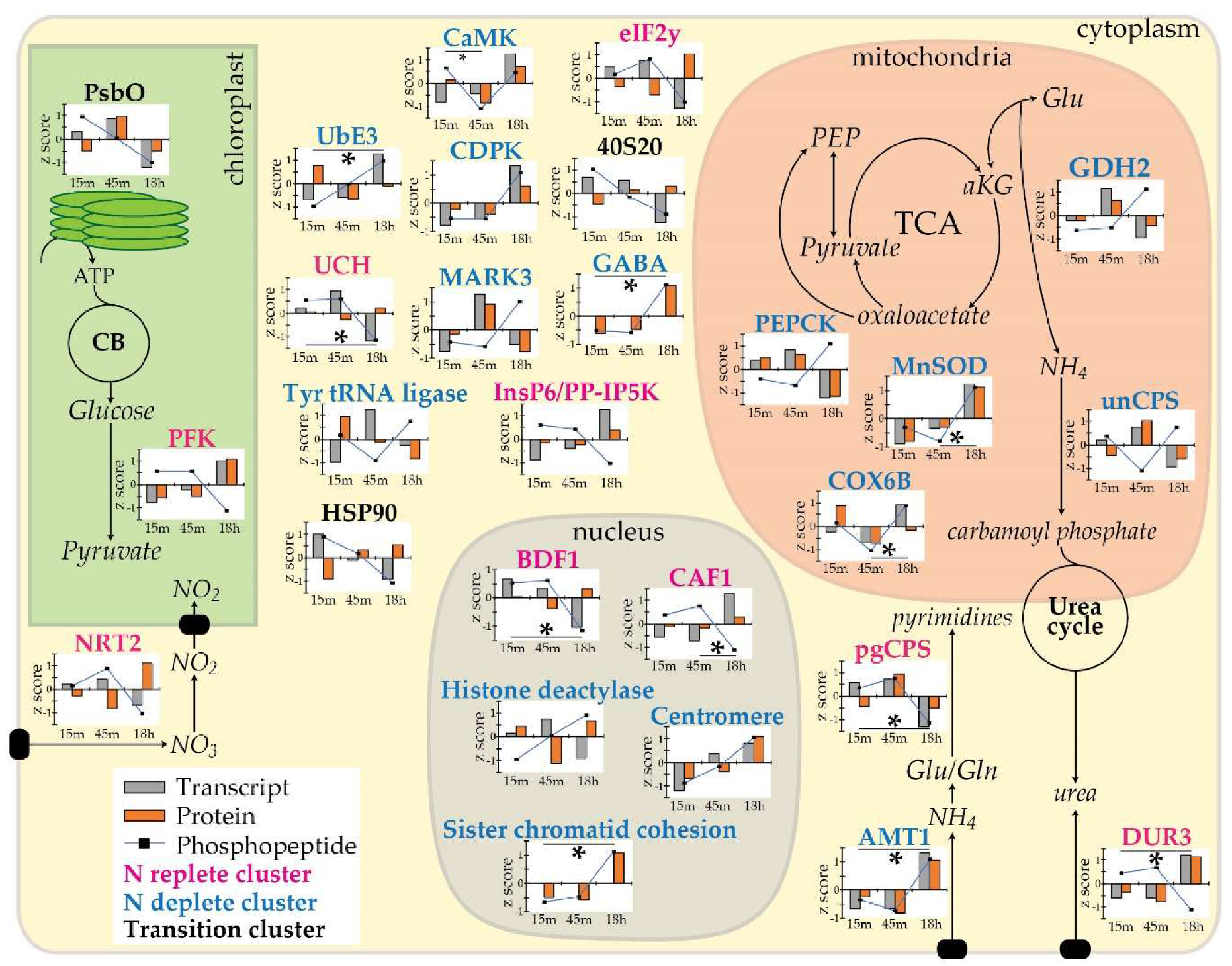
3.7. Phosphorylation in Response to Changes in Nitrogen
3.8. Inhibition of Transcription and Translation as an Early Response to N Depletion
3.9. Stress Related Signaling and Kinases Affected by N Status
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nelson, D.M.; Tréguer, P.; Brzezinski, M.A.; Leynaert, A.; Quéguiner, B. Production and dissolution of biogenic silica in the ocean, Revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem. Cycles 1995, 9, 359–372. [Google Scholar] [CrossRef]
- Kudo, I.; Yoshimura, T.; Yanada, M.; Matsunaga, K. Exhaustion of nitrate terminates a phytoplankton bloom in Funka Bay, Japan, change in SiO4, NO3 consumption rate during the bloom. Mar. Ecol. Prog. Ser. 2000, 193, 45–51. [Google Scholar] [CrossRef][Green Version]
- Krause, J.W.; Schulz, I.K.; Rowe, K.A.; Dobbins, W.; Winding, M.H.S.; Sejr, M.K.; Duarte, C.M.; Agustí, S. Silicic acid limitation drives bloom termination and potential carbon sequestration in an Arctic bloom. Sci. Rep. 2019, 9, 8149. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.K.; Doney, S.C.; Glover, D.M.; Fung, I.Y. Iron cycling and nutrient-limitation patterns in surface waters of the World Ocean. Deep Sea Res. Part 2 Top Stud Oceanogr. 2001, 49, 463–507. [Google Scholar] [CrossRef]
- Moore, J.K.; Doney, S.C.; Lindsay, K. Upper ocean ecosystem dynamics and iron cycling in a global three-dimensional model. Global Biogeochem. Cycles 2004, 18. [Google Scholar] [CrossRef]
- Moore, C.M.; Mills, M.M.; Arrigo, K.R.; Berman-Frank, I.; Bopp, L.; Boyd, P.W.; Geider, R.J.; Guieu, C.; Jaccard, S.L.; Jickells, T.D.; et al. Processes and patterns of oceanic nutrient limitation. Nat. Geosci. 2013, 6, 701. [Google Scholar] [CrossRef]
- Bruland, K.W.; Rue, E.L.; Smith, G.J. Iron and macronutrients in California coastal upwelling regimes, Implications for diatom blooms. Limnol. Oceanogr. 2001, 46, 1661–1674. [Google Scholar] [CrossRef]
- Bruland, K.W.; Rue, E.L.; Smith, G.J.; DiTullio, G.R. Iron, macronutrients and diatom blooms in the Peru upwelling regime, brown and blue waters of Peru. Mar. Chem. 2005, 93, 81–103. [Google Scholar] [CrossRef]
- Bertrand, E.M.; McCrow, J.P.; Moustafa, A.; Zheng, H.; McQuaid, J.B.; Delmont, T.O.; Post, A.F.; Sipler, R.E.; Spackeen, J.L.; Xu, K.; et al. Phytoplankton–bacterial interactions mediate micronutrient colimitation at the coastal Antarctic sea ice edge. Proc. Natl. Acad. Sci. USA 2015, 9938–9943. [Google Scholar] [CrossRef] [PubMed]
- Lommer, M.; Specht, M.; Roy, A.-S.; Kraemer, L.; Andreson, R.; Gutowska, M.A.; Wolf, J.; Bergner, S.V.; Schilhabel, M.B.; Klostermeier, U.C.; et al. Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation. Genome Biol. 2012, 13, R66. [Google Scholar] [CrossRef]
- Groussman, R.D.; Parker, M.S.; Armbrust, E.V. Diversity and Evolutionary History of Iron Metabolism Genes in Diatoms. PLoS ONE 2015, 10, e0129081. [Google Scholar] [CrossRef] [PubMed]
- Allen, A.E.; Laroche, J.; Maheswari, U.; Lommer, M.; Schauer, N.; Lopez, P.J.; Finazzi, G.; Fernie, A.R.; Bowler, C. Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc. Natl. Acad. Sci. USA 2008, 105, 10438–10443. [Google Scholar] [CrossRef] [PubMed]
- Nunn, B.L.; Faux, J.F.; Hippmann, A.A.; Maldonado, M.T.; Harvey, H.R.; Goodlett, D.R.; Boyd, P.W.; Strzepek, R.F. Diatom proteomics reveals unique acclimation strategies to mitigate Fe limitation. PLoS ONE 2013, 8, e75653. [Google Scholar] [CrossRef]
- Graff van Creveld, S.; Rosenwasser, S.; Levin, Y.; Vardi, A. Chronic Iron Limitation Confers Transient Resistance to Oxidative Stress in Marine Diatoms. Plant Physiol. 2016, 172, 968–979. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.R.; Gillard, J.T.F.; Kustka, A.B.; McCrow, J.P.; Badger, J.H.; Zheng, H.; New, A.M.; Dupont, C.L.; Obata, T.; Fernie, A.R.; et al. Transcriptional Orchestration of the Global Cellular Response of a Model Pennate Diatom to Diel Light Cycling under Iron Limitation. PLoS Genet. 2016, 12, e1006490. [Google Scholar] [CrossRef] [PubMed]
- Litchman, E.; Klausmeier, C.A.; Schofield, O.M.; Falkowski, P.G. The role of functional traits and trade-offs in structuring phytoplankton communities, scaling from cellular to ecosystem level. Ecol. Lett. 2007, 10, 1170–1181. [Google Scholar] [CrossRef] [PubMed]
- Lomas, M.W.; Glibert, P.M. Comparisons of nitrate uptake, storage, and reduction in marine diatoms and flagellates. J. Phycol. 2000, 36, 903–913. [Google Scholar] [CrossRef]
- Smith, S.R.; Dupont, C.L.; McCarthy, J.K.; Broddrick, J.T.; Oborník, M.; Horák, A.; Füssy, Z.; Cihlář, J.; Kleessen, S.; Zheng, H.; et al. Evolution and regulation of nitrogen flux through compartmentalized metabolic networks in a marine diatom. Nat. Commun. 2019, 10, 4552. [Google Scholar] [CrossRef]
- Olofsson, M.; Robertson, E.K.; Edler, L.; Arneborg, L.; Whitehouse, M.J.; Ploug, H. Nitrate and ammonium fluxes to diatoms and dinoflagellates at a single cell level in mixed field communities in the sea. Sci. Rep. 2019, 9, 1424. [Google Scholar] [CrossRef]
- Longworth, J.; Wu, D.; Huete-Ortega, M.; Wright, P.C.; Vaidyanathan, S. Proteome response of Phaeodactylum tricornutum, during lipid accumulation induced by nitrogen depletion. Algal Res. 2016, 18, 213–224. [Google Scholar] [CrossRef]
- Remmers, I.M.; D’Adamo, S.; Martens, D.E.; de Vos, R.C.H.; Mumm, R.; America, A.H.P.; Cordewener, H.G.; Bakker, L.V.; Peters, S.A.; Wijffels, R.H.; et al. Orchestration of transcriptome, proteome and metabolome in the diatom Phaeodactylum tricornutum during nitrogen limitation. Algal Res. 2018, 35, 33–49. [Google Scholar] [CrossRef]
- Deribe, Y.L.; Pawson, T.; Dikic, I. Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 2010, 17, 666–672. [Google Scholar] [CrossRef] [PubMed]
- Pinna, L.A.; Ruzzene, M. How do protein kinases recognize their substrates? Biochim. Biophys. Acta 1996, 1314, 191–225. [Google Scholar] [CrossRef]
- Cohen, P. The origins of protein phosphorylation. Nat. Cell Biol. 2002, 4, E127–E130. [Google Scholar] [CrossRef]
- Amoutzias, G.D.; He, Y.; Lilley, K.S.; Van de Peer, Y.; Oliver, S.G. Evaluation and properties of the budding yeast phosphoproteome. Mol. Cell Proteomics 2012, 11, M111.009555. [Google Scholar] [CrossRef] [PubMed]
- Sadowski, I.; Breitkreutz, B.-J.; Stark, C.; Su, T.-C.; Dahabieh, M.; Raithatha, S.; Bernhard, W.; Oughtred, R.; Dolinski, K.; Barreto, K.; et al. The PhosphoGRID Saccharomyces cerevisiae protein phosphorylation site database, version 2.0 update. Database 2013, 2013, bat026. [Google Scholar] [CrossRef]
- Vlastaridis, P.; Kyriakidou, P.; Chaliotis, A.; Van de Peer, Y.; Oliver, S.G.; Amoutzias, G.D. Estimating the total number of phosphoproteins and phosphorylation sites in eukaryotic proteomes. Gigascience 2017, 6, 1–11. [Google Scholar] [CrossRef]
- Guo, H.-B.; Ma, Y.; Tuskan, G.A.; Yang, X.; Guo, H. Classification of Complete Proteomes of Different Organisms and Protein Sets Based on Their Protein Distributions in Terms of Some Key Attributes of Proteins. Int. J. Genomics Proteomics 2018, 2018. [Google Scholar] [CrossRef]
- Wang, H.; Gau, B.; Slade, W.O.; Juergens, M.; Li, P.; Hicks, L.M. The global phosphoproteome of Chlamydomonas reinhardtii reveals complex organellar phosphorylation in the flagella and thylakoid membrane. Mol. Cell Proteomics 2014, 13, 2337–2353. [Google Scholar] [CrossRef]
- Chen, Z.; Yang, M.-K.; Li, C.-Y.; Wang, Y.; Zhang, J.; Wang, D.-B.; Zhang, X.; Ge, F. Phosphoproteomic analysis provides novel insights into stress responses in Phaeodactylum tricornutum, a model diatom. J. Proteome Res. 2014, 13, 2511–2523. [Google Scholar] [CrossRef]
- Cresswell, R.C.; Syrett, P.J. Ammonium inhibition of nitrate uptake by the diatom, Phaeodactylum tricornutum. Plant. Sci. Lett. 1979, 14, 321–325. [Google Scholar] [CrossRef]
- McCarthy, J.K.; Smith, S.R.; McCrow, J.P.; Tan, M.H.; Zheng, H.; Beeri, K.; Roth, R.; Lichtle, C.; Goodenough, U.; Bowler, C.P.; et al. Nitrate reductase knockout uncouples nitrate transport from nitrate assimilation and drives repartitioning of carbon flux in a model pennate diatom. Plant Cell 2017, 29, 2047–2070. [Google Scholar] [CrossRef]
- Cresswell, R.C.; Syrett, P.J. Uptake of nitrate by the diatom Phaeodactylum tricornutum. J. Exp. Bot. 1981, 19–25. [Google Scholar] [CrossRef]
- Levitan, O.; Dinamarca, J.; Zelzion, E.; Gorbunov, M.Y.; Falkowski, P.G. An RNA interference knock-down of nitrate reductase enhances lipid biosynthesis in the diatom Phaeodactylum tricornutum. Plant J. 2015, 84, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Matthijs, M.; Fabris, M.; Obata, T.; Foubert, I.; Franco-Zorrilla, J.M.; Solano, R.; Fernie, A.R.; Vyverman, W.; Goossens, A. The transcription factor bZIP14 regulates the TCA cycle in the diatom Phaeodactylum tricornutum. EMBO J. 2017, 36, 1559–1576. [Google Scholar] [CrossRef] [PubMed]
- Ficarro, S.B.; Adelmant, G.; Tomar, M.N.; Zhang, Y.; Cheng, V.J.; Marto, J.A. Magnetic bead processor for rapid evaluation and optimization of parameters for phosphopeptide enrichment. Anal. Chem. 2009, 81, 4566–4575. [Google Scholar] [CrossRef] [PubMed]
- Eng, J.K.; McCormack, A.L.; Yates, J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994, 5, 976–989. [Google Scholar] [CrossRef]
- Beausoleil, S.A.; Villén, J.; Gerber, S.A.; Rush, J.; Gygi, S.P. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006, 24, 1285–1292. [Google Scholar] [CrossRef]
- Kim, S.; Pevzner, P.A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 2014, 5, 5277. [Google Scholar] [CrossRef]
- Bai, X.; Song, H.; Lavoie, M.; Zhu, K.; Su, Y.; Ye, H.; Chen, S.; Fu, Z.; Qian, H. Proteomic analyses bring new insights into the effect of a dark stress on lipid biosynthesis in Phaeodactylum tricornutum. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Bai, X.; Lavoie, M.; Lu, H.; Fan, X.; Pan, X.; Fu, Z.; Qian, F. Analysis of the Proteome of the Marine Diatom Phaeodactylum tricornutum Exposed to Aluminum Providing Insights into Aluminum Toxicity Mechanisms. Environ. Sci. Technol. 2015, 49, 11182–11190. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhang, Z.; You, C.; Zhang, X.; Lu, J.; Ma, H. Abundant protein phosphorylation potentially regulates Arabidopsis anther development. J. Exp. Bot. 2016, 67, 4993–5008. [Google Scholar] [CrossRef] [PubMed]
- Ullmann, G.M.; Hauswald, M.; Jensen, A.; Knapp, E.-W. Structural alignment of ferredoxin and flavodoxin based on electrostatic potentials, Implications for their interactions with photosystem I and ferredoxin-NADP reductase. Proteins, Struct. Funct. Bioinf. 2000, 38, 301–309. [Google Scholar] [CrossRef]
- Hunter, T. The age of crosstalk, phosphorylation, ubiquitination, and beyond. Mol. Cell 2007, 28, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Elbirt, K.K.; Bonkovsky, H.L. Heme oxygenase, recent advances in understanding its regulation and role. Proc. Assoc. Am. Phys. 1999, 111, 438–447. [Google Scholar] [CrossRef]
- Van Oijen, T.; van Leeuwe, M.A.; Gieskes, W.W.C.; de Baar, H.J.W. Effects of iron limitation on photosynthesis and carbohydrate metabolism in the Antarctic diatom Chaetoceros brevis (Bacillariophyceae). Eur. J. Phycol. 2004, 39, 161–171. [Google Scholar] [CrossRef][Green Version]
- Li, Y.; Keller, D.M.; Scott, J.D.; Lu, H. CK2 phosphorylates SSRP1 and inhibits its DNA-binding activity. J. Biol. Chem. 2005, 280, 11869–11875. [Google Scholar] [CrossRef]
- Kisilevsky, R.; Treloar, M.A.; Weiler, L. Ribosome conformational changes associated with protein S6 phosphorylation. J. Biol. Chem. 1984, 259, 1351–1356. [Google Scholar] [PubMed]
- Hay, N.; Sonenberg, N. Upstream and downstream of mTOR. Genes Dev. 2004, 18, 1926–1945. [Google Scholar] [CrossRef] [PubMed]
- Ruvinsky, I.; Meyuhas, O. Ribosomal protein S6 phosphorylation, from protein synthesis to cell size. Trends Biochem. Sci. 2006, 31, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Jefferies, H.B.J.; Fumagalli, S.; Dennis, P.B.; Reinhard, C.; Pearson, R.B.; Thomas, G. Rapamycin suppresses 5′ TOP mRNA translation through inhibition of p70s6k. EMBO J. 1997, 16, 3693–3704. [Google Scholar] [CrossRef]
- Fumagalli, S.; Thomas, G. S6 phosphorylation and signal transduction. Cold Spring Harbor Monograph Series. 2000, 39, 695–718. [Google Scholar]
- Dedon, P.C.; Begley, T.J. A system of RNA modifications and biased codon use controls cellular stress response at the level of translation. Chem. Res. Toxicol. 2014, 27, 330–337. [Google Scholar] [CrossRef]
- Li, Q.; Fazly, A.M.; Zhou, H.; Huang, S.; Zhang, Z.; Stillman, B. The elongator complex interacts with PCNA and modulates transcriptional silencing and sensitivity to DNA damage agents. PLoS Genet. 2009, 5, e1000684. [Google Scholar] [CrossRef]
- Abdel-Fattah, W.; Jablonowski, D.; Di Santo, R.; Thüring, K.L.; Scheidt, V.; Hammermeister, A.; Have, S.; Helm, M.; Schaffrath, R.; Stark, M.J.R. Phosphorylation of Elp1 by Hrr25 is required for elongator-dependent tRNA modification in yeast. PLoS Genet. 2015, 11, e1004931. [Google Scholar] [CrossRef] [PubMed]
- Mehlgarten, C.; Jablonowski, D.; Wrackmeyer, U.; Tschitschmann, S.; Sondermann, D.; Jäger, G.; Gong, Z.; Byström, A.S.; Schaffrath, R.; Breunig, K.D. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol. Microbiol. 2010, 76, 1082–1094. [Google Scholar] [CrossRef]
- Frenkel, A. Light induced phosphorylation by cell-free preparations of photosynthetic bacteria. J. Am. Chem. Soc. 1954, 76, 5568–5569. [Google Scholar] [CrossRef]
- Shen, H.; Zhu, L.; Castillon, A.; Majee, M.; Downie, B.; Huq, E. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 2008, 20, 1586–1602. [Google Scholar] [CrossRef] [PubMed]
- Carlberg, I.; Hansson, M.; Kieselbach, T.; Schröder, W.P.; Andersson, B.; Vener, A.V. A novel plant protein undergoing light-induced phosphorylation and release from the photosynthetic thylakoid membranes. Proc. Natl. Acad. Sci. USA 2003, 100, 757–762. [Google Scholar] [CrossRef] [PubMed]
- McDowell, J.H.; Kühn, H. Light-induced phosphorylation of rhodopsin in cattle photoreceptor membranes: Substrate activation and inactivation. Biochemistry 1977, 16, 4054–4060. [Google Scholar] [CrossRef]
- Reiland, S.; Messerli, G.; Baerenfaller, K.; Gerrits, B.; Endler, A.; Grossmann, J.; Gruissem, W.; Baginsky, S. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 2009, 150, 889–903. [Google Scholar] [CrossRef] [PubMed]
- Bustos, D.M.; Iglesias, A.A. Phosphorylated non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from heterotrophic cells of wheat interacts with 14-3-3 proteins. Plant Physiol. 2003, 133, 2081–2088. [Google Scholar] [CrossRef] [PubMed]
- Numa, S.; Nakanishi, S.; Hashimoto, T.; Iritani, N.; Okazaki, T. Role of acetyl coenzyme A carboxylase in the control of fatty acid synthesis. Vitam. Horm. 1970, 28, 213–243. [Google Scholar] [PubMed]
- Fullerton, M.D.; Galic, S.; Marcinko, K.; Sikkema, S.; Pulinilkunnil, T.; Chen, Z.-P.; Palanivel, R.; O’Brien, M.; Hardie, D.G.; Macaulay, S.L.; et al. Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat. Med. 2013, 19, 1649–1654. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Jedrychowski, M.P.; Elias, J.E.; Goswami, T.; Rad, R.; Beausoleil, S.A.; Villén, J.; Haas, W.; Sowa, M.E.; Gygi, S.P. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 2010, 143, 1174–1189. [Google Scholar] [CrossRef]
- Munday, M.R.; Campbell, D.G.; Carling, D.; Hardie, D.G. Identification by amino acid sequencing of three major regulatory phosphorylation sites on rat acetyl-CoA carboxylase. Eur. J. Biochem. 1988, 175, 331–338. [Google Scholar] [CrossRef]
- Chen, Y.-B.; Lu, T.-C.; Wang, H.-X.; Shen, J.; Bu, T.-T.; Chao, Q.; Gao, Z.-F.; Zhu, X.-G.; Wang, Y.-F.; Wang, B.-C. Posttranslational Modification of Maize Chloroplast Pyruvate Orthophosphate Dikinase Reveals the Precise Regulatory Mechanism of Its Enzymatic Activity. Plant Physiol. 2014, 165, 534–549. [Google Scholar] [CrossRef] [PubMed]
- Chastain, C.J.; Botschner, M.; Harrington, G.E.; Thompson, B.J.; Mills, S.E.; Sarath, G.; Chollet, R. Further analysis of maize C4 pyruvate, orthophosphate dikinase phosphorylation by its bifunctional regulatory protein using selective substitutions of the regulatory Thr-456 and catalytic His-458 residues. Arch. Biochem. Biophys. 2000, 375, 165–170. [Google Scholar] [CrossRef]
- Minard, K.I.; McAlister-Henn, L. Glucose-induced phosphorylation of the MDH2 isozyme of malate dehydrogenase in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 1994, 315, 302–309. [Google Scholar] [CrossRef]
- Sauer, U.; Eikmanns, B.J. The PEP—pyruvate—oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 2005, 29, 765–794. [Google Scholar] [CrossRef]
- Bender, S.J.; Parker, M.S.; Armbrust, E.V. Coupled effects of light and nitrogen source on the urea cycle and nitrogen metabolism over a diel cycle in the marine diatom Thalassiosira pseudonana. Protist 2012, 163, 232–251. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Choi, H.-J.; Park, E.-J.; Park, H.-J.; Kwon, T.-H. Depletion of vacuolar protein sorting-associated protein 35 is associated with increased lysosomal degradation of aquaporin-2. Am. J. Physiol. Renal Physiol. 2016, 311, F1294–F1307. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, M.; Shimada, T.; Takahashi, H.; Tamura, K.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. Arabidopsis VPS35, a retromer component, is required for vacuolar protein sorting and involved in plant growth and leaf senescence. Plant Cell Physiol. 2008, 49, 142–156. [Google Scholar] [CrossRef] [PubMed]
- Chotewutmontri, P.; Barkan, A. Multilevel effects of light on ribosome dynamics in chloroplasts program genome-wide and psbA-specific changes in translation. PLoS Genet. 2019, 15, e1007907. [Google Scholar] [CrossRef]
- Piques, M.; Schulze, W.X.; Höhne, M.; Usadel, B.; Gibon, Y.; Rohwer, J.; Stitt, M. Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol. Syst. Biol. 2009, 5, 314. [Google Scholar] [CrossRef]
- Zhou, H.; Di Palma, S.; Preisinger, C.; Peng, M.; Polat, A.N.; Heck, A.J.R.; Mohammed, S. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 2013, 12, 260–271. [Google Scholar] [CrossRef]
- Remacha, M.; Jimenez-Diaz, A.; Santos, C.; Briones, E.; Zambrano, R.; Rodriguez Gabriel, M.A.; Guarinos, E.; Ballesta, J.P.G. Proteins P1, P2, and P0, components of the eukaryotic ribosome stalk. New structural and functional aspects. Biochem. Cell Biol. 1995, 73, 959–968. [Google Scholar] [CrossRef]
- Ballesta, J.P.; Rodriguez-Gabriel, M.A.; Bou, G.; Briones, E.; Zambrano, R.; Remacha, M. Phosphorylation of the yeast ribosomal stalk. Functional effects and enzymes involved in the process. FEMS Microbiol. Rev. 1999, 23, 537–550. [Google Scholar] [CrossRef][Green Version]
- Belfield, G.P.; Tuite, M.F. Translation elongation factor 3, a fungus-specific translation factor? Mol. Microbiol. 1993, 9, 411–418. [Google Scholar] [CrossRef]
- Belfield, G.P.; Ross-Smith, N.J.; Tuite, M.F. Translation elongation factor-3 (EF-3), an evolving eukaryotic ribosomal protein? J. Mol. Evol. 1995, 41, 376–387. [Google Scholar] [CrossRef]
- Tavares, C.D.J.; Giles, D.H.; Stancu, G.; Chitjian, C.A.; Ferguson, S.B.; Wellmann, R.M.; Kaoud, T.S.; Ghose, R.; Dalby, K.N. Signal Integration at Elongation Factor 2 Kinase, the roles of calcium, calmodulin, and Ser-500 phosphorylation. J. Biol. Chem. 2017, 292, 2032–2045. [Google Scholar] [CrossRef]
- Browne, G.J.; Finn, S.G.; Proud, C.G. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J. Biol. Chem. 2004, 279, 12220–12231. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, J.M.; Mahoney, S.J.; Blenis, J. mTORC1-Mediated Control of Protein Translation. In The Enzymes; Academic Press: Cambridge, MA, USA, 2010; pp. 1–20. [Google Scholar]
- Pelletier, J.; Graff, J.; Ruggero, D.; Sonenberg, N. Targeting the eIF4F Translation Initiation Complex, A Critical Nexus for Cancer Development. Cancer Res. 2015, 250–263. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Liu, Q.; Miller, W.A.; Goss, D.J. Eukaryotic translation initiation factor 4G (eIF4G) coordinates interactions with eIF4A, eIF4B, and eIF4E in binding and translation of the barley yellow dwarf virus 3′ cap-independent translation element (BTE). J. Biol. Chem. 2017, 292, 5921–5931. [Google Scholar] [CrossRef]
- Chappell, P.D.; Whitney, L.P.; Wallace, J.R.; Darer, A.I.; Jean-Charles, S.; Jenkins, B.D. Genetic indicators of iron limitation in wild populations of Thalassiosira oceanica from the northeast Pacific Ocean. ISME J. 2015, 9, 592–602. [Google Scholar] [CrossRef]
- Cohen, N.R.; Ellis, K.A.; Lampe, R.H.; McNair, H.; Twining, B.S.; Maldonado, M.T.; Brzezinski, M.A.; Kuzminov, F.I.; Thamatrakoln, K.; Till, C.P.; et al. Diatom Transcriptional and Physiological Responses to Changes in Iron Bioavailability across Ocean Provinces. Front. Mar. Sci. 2017. [Google Scholar] [CrossRef]
- Marchetti, A.; Schruth, D.M.; Durkin, C.A.; Parker, M.S.; Kodner, R.B.; Berthiaume, C.T.; Morales, R.; Allen, A.E.; Armbrust, E.V. Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc. Natl. Acad. Sci. USA 2012, 109, E317–E325. [Google Scholar] [CrossRef]
- Coale, T.H.; Moosburner, M.; Horák, A.; Oborník, M.; Barbeau, K.A.; Allen, A.E. Reduction-dependent siderophore assimilation in a model pennate diatom. Proc. Natl. Acad. Sci. USA 2019, 116, 23609–23617. [Google Scholar] [CrossRef]
- Kazamia, E.; Sutak, R.; Paz-Yepes, J.; Dorrell, R.G.; Vieira, F.R.J.; Mach, J.; Leon, S.; Lam, F.; Pelletier, E.; Camadro, J.-M.; et al. Endocytosis-mediated siderophore uptake as a strategy for Fe acquisition in diatoms. Sci. Adv. 2018, 4, eaar4536. [Google Scholar] [CrossRef]
- McQuaid, J.B.; Kustka, A.B.; Oborník, M.; Horák, A.; McCrow, J.P.; Karas, B.J.; Zheng, H.; Kindeberg, T.; Andersson, A.J.; Barbeau, K.A.; et al. Carbonate-sensitive phytotransferrin controls high-affinity iron uptake in diatoms. Nature 2018, 555, 534–537. [Google Scholar] [CrossRef]
- Morrissey, J.; Sutak, R.; Paz-Yepes, J.; Tanaka, A.; Moustafa, A.; Veluchamy, A.; Thomas, Y.; Botebol, H.; Bouget, F.-Y.; McQuaid, J.B.; et al. A novel protein, ubiquitous in marine phytoplankton, concentrates iron at the cell surface and facilitates uptake. Curr. Biol. 2015, 25, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Jacquot, A.; Li, Z.; Gojon, A.; Schulze, W.; Lejay, L. Post-translational regulation of nitrogen transporters in plants and microorganisms. J. Exp. Bot. 2017, 68, 2567–2580. [Google Scholar] [CrossRef] [PubMed]
- Durek, P.; Schmidt, R.; Heazlewood, J.L.; Jones, A.; MacLean, D.; Nagel, A.; Kersten, B.; Schulze, W.X. PhosPhAt, the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 2010, 38, D828–D834. [Google Scholar] [CrossRef] [PubMed]
- Martín, Y.; González, Y.V.; Cabrera, E.; Rodríguez, C.; Siverio, J.M. Npr1 Ser/Thr protein kinase links nitrogen source quality and carbon availability with the yeast nitrate transporter (Ynt1) levels. J. Biol. Chem. 2011, 286, 27225–27235. [Google Scholar] [CrossRef]
- Navarro, F.J.; Martín, Y.; Siverio, J.M. Phosphorylation of the yeast nitrate transporter Ynt1 is essential for delivery to the plasma membrane during nitrogen limitation. J. Biol. Chem. 2008, 283, 31208–31217. [Google Scholar] [CrossRef]
- Smith, S.; Horak, A.; Füssy, Z.; Cihlář, J.; Oborník, M.; Allen, A. Figshare: Phylogenies of nitrogen acquisition and metabolism genes in the model marine diatom Phaeodactylum. tricornutum. 2019. [Google Scholar] [CrossRef]
- Walker, R.P.; Chen, Z.-H.; Acheson, R.M.; Leegood, R.C. Effects of phosphorylation on phosphoenolpyruvate carboxykinase from the C4 plant Guinea grass. Plant Physiol. 2002, 128, 165–172. [Google Scholar] [CrossRef]
- Bailey, K.J.; Gray, J.E.; Walker, R.P.; Leegood, R.C. Coordinate regulation of phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxykinase by light and CO2 during C4 photosynthesis. Plant Physiol. 2007, 144, 479–486. [Google Scholar] [CrossRef]
- Hemmings, B.A. Phosphorylation of NAD-dependent glutamate dehydrogenase from yeast. J. Biol. Chem. 1978, 253, 5255–5258. [Google Scholar]
- Shaw, S.M.; Carrey, E.A. Regulation of the mammalian carbamoyl-phosphate synthetase II by effectors and phosphorylation, altered affinity for ATP and magnesium ions measured using the ammonia-dependent part reaction. Eur. J. Biochem. 1992, 207, 957–965. [Google Scholar] [CrossRef]
- Graves, L.M.; Guy, H.I.; Kozlowski, P.; Huang, M.; Lazarowski, E.; Pope, R.M.; Dahlstrand, R.N.; Earp, H.S., III; Evans, D.R. Regulation of carbamoyl phosphate synthetase by MAP kinase. Nature 2000, 403, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Plate, L.; Marletta, M.A. Phosphorylation-dependent derepression by the response regulator HnoC in the Shewanella oneidensis nitric oxide signaling network. Proc. Natl. Acad. Sci. USA 2013, 110, E4648–E4657. [Google Scholar] [CrossRef]
- Clemens, M.J. Initiation factor eIF2 alpha phosphorylation in stress responses and apoptosis. Prog. Mol. Subcell Biol. 2001, 27, 57–89. [Google Scholar] [PubMed]
- Rajesh, K.; Krishnamoorthy, J.; Kazimierczak, U.; Tenkerian, C.; Papadakis, A.I.; Wang, S.; Huang, S.; Koromilas, A.E. Phosphorylation of the translation initiation factor eIF2α at serine 51 determines the cell fate decisions of Akt in response to oxidative stress. Cell Death Dis. 2015, 6, 1591. [Google Scholar] [CrossRef] [PubMed]
- Karki, S.; Castillo, K.; Ding, Z.; Kerr, O.; Lamb, T.M.; Wu, C.; Sachs, M.S.; Bell-Pedersen, D. Circadian clock control of eIF2α phosphorylation is necessary for rhythmic translation initiation. Proc. Natl. Acad. Sci. USA 2020, 117, 10935–10945. [Google Scholar] [CrossRef]
- Piao, X.; Zhang, X.; Wu, L.; Belasco, J.G. CCR4-NOT deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol. Cell Biol. 2010, 30, 1486–1494. [Google Scholar] [CrossRef]
- Cooke, A.; Prigge, A.; Wickens, M. Translational repression by deadenylases. J. Biol. Chem. 2010, 285, 28506–28513. [Google Scholar] [CrossRef]
- Matangkasombut, O.; Buratowski, R.M.; Swilling, N.W.; Buratowski, S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 2000, 14, 951–962. [Google Scholar]
- Josling, G.A.; Selvarajah, S.A.; Petter, M.; Duffy, M.F. The role of bromodomain proteins in regulating gene expression. Genes 2012, 3, 320–343. [Google Scholar] [CrossRef]
- Sawa, C.; Nedea, E.; Krogan, N.; Wada, T.; Handa, H.; Greenblatt, J.; Buratowski, S. Bromodomain factor 1 (Bdf1) is phosphorylated by protein kinase CK2. Mol. Cell Biol. 2004, 24, 4734–4742. [Google Scholar] [CrossRef]
- Soroka, J.; Wandinger, S.K.; Mäusbacher, N.; Schreiber, T.; Richter, K.; Daub, H.; Buchner, J. Conformational switching of the molecular chaperone Hsp90 via regulated phosphorylation. Mol. Cell 2012, 45, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Candas, D.; Fan, M.; Nantajit, D.; Vaughan, A.T.; Murley, J.S.; Woloschak, G.E.; Grdina, D.J.; Li, J.J. CyclinB1/Cdk1 phosphorylates mitochondrial antioxidant MnSOD in cell adaptive response to radiation stress. J. Mol. Cell Biol. 2013, 5, 166–175. [Google Scholar] [CrossRef]
- Larsen, C.N.; Krantz, B.A.; Wilkinson, K.D. Substrate specificity of deubiquitinating enzymes, ubiquitin C-terminal hydrolases. Biochemistry 1998, 37, 3358–3368. [Google Scholar] [CrossRef] [PubMed]
- Fraile, J.M.; Quesada, V.; Rodríguez, D.; Freije, J.M.P.; López-Otín, C. Deubiquitinases in cancer, new functions and therapeutic options. Oncogene 2012, 31, 2373–2388. [Google Scholar] [CrossRef]
- Gallagher, E.; Gao, M.; Liu, Y.-C.; Karin, M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc. Natl. Acad. Sci. USA 2006, 103, 1717–1722. [Google Scholar] [CrossRef]
- Ichimura, T.; Yamamura, H.; Sasamoto, K.; Tominaga, Y.; Taoka, M.; Kakiuchi, K.; Shinkawa, T.; Takahashi, N.; Shimada, S.; Toshiaki Isobe, T. 14-3-3 proteins modulate the expression of epithelial Na+ channels by phosphorylation-dependent interaction with Nedd4-2 ubiquitin ligase. J. Biol. Chem. 2005, 280, 13187–13194. [Google Scholar] [CrossRef] [PubMed]
- Schulz, P.; Herde, M.; Romeis, T. Calcium-dependent protein kinases, hubs in plant stress signaling and development. Plant Physiol. 2013, 163, 523–530. [Google Scholar] [CrossRef]
- Saiardi, A.; Caffrey, J.J.; Snyder, S.H.; Shears, S.B. The inositol hexakisphosphate kinase family. Catalytic flexibility and function in yeast vacuole biogenesis. J. Biol. Chem. 2000, 275, 24686–24692. [Google Scholar] [CrossRef]
- Lopez, P.J.; Desclés, J.; Allen, A.E.; Bowler, C. Prospects in diatom research. Curr. Opin. Biotechnol. 2005, 16, 180–186. [Google Scholar] [CrossRef]
- Ho, C.-H.; Lin, S.-H.; Hu, H.-C.; Tsay, Y.-F. CHL1 functions as a nitrate sensor in plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef]
- Koltermann, M.; Moroni, A.; Gazzarini, S.; Nowara, D.; Tischner, R. Cloning, functional expression and expression studies of the nitrate transporter gene from Chlorella sorokiniana (strain 211-8k). Plant Mol. Biol. 2003, 52, 855–864. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.I.; Delgado, A.; Gunsalus, R.P. Signal-dependent phosphorylation of the membrane-bound NarX two-component sensor-transmitter protein of Escherichia coli, nitrate elicits a superior anion ligand response compared to nitrite. J. Bacteriol. 1999, 181, 5309–5316. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, M.H.; Smith, S.R.; Hixson, K.K.; Tan, J.; McCarthy, J.K.; Kustka, A.B.; Allen, A.E. The Importance of Protein Phosphorylation for Signaling and Metabolism in Response to Diel Light Cycling and Nutrient Availability in a Marine Diatom. Biology 2020, 9, 155. https://doi.org/10.3390/biology9070155
Tan MH, Smith SR, Hixson KK, Tan J, McCarthy JK, Kustka AB, Allen AE. The Importance of Protein Phosphorylation for Signaling and Metabolism in Response to Diel Light Cycling and Nutrient Availability in a Marine Diatom. Biology. 2020; 9(7):155. https://doi.org/10.3390/biology9070155
Chicago/Turabian StyleTan, Maxine H., Sarah R. Smith, Kim K. Hixson, Justin Tan, James K. McCarthy, Adam B. Kustka, and Andrew E. Allen. 2020. "The Importance of Protein Phosphorylation for Signaling and Metabolism in Response to Diel Light Cycling and Nutrient Availability in a Marine Diatom" Biology 9, no. 7: 155. https://doi.org/10.3390/biology9070155
APA StyleTan, M. H., Smith, S. R., Hixson, K. K., Tan, J., McCarthy, J. K., Kustka, A. B., & Allen, A. E. (2020). The Importance of Protein Phosphorylation for Signaling and Metabolism in Response to Diel Light Cycling and Nutrient Availability in a Marine Diatom. Biology, 9(7), 155. https://doi.org/10.3390/biology9070155




