Curcumin Sensitizes Kidney Cancer Cells to TRAIL-Induced Apoptosis via ROS Mediated Activation of JNK-CHOP Pathway and Upregulation of DR4
Abstract
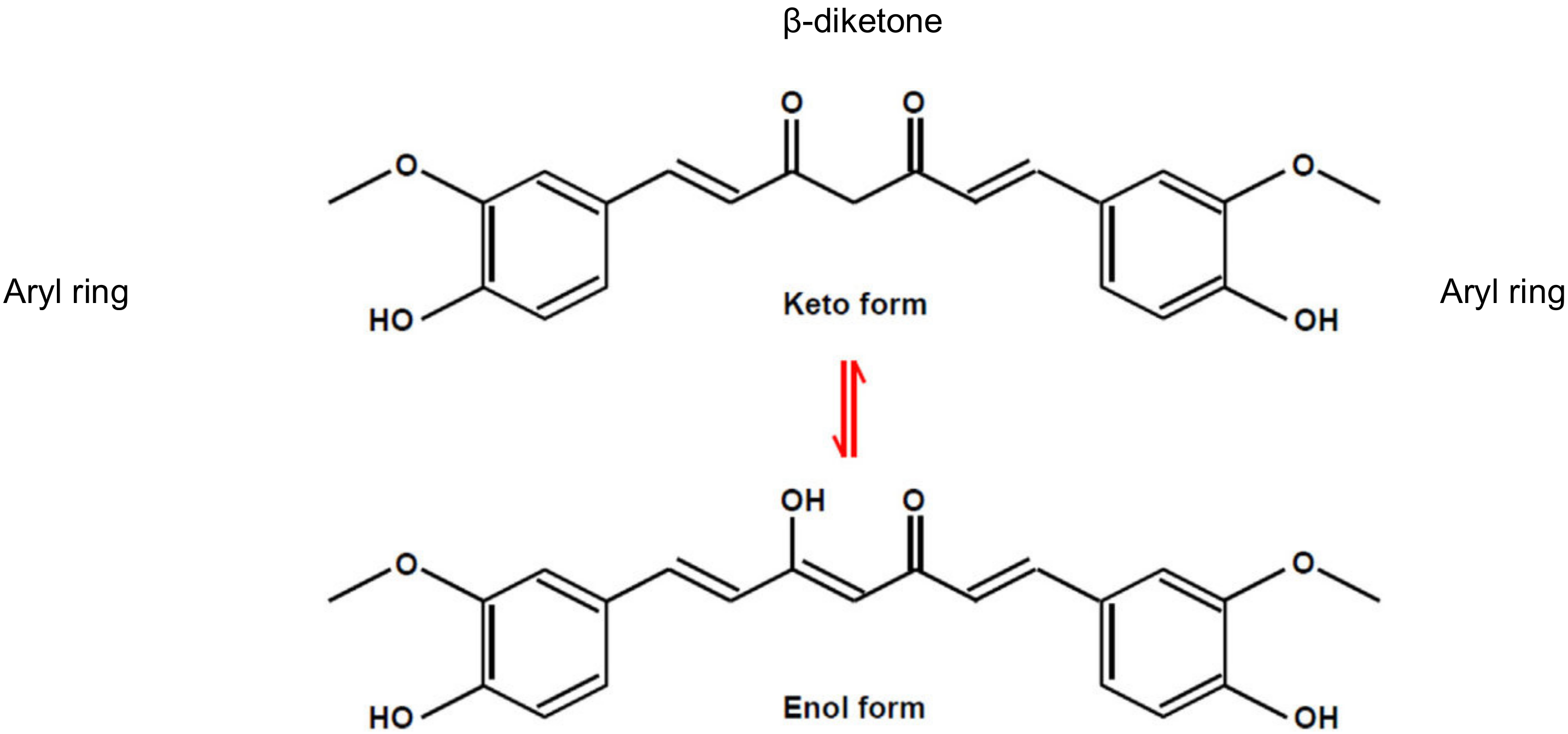
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Cell Viability Assays
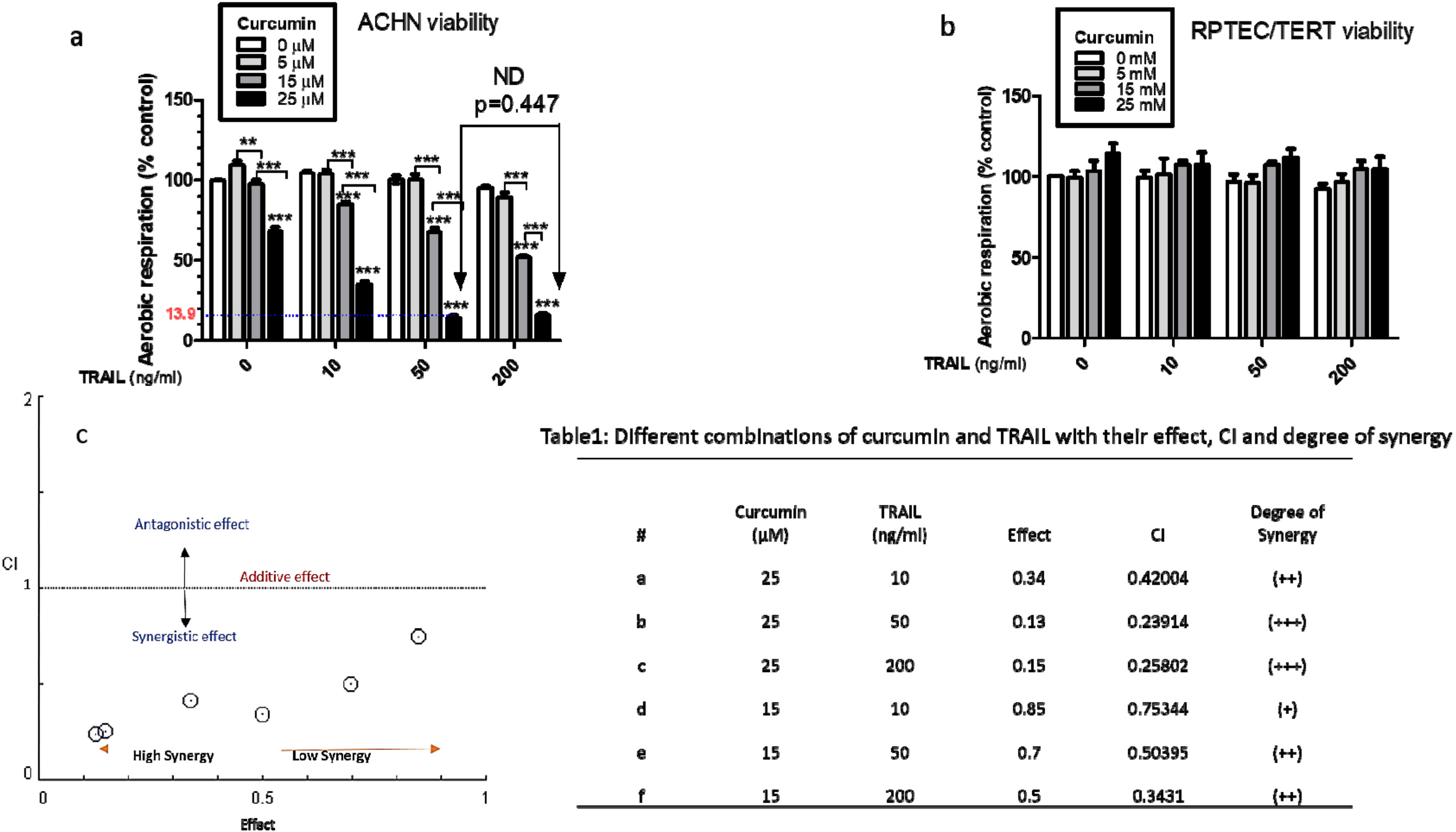
2.3. Combination Index Analysis of Synergy
2.4. Phase Contrast Microscopy
2.5. Flow Cytometry
2.6. Caspase Activity Assays
2.7. Proteasome Assay
2.8. RNA Extraction, cDNA Extraction and qRT-PCR
2.9. Western blot Analysis
2.10. Zebrafish Toxicity Testing and Xenotransplantation
2.11. Statistical Analysis
3. Results
3.1. Combination Treatment of Curcumin and TRAIL Increases Cell Death in the Cancerous Renal ACHN Cells without Affecting the Normal Epithelial RPTEC/TERT1 Cells
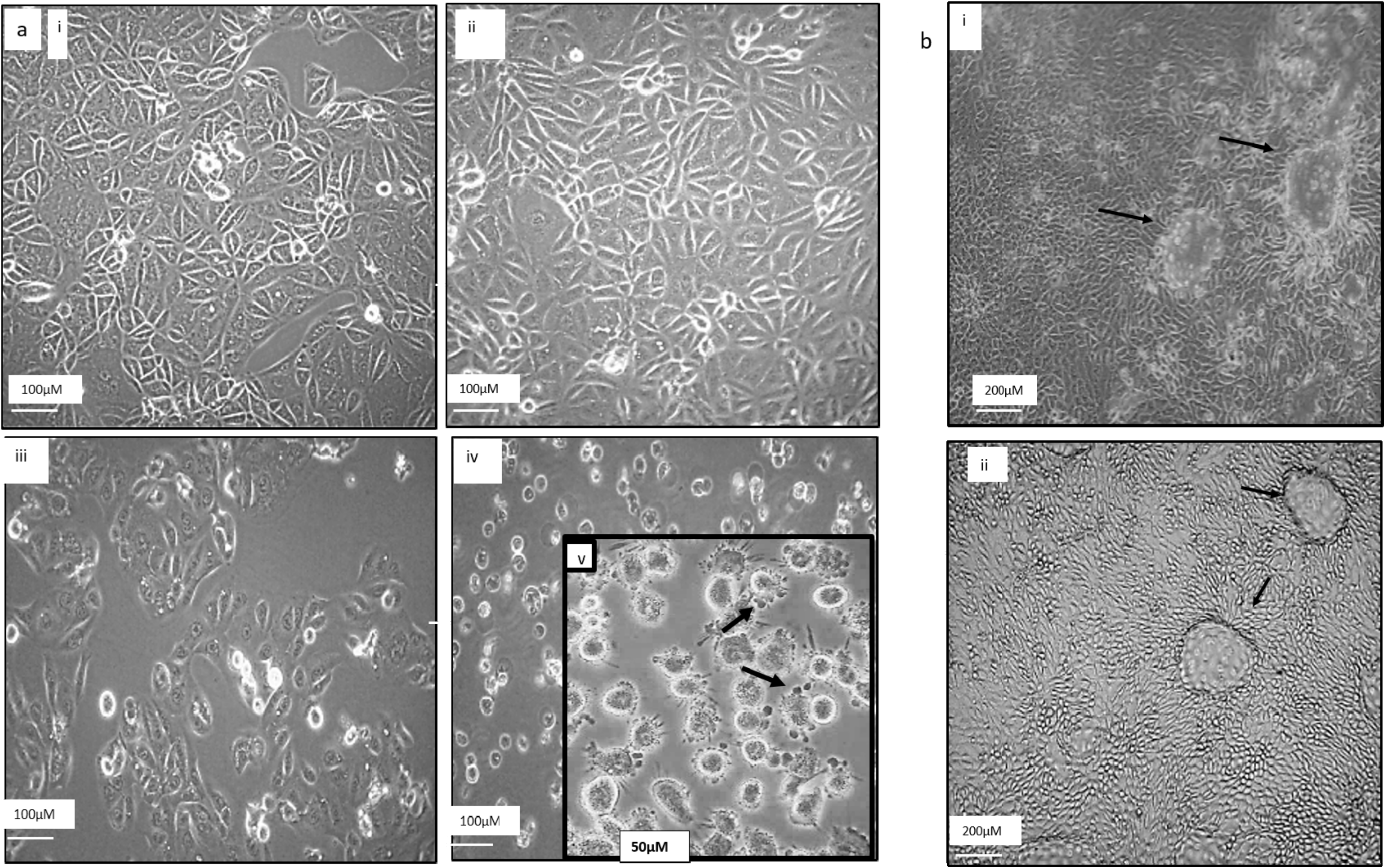
3.2. Morphological Comparison between RPTEC/TERT1 and ACHN Cells upon Exposure to Curcumin, TRAIL and Curcumin/TRAIL Co-Treatment
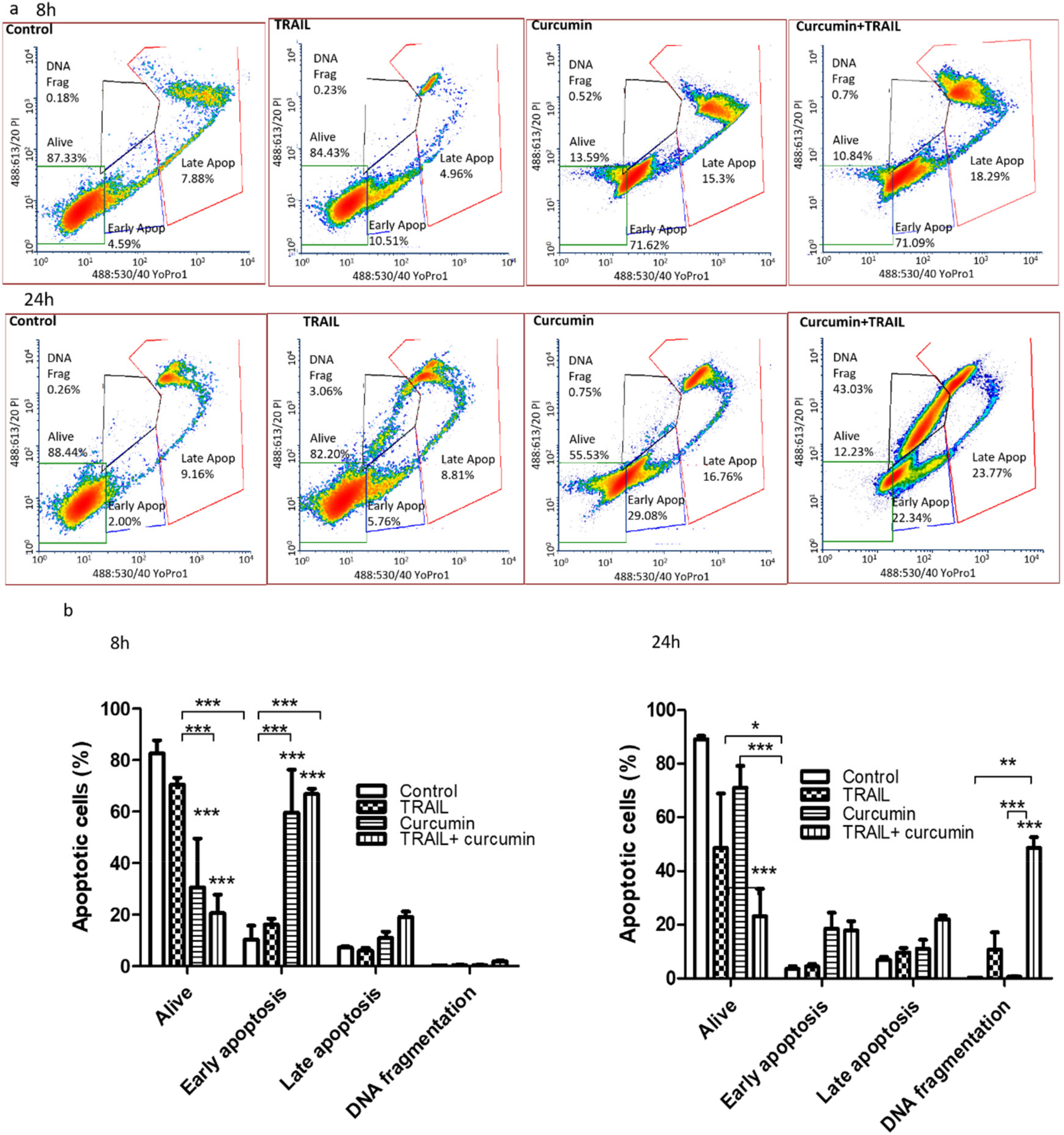
3.3. Curcumin Alone or in a Combination with TRAIL Induced Apoptosis in Renal ACHN Cells
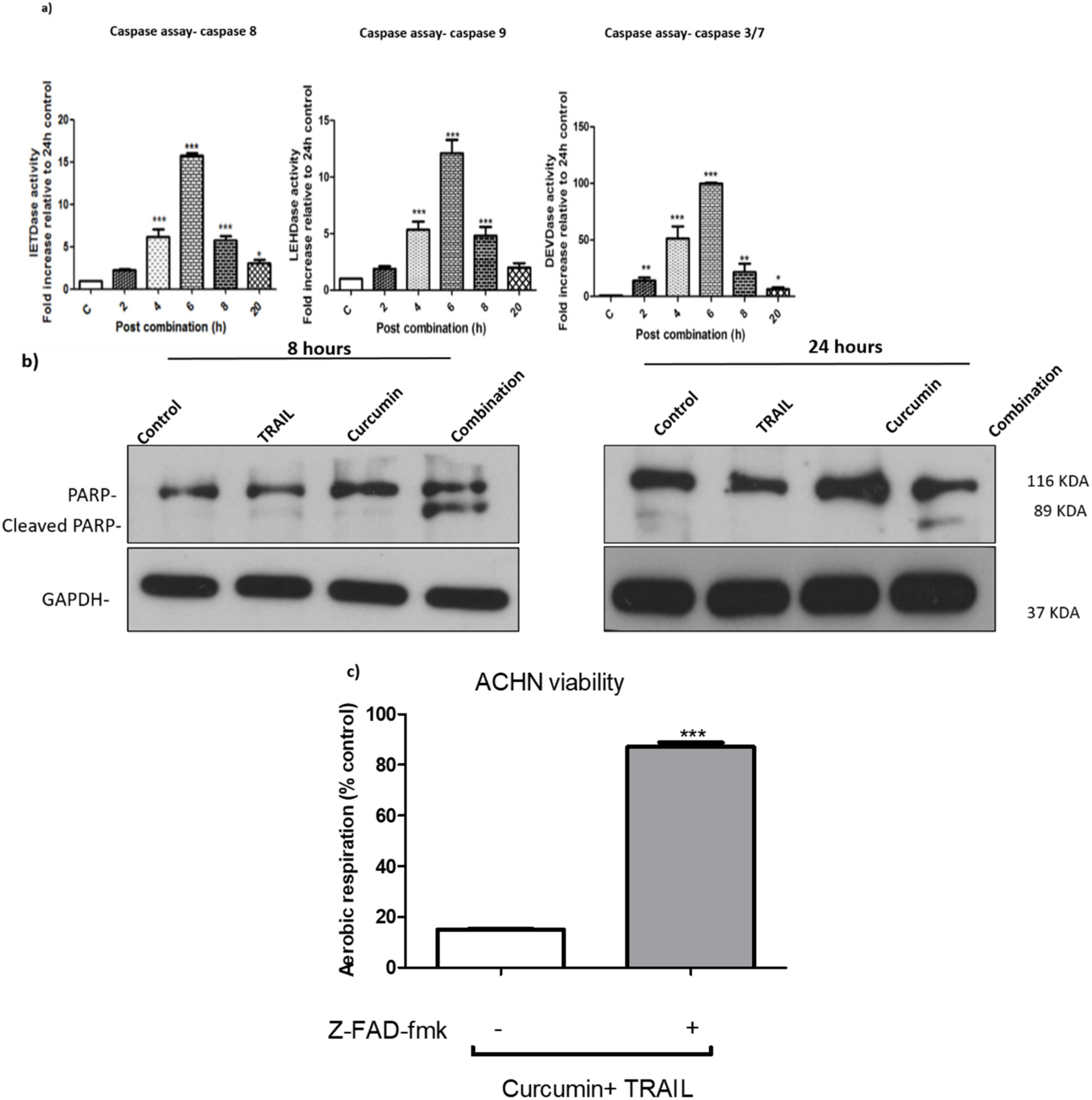
3.4. Curcumin/TRAIL Combination Treatment Induces a Caspase-Dependent Apoptosis
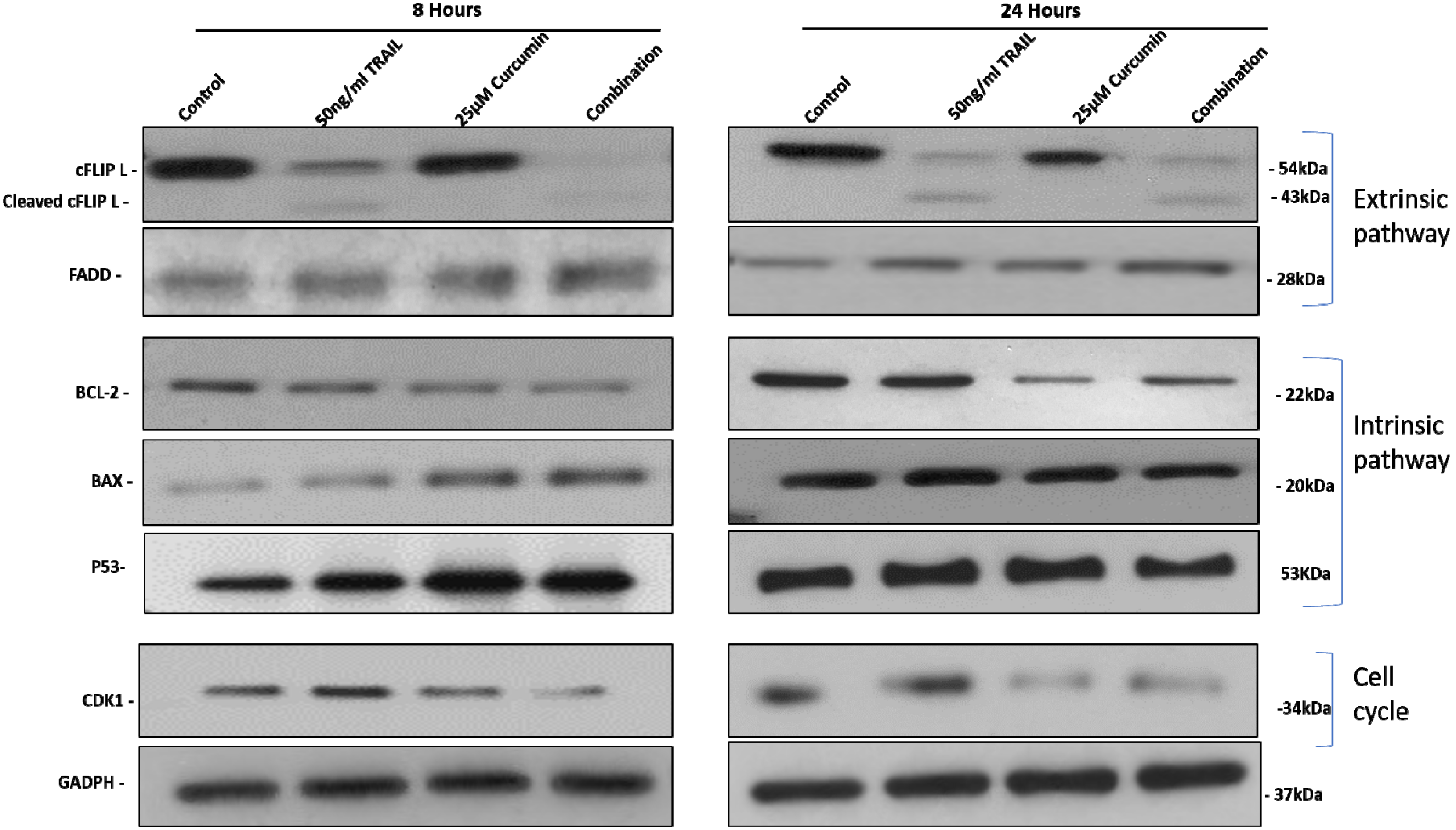
3.5. The Effect of Curcumin, TRAIL, and Curcumin/TRAIL Combination on Pro- and Anti-Apoptotic PROTEIN Expression
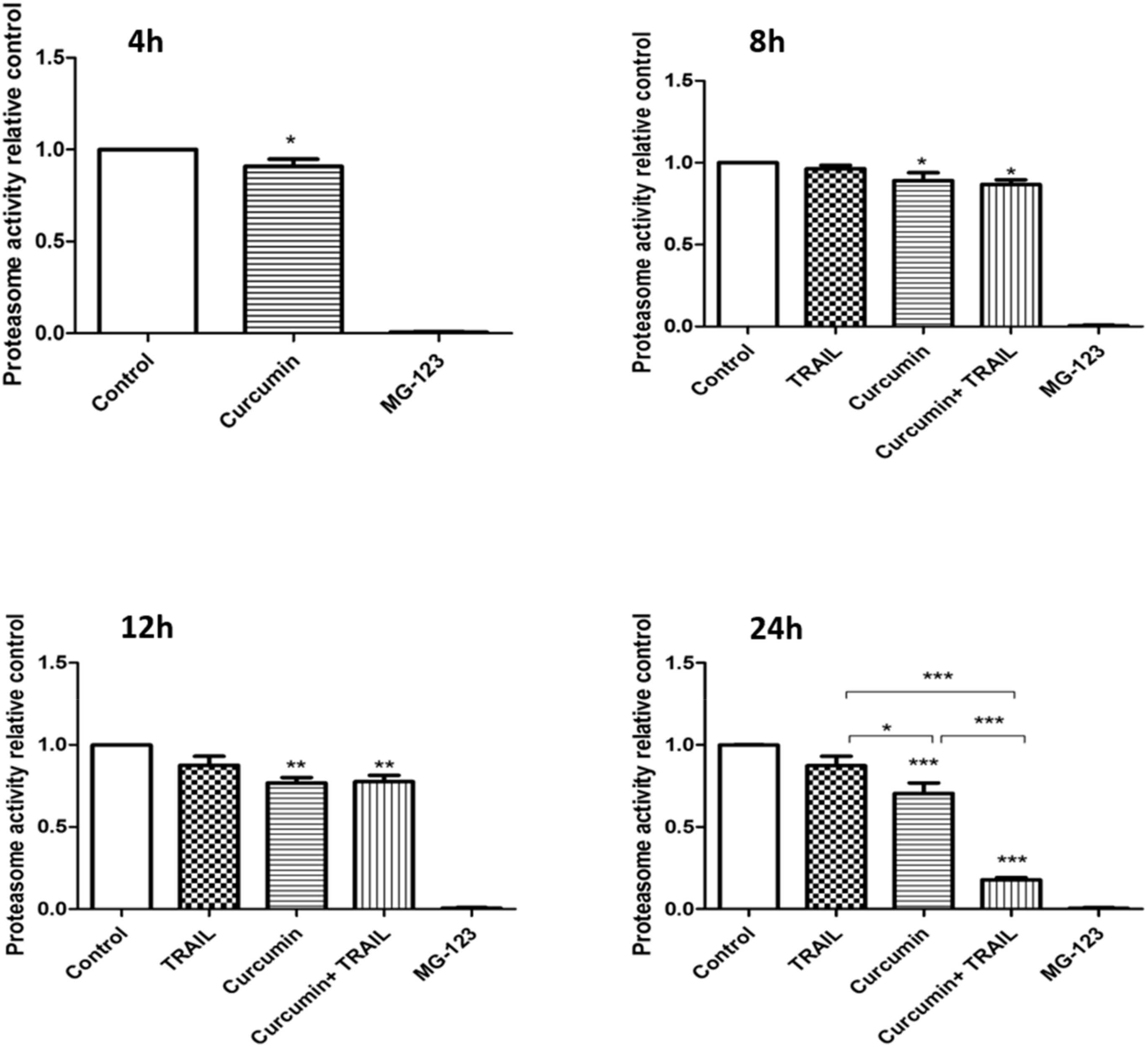
3.6. Curcumin, Alone or in a Combination with TRAIL, Inhibited Proteasome Activity
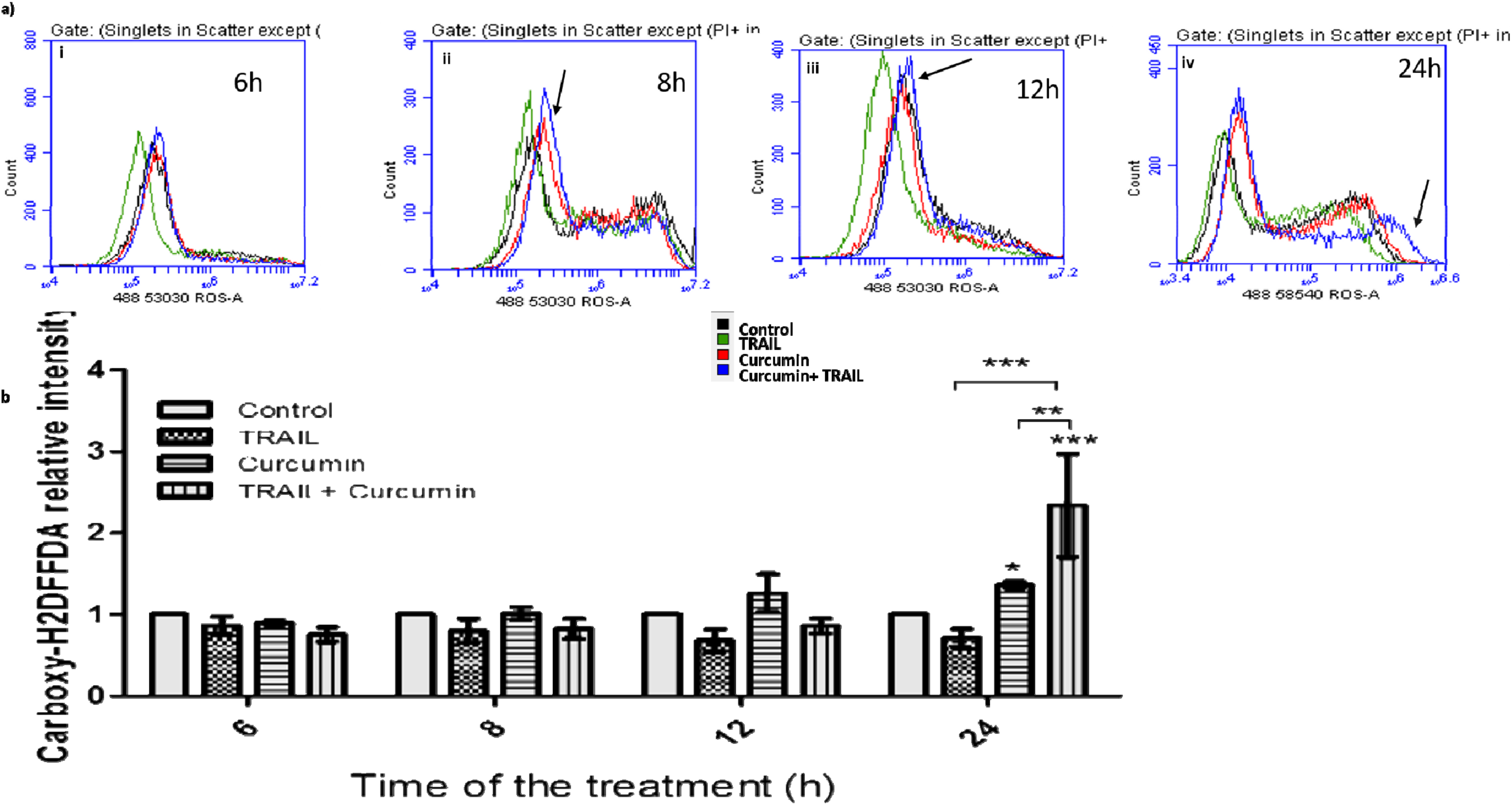
3.7. Curcumin/TRAIL Combination Treatment Induced ROS Induction
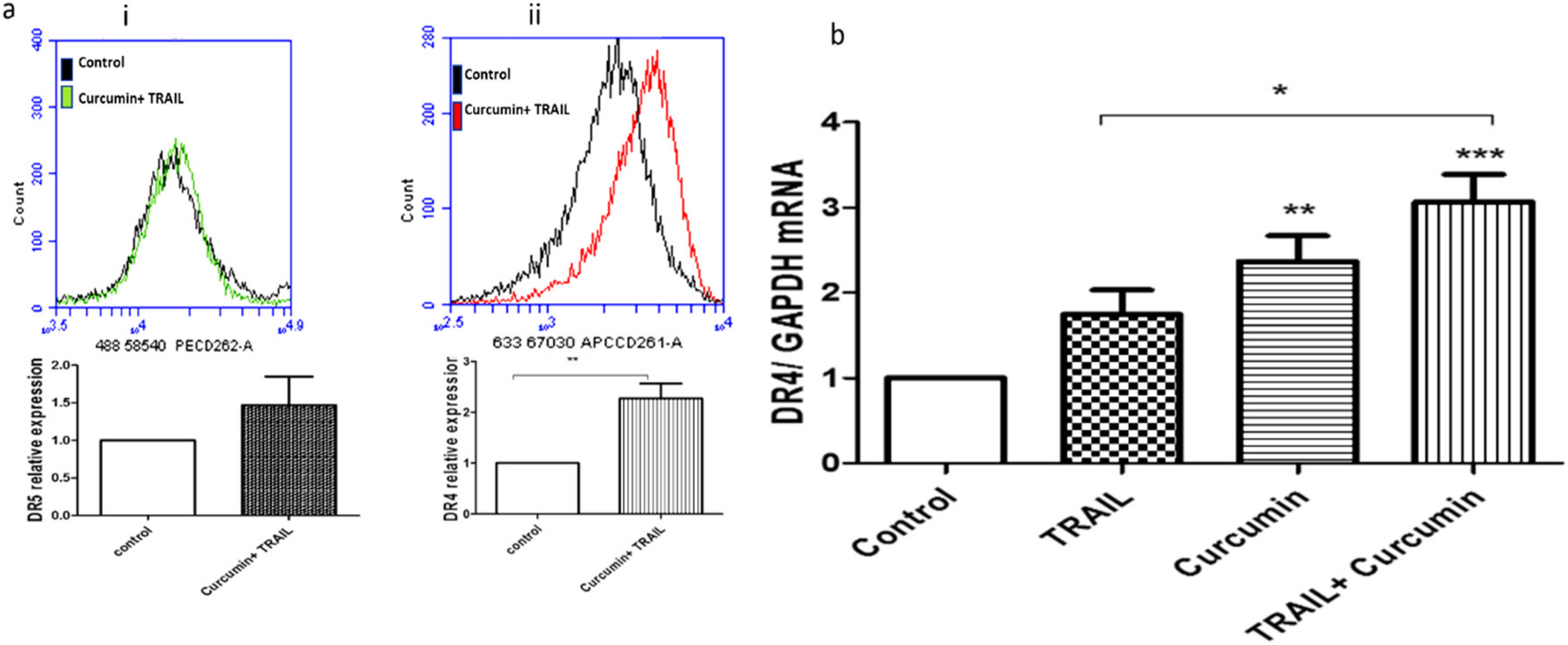
3.8. Curcumin/TRAIL Combination Treatment Induced DR4 Upregulation
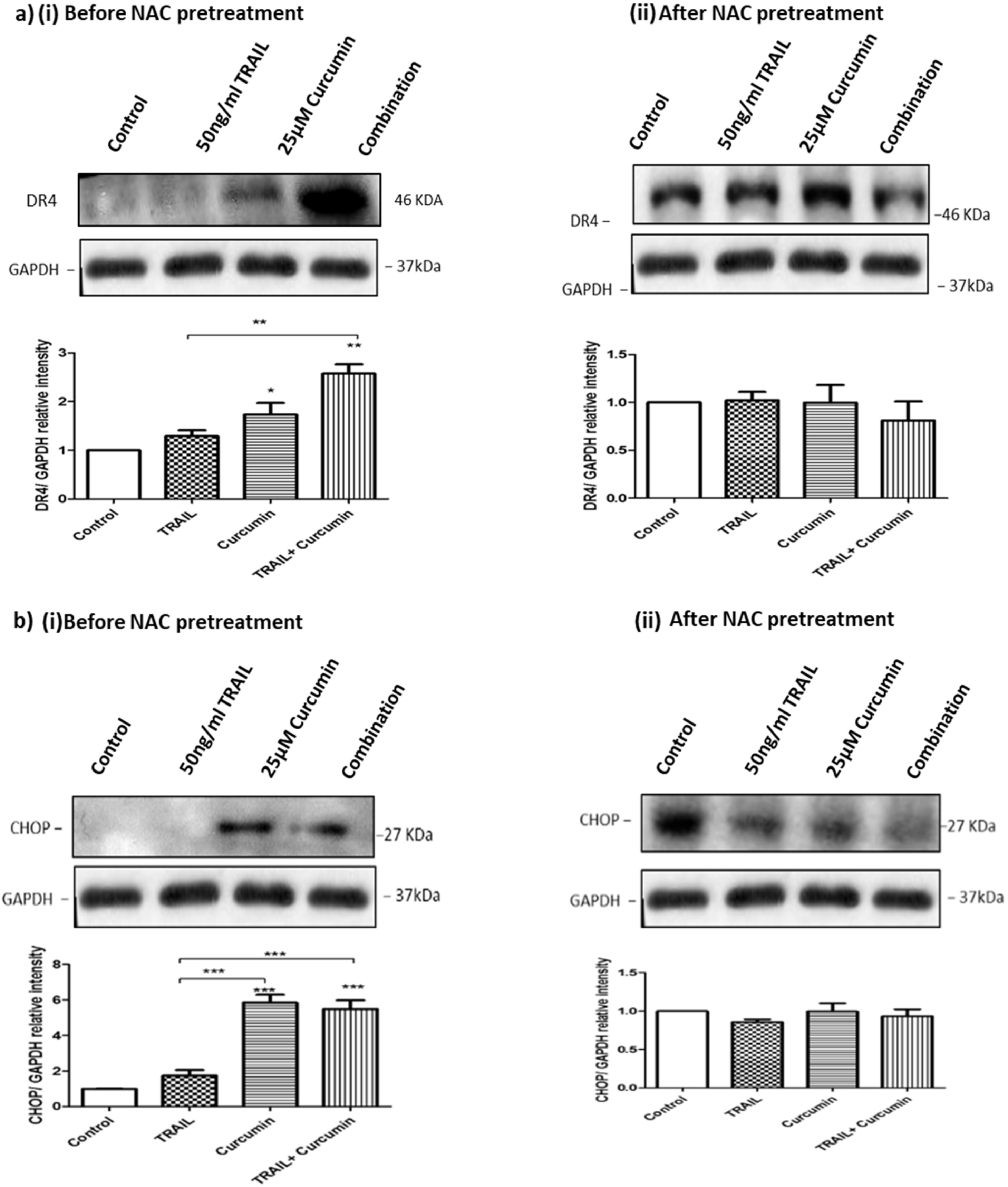
3.9. ROS Regulates Curcumin or Curcumin/TRAIL Combination-Induced CHOP Activation and DR4 Upregulation
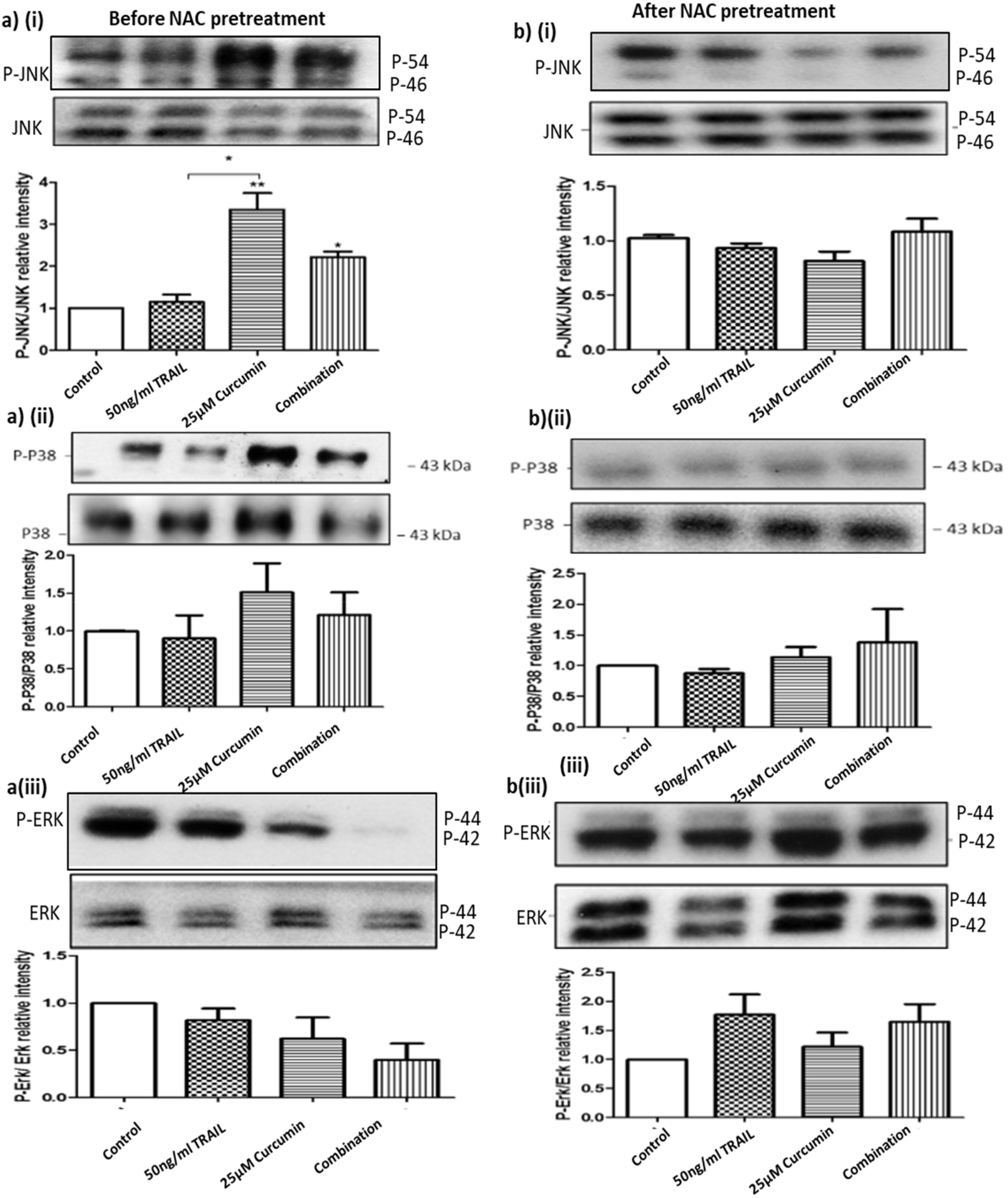
3.10. ROS Regulates Curcumin or Curcumin/TRAIL Combination-Induced MAPK Dysregulation
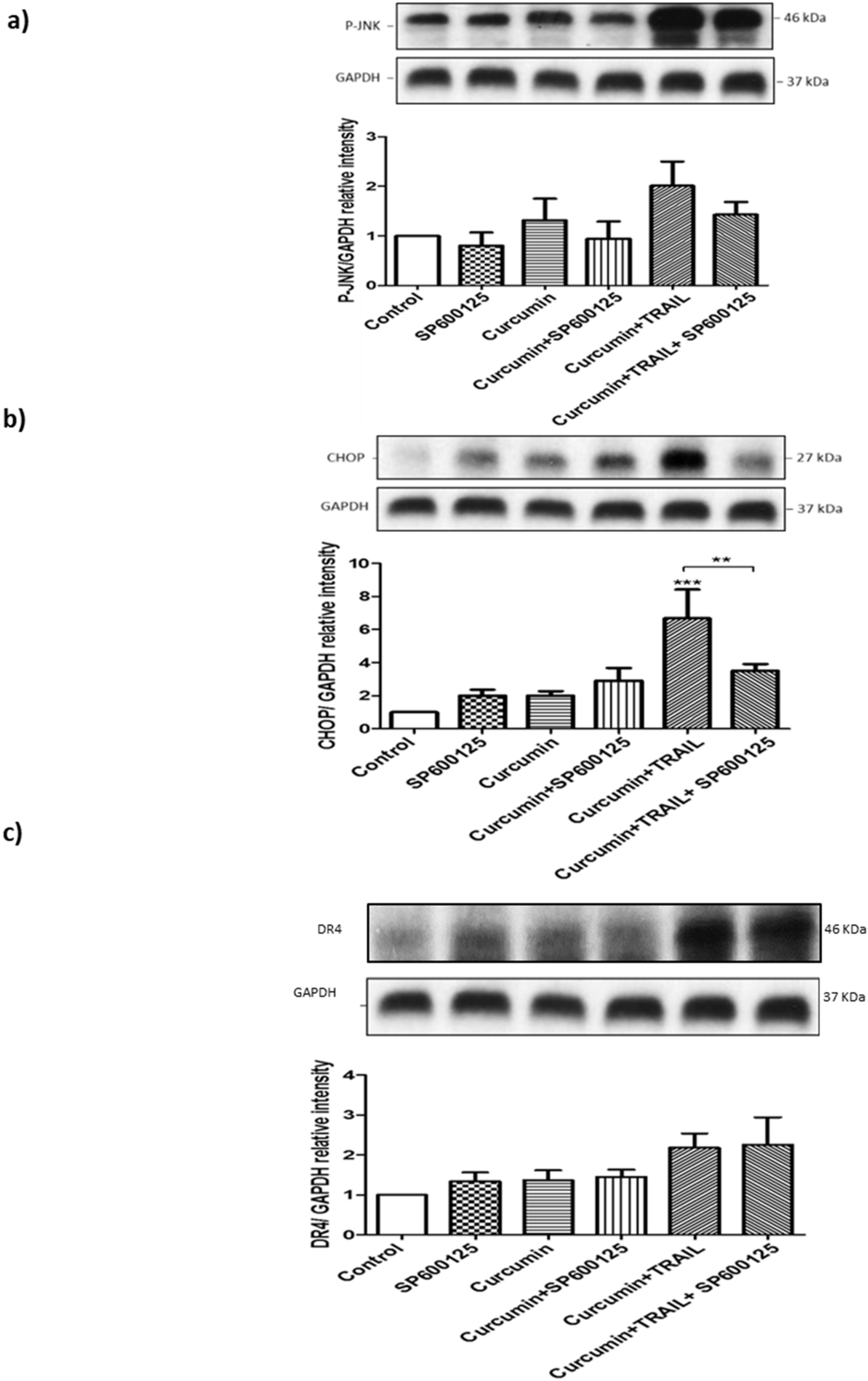
3.11. JNK Regulated CHOP, but Not DR4 Activation
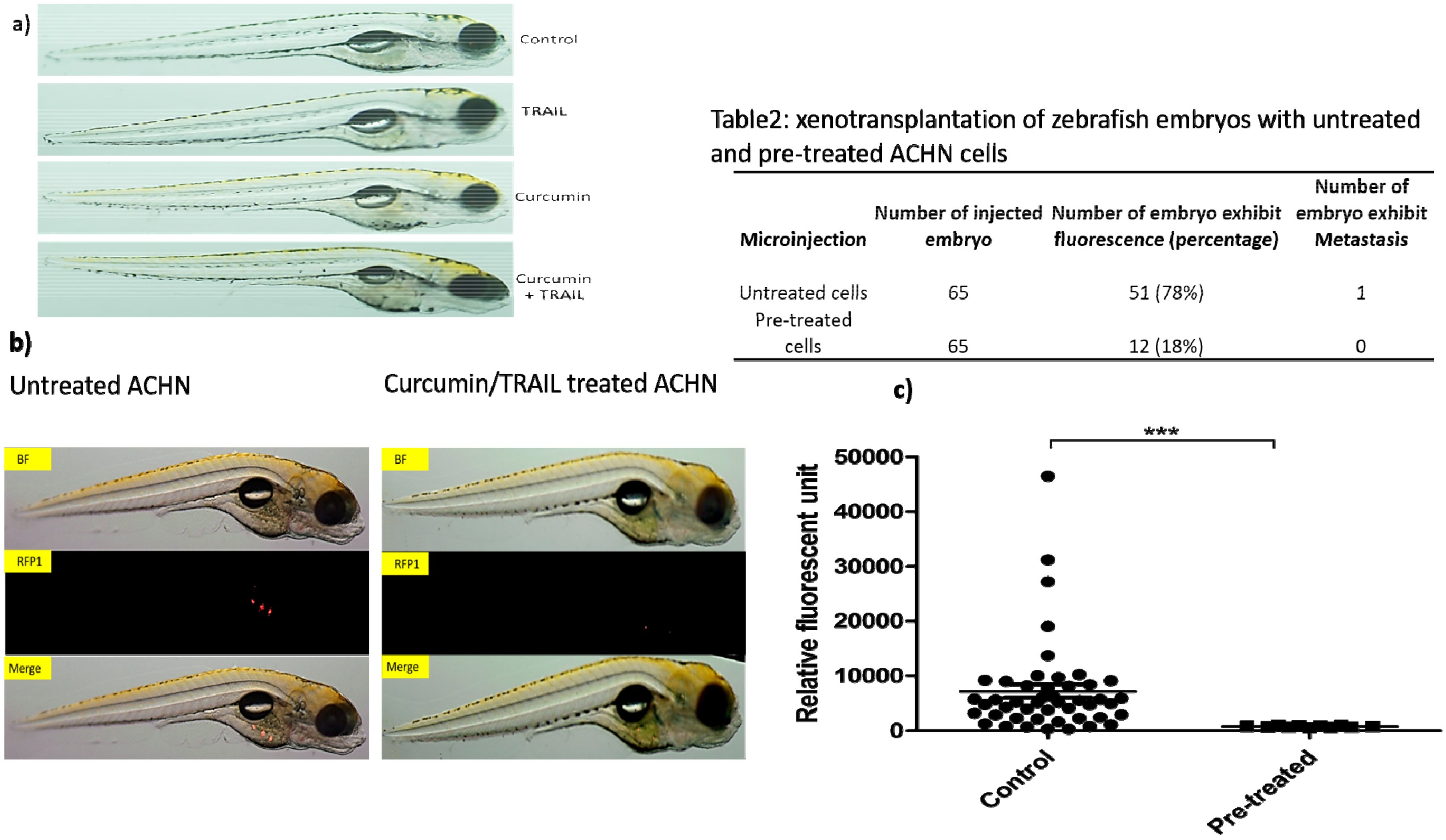
3.12. Assessment of Treatments Toxicities and Effectiveness Using Zebrafish In Vivo Testing Model
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Hutson, T.E. Renal cell carcinoma: Diagnosis and treatment 1994–2003. In Proceedings of the Baylor University Medical Center Proceedings, 2005; Taylor & Francis: Abingdon, UK, 2005; Volume 18, pp. 337–340. [Google Scholar]
- Ozoren, N.; El-Deiry, W.S. Defining characteristics of Types I and II apoptotic cells in response to TRAIL. Neoplasia 2002, 4, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Hassanzadeh, A.; Farshdousti Hagh, M.; Alivand, M.R. Down-regulation of intracellular anti-apoptotic proteins, particularly c-FLIP by therapeutic agents; the novel view to overcome resistance to TRAIL. J. Cell Physiol. 2018, 233, 6470–6485. [Google Scholar] [CrossRef] [PubMed]
- Dimberg, L.Y.; Anderson, C.K.; Camidge, R.; Behbakht, K.; Thorburn, A.; Ford, H.L. On the TRAIL to successful cancer therapy? Predicting and counteracting resistance against TRAIL-based therapeutics. Oncogene 2013, 32, 1341–1350. [Google Scholar] [CrossRef]
- Brooks, A.D.; Jacobsen, K.M.; Li, W.; Shanker, A.; Sayers, T.J. Bortezomib sensitizes human renal cell carcinomas to TRAIL apoptosis through increased activation of caspase-8 in the death-inducing signaling complex. Mol. Cancer Res. 2010, 8, 729–738. [Google Scholar] [CrossRef] [PubMed]
- de Wilt, L.H.; Kroon, J.; Jansen, G.; de Jong, S.; Peters, G.J.; Kruyt, F.A. Bortezomib and TRAIL: A perfect match for apoptotic elimination of tumour cells? Crit. Rev. Oncol. Hematol. 2013, 85, 363–372. [Google Scholar] [CrossRef]
- Lemke, J.; von Karstedt, S.; Zinngrebe, J.; Walczak, H. Getting TRAIL back on track for cancer therapy. Cell Death Differ. 2014, 21, 1350–1364. [Google Scholar] [CrossRef]
- Henrich, C.J.; Brooks, A.D.; Erickson, K.L.; Thomas, C.L.; Bokesch, H.R.; Tewary, P.; Thompson, C.R.; Pompei, R.J.; Gustafson, K.R.; McMahon, J.B.; et al. Withanolide E sensitizes renal carcinoma cells to TRAIL-induced apoptosis by increasing cFLIP degradation. Cell Death Dis. 2015, 6, e1666. [Google Scholar] [CrossRef]
- Lee, H.-P.; Li, T.M.; Tsao, J.Y.; Fong, Y.C.; Tang, C.H. Curcumin induces cell apoptosis in human chondrosarcoma through extrinsic death receptor pathway. Int. Immunopharmacol. 2012, 13, 163–169. [Google Scholar] [CrossRef]
- Bush, J.A.; Cheung, K.J.J.J.; Li, G. Curcumin Induces Apoptosis in Human Melanoma Cells through a Fas Receptor/Caspase-8 Pathway Independent of p53. Exp. Cell Res. 2001, 271, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; He, B.; Lin, L.; Malhotra, A.; Yuan, N. Potential of curcumin and resveratrol as biochemical and biophysical modulators during lung cancer in rats. Drug Chem. Toxicol. 2019, 42, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, S.; Inostroza-Riquelme, M.; Contreras-Orellana, P.; Diaz-Garcia, V.; Lara, P.; Vivanco-Palma, A.; Cardenas, A.; Miranda, V.; Robert, P.; Leyton, L.; et al. Curcumin-loaded nanoemulsion: A new safe and effective formulation to prevent tumor reincidence and metastasis. Nanoscale 2018, 10, 22612–22622. [Google Scholar] [CrossRef] [PubMed]
- Kundur, S.; Prayag, A.; Selvakumar, P.; Nguyen, H.; McKee, L.; Cruz, C.; Srinivasan, A.; Shoyele, S.; Lakshmikuttyamma, A. Synergistic anticancer action of quercetin and curcumin against triple-negative breast cancer cell lines. J. Cell. Physiol. 2019, 234, 11103–11118. [Google Scholar] [CrossRef]
- Seyed Hosseini, E.; Alizadeh Zarei, M.; Babashah, S.; Nakhaei Sistani, R.; Sadeghizadeh, M.; Haddad Kashani, H.; Amini Mahabadi, J.; Izadpanah, F.; Atlasi, M.A.; Nikzad, H. Studies on combination of oxaliplatin and dendrosomal nanocurcumin on proliferation, apoptosis induction, and long non-coding RNA expression in ovarian cancer cells. Cell Biol. Toxicol. 2019, 35, 247–266. [Google Scholar] [CrossRef]
- Li, H.; Krstin, S.; Wink, M. Modulation of multidrug resistant in cancer cells by EGCG, tannic acid and curcumin. Phytomedicine 2018, 50, 213–222. [Google Scholar] [CrossRef]
- Thongnopkoon, T.; Chittasupho, C. Curcumin composite particles prepared by spray drying and in vitro anti-cancer activity on lung cancer cell line. J. Drug Deliv. Sci. Technol. 2018, 45, 397–407. [Google Scholar] [CrossRef]
- Bhatia, N.K.; Kishor, S.; Katyal, N.; Gogoi, P.; Narang, P.; Deep, S. Effect of pH and temperature on conformational equilibria and aggregation behaviour of curcumin in aqueous binary mixtures of ethanol. RSC Adv. 2016, 6, 103275–103288. [Google Scholar] [CrossRef]
- Chou, T.C.; Talalay, P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984, 22, 27–55. [Google Scholar] [CrossRef]
- Rio, D.C.; Ares, M., Jr.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 2010, pdb.prot5439. [Google Scholar] [CrossRef]
- Gangisetty, O.; Reddy, D.S. The optimization of TaqMan real-time RT-PCR assay for transcriptional profiling of GABA-A receptor subunit plasticity. J. Neurosci. Methods 2009, 181, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Obaidi, I.; Higgins, M.; Bahar, B.; Davis, J.L.; McMorrow, T. Identification of the Multifaceted Chemopreventive Activity of Curcumin Against the Carcinogenic Potential of the Food Additive, KBrO3. Curr. Pharm. Des. 2018, 24, 595–614. [Google Scholar] [CrossRef] [PubMed]
- Buchmann, K.; Pedersen, L.; Glamann, J. Humoral immune response of European eel Anguilla anguilla to a major antigen in Anguillicola crassus (Nematoda). Dis. Aquat. Organ. 1991, 12, 55–57. [Google Scholar] [CrossRef]
- Westerfield, M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio); University of Oregon Press: Eugene, OR, USA, 1995. [Google Scholar]
- Nalli, A.D.; Brown, L.E.; Thomas, C.L.; Sayers, T.J. Sensitization of renal carcinoma cells to TRAIL-induced apoptosis by rocaglamide and analogs. Sci. Rep. 2018, 8, 17519. [Google Scholar] [CrossRef]
- Clark, P.E.; Polosukhina, D.A.; Gyabaah, K.; Moses, H.L.; Thorburn, A.; Zent, R. TRAIL and IFNα act synergistically to induce renal cell carcinoma apoptosis. J. Urol. 2010, 184, 1166–1174. [Google Scholar] [CrossRef]
- Zhao, L.; Au, J.L.S.; Wientjes, M.G. Comparison of methods for evaluating drug-drug interaction. Front. Biosci. 2010, 2, 241–249. [Google Scholar]
- Kunwar, A.; Barik, A.; Mishra, B.; Rathinasamy, K.; Pandey, R.; Priyadarsini, K.I. Quantitative cellular uptake, localization and cytotoxicity of curcumin in normal and tumor cells. Biochim. Biophys. Acta (BBA) Gen. Subj. 2008, 1780, 673–679. [Google Scholar] [CrossRef]
- Jaruga, E.; Salvioli, S.; Dobrucki, J.; Chrul, S.; Bandorowicz-Pikuła, J.; Sikora, E.; Franceschi, C.; Cossarizza, A.; Bartosz, G. Apoptosis-like, reversible changes in plasma membrane asymmetry and permeability, and transient modifications in mitochondrial membrane potential induced by curcumin in rat thymocytes. FEBS Lett. 1998, 433, 287–293. [Google Scholar] [CrossRef]
- Jaruga, E.; Sokal, A.; Chrul, S.; Bartosz, G. Apoptosis-independent alterations in membrane dynamics induced by curcumin. Exp. Cell Res. 1998, 245, 303–312. [Google Scholar] [CrossRef]
- Singh, R.; Tønnesen, H.H.; Vogensen, S.B.; Loftsson, T.; Másson, M. Studies of curcumin and curcuminoids. XXXVI. The stoichiometry and complexation constants of cyclodextrin complexes as determined by the phase-solubility method and UV–Vis titration. J. Incl. Phenom. Macrocycl. Chem. 2010, 66, 335–348. [Google Scholar] [CrossRef]
- Tønnesen, H.; Arrieta, A.; Lerner, D. Studies on curcumin and curcuminoids. XXIV: Characterization of the spectroscopic properties of the naturally occurring curcuminoids and selected derivatives. Pharmazie 1995, 50, 689–693. [Google Scholar]
- Cao, J.; Jia, L.; Zhou, H.M.; Liu, Y.; Zhong, L.F. Mitochondrial and Nuclear DNA Damage Induced by Curcumin in Human Hepatoma G2 Cells. Toxicol. Sci. 2006, 91, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.H.; Lee, J.H.; Park, J.W.; Moon, S.H.; Cho, Y.S.; Choe, Y.S.; Lee, K.H. Effects of curcumin on cancer cell mitochondrial function and potential monitoring with (1)(8)F-FDG uptake. Oncol. Rep. 2016, 35, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Kesharwani, R.K.; Misra, K.; Rizvi, S.I. The modulation of erythrocyte Na+/K+-ATPase activity by curcumin. J. Adv. Res. 2015, 6, 1023–1030. [Google Scholar] [CrossRef]
- Mahmmoud, Y.A. Modulation of protein kinase C by curcumin; inhibition and activation switched by calcium ions. Br. J. Pharmacol. 2007, 150, 200–208. [Google Scholar] [CrossRef]
- Tyagi, P.; Singh, M.; Kumari, H.; Kumari, A.; Mukhopadhyay, K. Bactericidal activity of curcumin I is associated with damaging of bacterial membrane. PLoS ONE 2015, 10, e0121313. [Google Scholar] [CrossRef]
- Ligeret, H.; Barthelemy, S.; Zini, R.; Tillement, J.P.; Labidalle, S.; Morin, D. Effects of curcumin and curcumin derivatives on mitochondrial permeability transition pore. Free Radic. Biol. Med. 2004, 36, 919–929. [Google Scholar] [CrossRef]
- Alsop, R.J.; Dhaliwal, A.; Rheinstädter, M.C. Curcumin Protects Membranes through a Carpet or Insertion Model Depending on Hydration. Langmuir 2017, 33, 8516–8524. [Google Scholar] [CrossRef]
- Leite, N.B.; Martins, D.B.; Fazani, V.E.; Vieira, M.R.; dos Santos Cabrera, M.P. Cholesterol modulates curcumin partitioning and membrane effects. Biochim. Biophys. Acta (BBA) Biomembr. 2018, 1860, 2320–2328. [Google Scholar] [CrossRef]
- Diril, M.K.; Ratnacaram, C.K.; Padmakumar, V.C.; Du, T.; Wasser, M.; Coppola, V.; Tessarollo, L.; Kaldis, P. Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proc. Natl. Acad. Sci. USA 2012, 109, 3826. [Google Scholar] [CrossRef]
- Zhang, L.; Tao, X.; Fu, Q.; Ge, C.; Li, R.; Li, Z.; Zhu, Y.; Tian, H.; Li, Q.; Liu, M.; et al. Curcumin inhibits cell proliferation and migration in NSCLC through a synergistic effect on the TLR4/MyD88 and EGFR pathways. Oncol. Rep. 2019, 42, 1843–1855. [Google Scholar] [CrossRef] [PubMed]
- Debata, P.R.; Begum, S.; Mata, A.; Genzer, O.; Kleiner, M.J.; Banerjee, P.; Castellanos, M.R. Curcumin potentiates the ability of sunitinib to eliminate the VHL-lacking renal cancer cells 786-O: Rapid inhibition of Rb phosphorylation as a preamble to cyclin D1 inhibition. Anti Cancer Agents Med. Chem. (Former Curr. Med. Chem. Anti Cancer Agents) 2013, 13, 1508–1513. [Google Scholar] [CrossRef] [PubMed]
- Sa, G.; Das, T. Anti cancer effects of curcumin: Cycle of life and death. Cell Div. 2008, 3, 14. [Google Scholar] [CrossRef] [PubMed]
- Milacic, V.; Banerjee, S.; Landis-Piwowar, K.R.; Sarkar, F.H.; Majumdar, A.P.N.; Dou, Q.P. Curcumin inhibits the proteasome activity in human colon cancer cells in vitro and in vivo. Cancer Res. 2008, 68, 7283–7292. [Google Scholar] [CrossRef]
- Voutsadakis, I.A. Proteasome expression and activity in cancer and cancer stem cells. Tumour. Biol. 2017, 39, 1010428317692248. [Google Scholar] [CrossRef]
- Soave, C.L.; Guerin, T.; Liu, J.; Dou, Q.P. Targeting the ubiquitin-proteasome system for cancer treatment: Discovering novel inhibitors from nature and drug repurposing. Cancer Metastasis Rev. 2017, 36, 717–736. [Google Scholar] [CrossRef]
- Khan, A.Q.; Siveen, K.S.; Prabhu, K.S.; Kuttikrishnan, S.; Akhtar, S.; Shaar, A.; Raza, A.; Mraiche, F.; Dermime, S.; Uddin, S. Curcumin-Mediated Degradation of S-Phase Kinase Protein 2 Induces Cytotoxic Effects in Human Papillomavirus-Positive and Negative Squamous Carcinoma Cells. Front. Oncol. 2018, 8, 399. [Google Scholar] [CrossRef]
- Banerjee, S.; Ji, C.; Mayfield, J.E.; Goel, A.; Xiao, J.; Dixon, J.E.; Guo, X. Ancient drug curcumin impedes 26S proteasome activity by direct inhibition of dual-specificity tyrosine-regulated kinase 2. Proc. Natl. Acad. Sci. USA 2018, 115, 8155–8160. [Google Scholar] [CrossRef]
- Si, X.; Wang, Y.; Wong, J.; Zhang, J.; McManus, B.M.; Luo, H. Dysregulation of the ubiquitin-proteasome system by curcumin suppresses coxsackievirus B3 replication. J. Virol. 2007, 81, 3142–3150. [Google Scholar] [CrossRef]
- Helson, L. Curcumin (diferuloylmethane) delivery methods: A review. Biofactors 2013, 39, 21–26. [Google Scholar] [CrossRef]
- Yoon, M.J.; Kang, Y.J.; Lee, J.A.; Kim, I.Y.; Kim, M.A.; Lee, Y.S.; Park, J.H.; Lee, B.Y.; Kim, I.A.; Kim, H.S.; et al. Stronger proteasomal inhibition and higher CHOP induction are responsible for more effective induction of paraptosis by dimethoxycurcumin than curcumin. Cell Death Dis. 2014, 5, e1112. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, B.; Ghildiyal, A.; Sahabjada, A.; Singh, S.; Arshad, M.; Mahdi, A.A.; Tiwari, S. Antiproliferative and Apoptotic Effect of Curcumin and TRAIL (TNF Related Apoptosis inducing Ligand) in Chronic Myeloid Leukaemic Cells. J. Clin. Diagn. Res. JCDR 2016, 10, XC01–XC05. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.; Zhang, X.; Xue, W.; Zhao, S.; Zhang, X.; Pei, J. Curcumin Induced Human Gastric Cancer BGC-823 Cells Apoptosis by ROS-Mediated ASK1-MKK4-JNK Stress Signaling Pathway. Int. J. Mol. Sci. 2014, 15, 15754–15765. [Google Scholar] [CrossRef]
- Feng, C.; Xia, Y.; Zou, P.; Shen, M.; Hu, J.; Ying, S.; Pan, J.; Liu, Z.; Dai, X.; Zhuge, W.; et al. Curcumin analog L48H37 induces apoptosis through ROS-mediated endoplasmic reticulum stress and STAT3 pathways in human lung cancer cells. Mol. Carcinog. 2017, 56, 1765–1777. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.Z.; Li, A.F.; Sun, Y.H.; Sun, G.C. A novel synthetic curcumin derivative MHMM-41 induces ROS-mediated apoptosis and migration blocking of human lung cancer cells A549. Biomed. Pharmacother. 2018, 103, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Yang, Y.; Liu, W.; Li, Z. T59, a New Compound Reconstructed from Curcumin, Induces Cell Apoptosis through Reactive Oxygen Species Activation in Human Lung Cancer Cells. Molecules 2018, 23, 1251. [Google Scholar] [CrossRef] [PubMed]
- Kalashnikova, I.; Mazar, J. Nanoparticle delivery of curcumin induces cellular hypoxia and ROS-mediated apoptosis via modulation of Bcl-2/Bax in human neuroblastoma. Nanoscale 2017, 9, 10375–10387. [Google Scholar] [CrossRef]
- Priyadarsini, K.I. Chemical and structural features influencing the biological activity of curcumin. Curr. Pharm. Des. 2013, 19, 2093–2100. [Google Scholar]
- Priyadarsini, K.I. The chemistry of curcumin: From extraction to therapeutic agent. Molecules 2014, 19, 20091–20112. [Google Scholar] [CrossRef]
- Wondrak, G.T. Redox-directed cancer therapeutics: Molecular mechanisms and opportunities. Antioxid. Redox Signal. 2009, 11, 3013–3069. [Google Scholar] [CrossRef]
- Fang, J.; Lu, J.; Holmgren, A. Thioredoxin reductase is irreversibly modified by curcumin a novel molecular mechanism for its anticancer activity. J. Biol. Chem. 2005, 280, 25284–25290. [Google Scholar] [CrossRef] [PubMed]
- Dufour, F.; Rattier, T.; Constantinescu, A.A.; Zischler, L.; Morle, A.; Ben Mabrouk, H.; Humblin, E.; Jacquemin, G.; Szegezdi, E.; Delacote, F.; et al. TRAIL receptor gene editing unveils TRAIL-R1 as a master player of apoptosis induced by TRAIL and ER stress. Oncotarget 2016, 8, 9974–9985. [Google Scholar] [CrossRef] [PubMed]
- Prasad, S.; Yadav, V.R.; Ravindran, J.; Aggarwal, B.B. ROS and CHOP are Critical for Dibenzylideneacetone to Sensitize Tumor Cells to TRAIL Through Induction of Death Receptors and Downregulation of Cell Survival Proteins. Cancer Res. 2011, 71, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Sung, B.; Prasad, S.; Ravindran, J.; Yadav, V.R.; Aggarwal, B.B. Capsazepine, a TRPV1 antagonist, sensitizes colorectal cancer cells to apoptosis by TRAIL through ROS-JNK-CHOP-mediated upregulation of death receptors. Free Radic. Biol. Med. 2012, 53, 1977–1987. [Google Scholar] [CrossRef]
- Tucker-Kellogg, L.; Shi, Y.; White, J.K.; Pervaiz, S. Reactive oxygen species (ROS) and sensitization to TRAIL-induced apoptosis, in Bayesian network modelling of HeLa cell response to LY303511. Biochem. Pharmacol. 2012, 84, 1307–1317. [Google Scholar] [CrossRef]
- Wang, X.; Thomas, B.; Sachdeva, R.; Arterburn, L.; Frye, L.; Hatcher, P.G.; Cornwell, D.G.; Ma, J. Mechanism of arylating quinone toxicity involving Michael adduct formation and induction of endoplasmic reticulum stress. Proc. Natl. Acad. Sci. USA 2006, 103, 3604–3609. [Google Scholar] [CrossRef]
- Li, Y.; Guo, Y.; Tang, J.; Jiang, J.; Chen, Z. New insights into the roles of CHOP-induced apoptosis in ER stress. Acta Biochim. Biophys. Sin. 2014, 46, 629–640. [Google Scholar] [CrossRef]
- Deng, L.; Gao, X.; Liu, B.; He, X.; Xu, J.; Qiang, J.; Wu, Q.; Liu, S. NMT1 inhibition modulates breast cancer progression through stress-triggered JNK pathway. Cell Death Dis. 2018, 9, 1143. [Google Scholar] [CrossRef]
- Xin, H.; Deng, Y.; Cao, J. Proviral insertion in murine lymphomas 2 promotes stomach cancer progression by regulating apoptosis via reactive oxygen species-triggered endoplasmic reticulum stress. Biochem. Biophys. Res. Commun. 2018, 506, 145–152. [Google Scholar] [CrossRef]
- Yang, Y.; Yuan, P.; Wei, X.; Fu, C.; Li, J.; Wang, W.; Wang, X.; Li, Y.; Li, J. Cultivated and wild Pleurotus ferulae ethanol extracts inhibit hepatocellular carcinoma cell growth via inducing endoplasmic reticulum stress- and mitochondria-dependent apoptosis. Sci. Rep. 2018, 8, 13984. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, X.; Liu, H.; Li, Z.; Chen, F.; Wang, H.; Zheng, Z.; Wang, J. TNF-alpha enhances apoptosis by promoting CHOP expression in nucleus pulposus cells: Role of the MAPK and NF-kappaB pathways. J. Orthop. Res. 2019, 37, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Xu, A.; Wu, X.; Zhang, Y.; Guo, Y.; Guo, F.; Pan, Z.; Kong, L. Japanese encephalitis virus induces apoptosis by the IRE1/JNK pathway of ER stress response in BHK-21 cells. Arch. Virol. 2016, 161, 699–703. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Lovera, C.; Vazquez-Rios, A.J.; Guerra-Varela, J.; Sanchez, L.; de la Fuente, M. The Potential of Zebrafish as a Model Organism for Improving the Translation of Genetic Anticancer Nanomedicines. Genes 2017, 8, 349. [Google Scholar] [CrossRef] [PubMed]
- Tabassum, N.; Tai, H.; Jung, D.W.; Williams, D.R. Fishing for nature’s hits: Establishment of the zebrafish as a model for screening antidiabetic natural products. Evid. Based Complement. Altern. Med. 2015, 2015, 287847. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Obaidi, I.; Cassidy, H.; Ibáñez Gaspar, V.; McCaul, J.; Higgins, M.; Halász, M.; Reynolds, A.L.; Kennedy, B.N.; McMorrow, T. Curcumin Sensitizes Kidney Cancer Cells to TRAIL-Induced Apoptosis via ROS Mediated Activation of JNK-CHOP Pathway and Upregulation of DR4. Biology 2020, 9, 92. https://doi.org/10.3390/biology9050092
Obaidi I, Cassidy H, Ibáñez Gaspar V, McCaul J, Higgins M, Halász M, Reynolds AL, Kennedy BN, McMorrow T. Curcumin Sensitizes Kidney Cancer Cells to TRAIL-Induced Apoptosis via ROS Mediated Activation of JNK-CHOP Pathway and Upregulation of DR4. Biology. 2020; 9(5):92. https://doi.org/10.3390/biology9050092
Chicago/Turabian StyleObaidi, Ismael, Hilary Cassidy, Verónica Ibáñez Gaspar, Jasmin McCaul, Michael Higgins, Melinda Halász, Alison L. Reynolds, Breandan N. Kennedy, and Tara McMorrow. 2020. "Curcumin Sensitizes Kidney Cancer Cells to TRAIL-Induced Apoptosis via ROS Mediated Activation of JNK-CHOP Pathway and Upregulation of DR4" Biology 9, no. 5: 92. https://doi.org/10.3390/biology9050092
APA StyleObaidi, I., Cassidy, H., Ibáñez Gaspar, V., McCaul, J., Higgins, M., Halász, M., Reynolds, A. L., Kennedy, B. N., & McMorrow, T. (2020). Curcumin Sensitizes Kidney Cancer Cells to TRAIL-Induced Apoptosis via ROS Mediated Activation of JNK-CHOP Pathway and Upregulation of DR4. Biology, 9(5), 92. https://doi.org/10.3390/biology9050092





