Comparative Analysis of Porcine Follicular Fluid Proteomes of Small and Large Ovarian Follicles
Abstract
1. Introduction
2. Materials and Methods
2.1. Follicle Categorization, Follicular Fluids Aspirations, Estradiol (E2), and Protein Assays
2.2. Proteomic Analyses
2.3. Database Search, Protein Function, and Pathway Identification
2.4. Statistical Analyses
3. Results
3.1. Estradiol (E2) and Protein Concentration
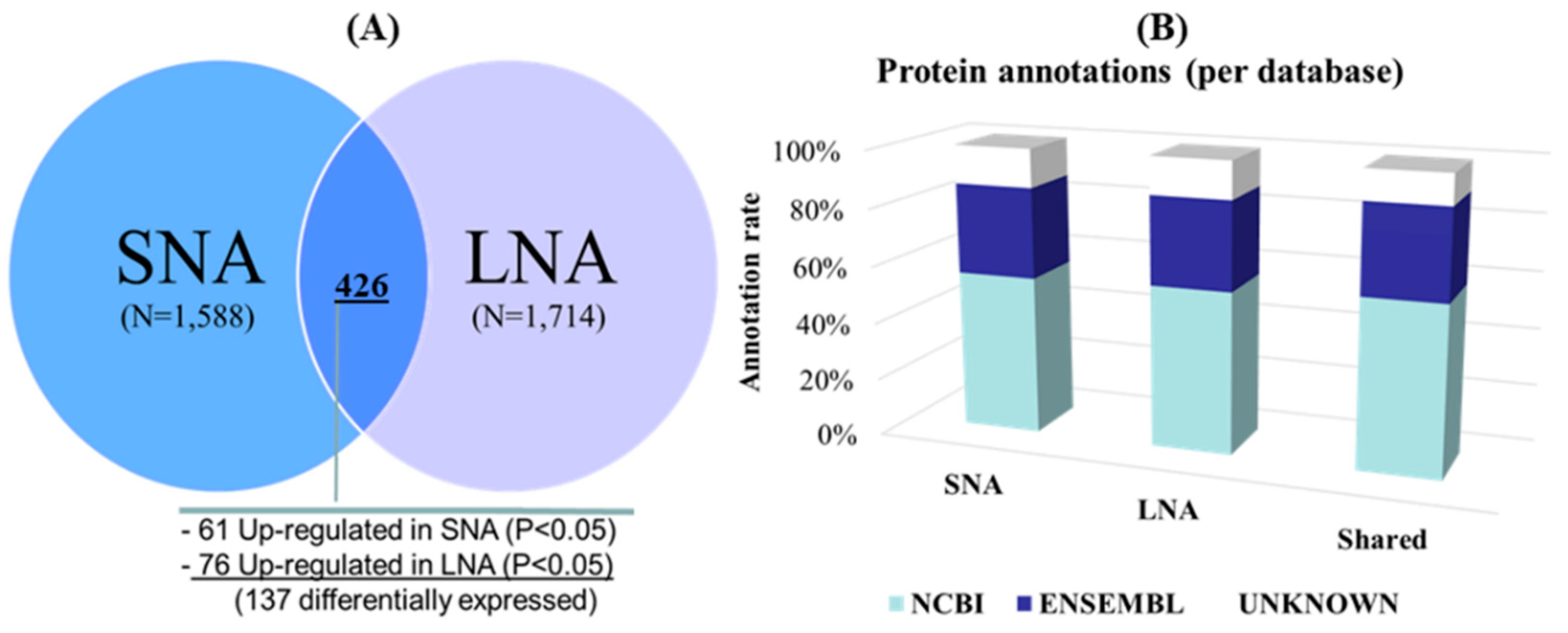
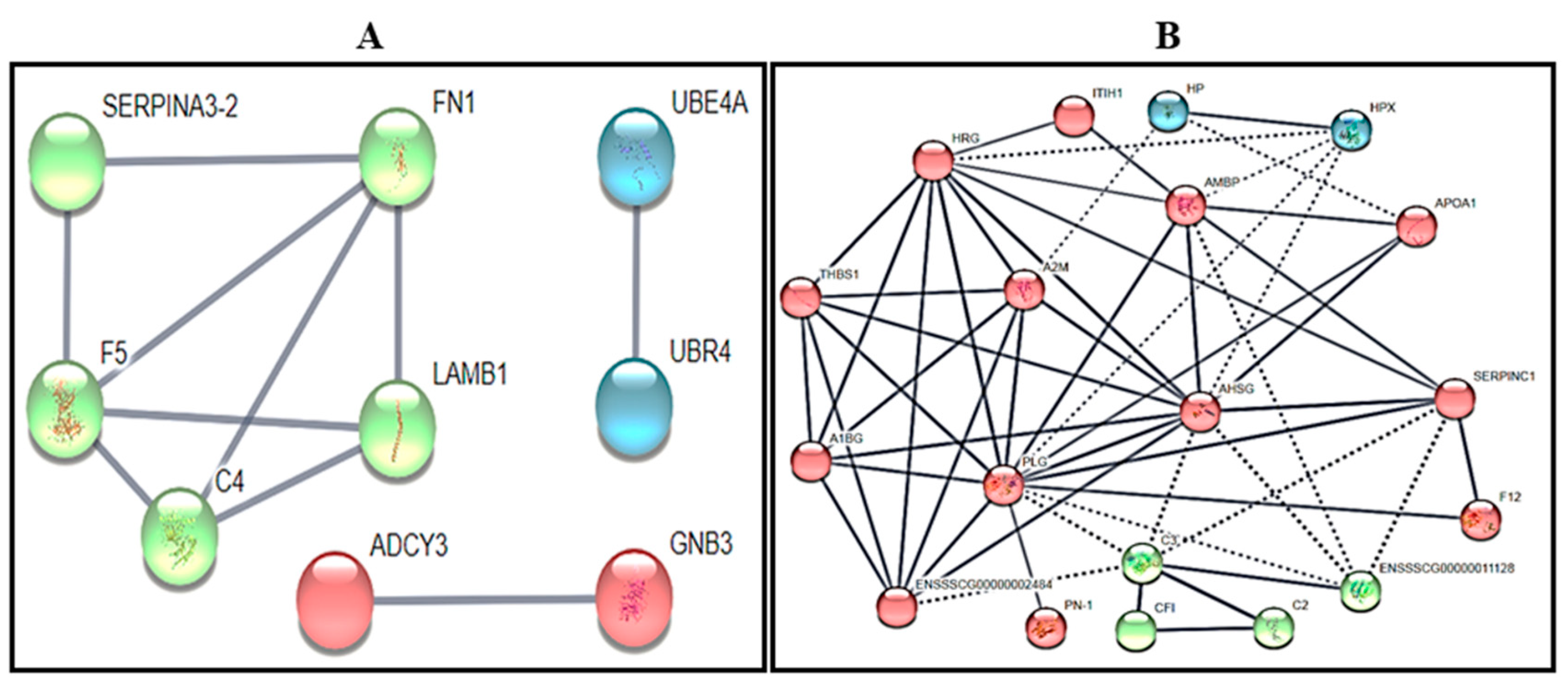
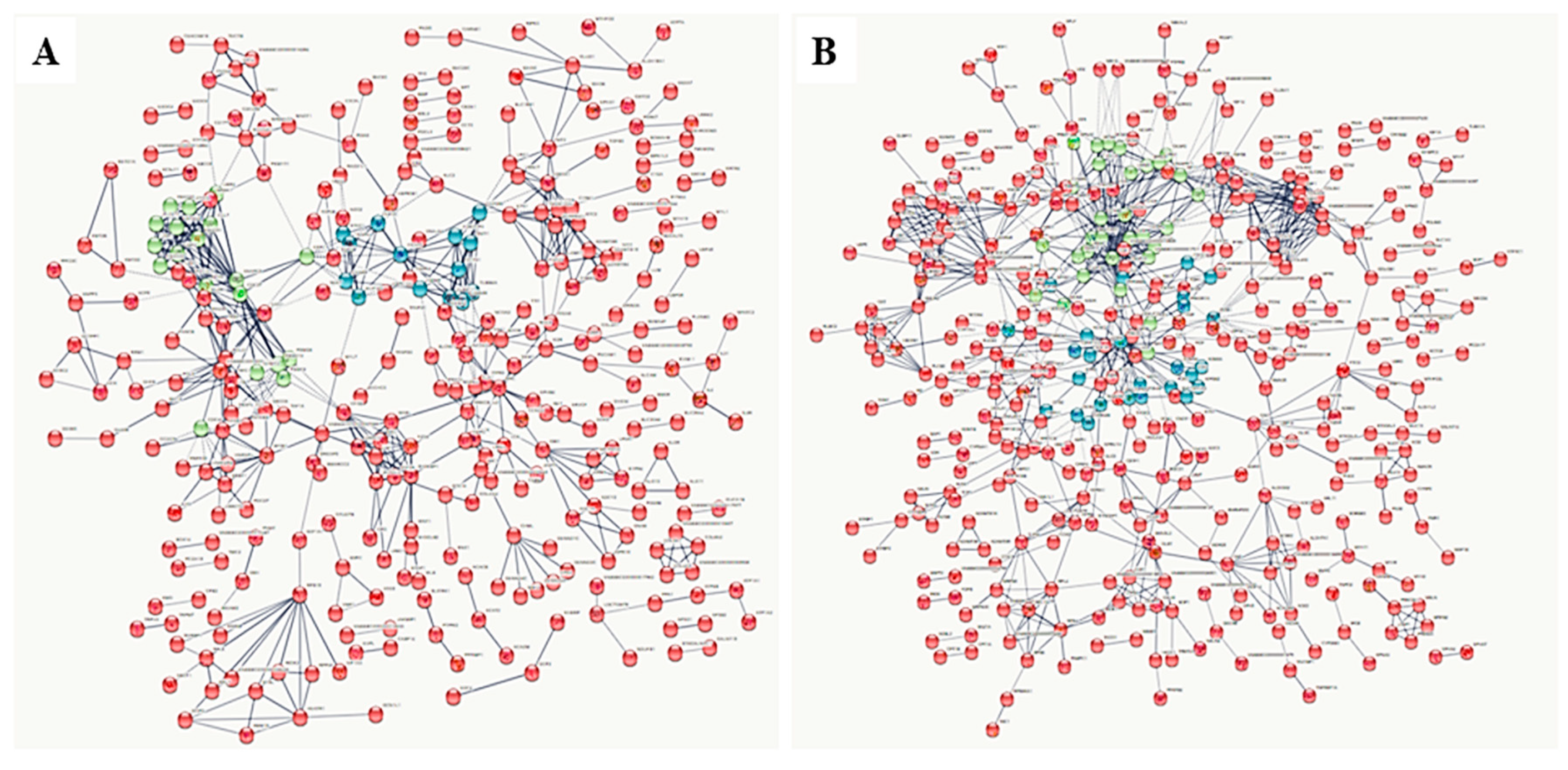
3.2. Gel-Free (Shotgun) Proteome Identification and Functional Analyses
3.3. Detection of Male-Known Proteins
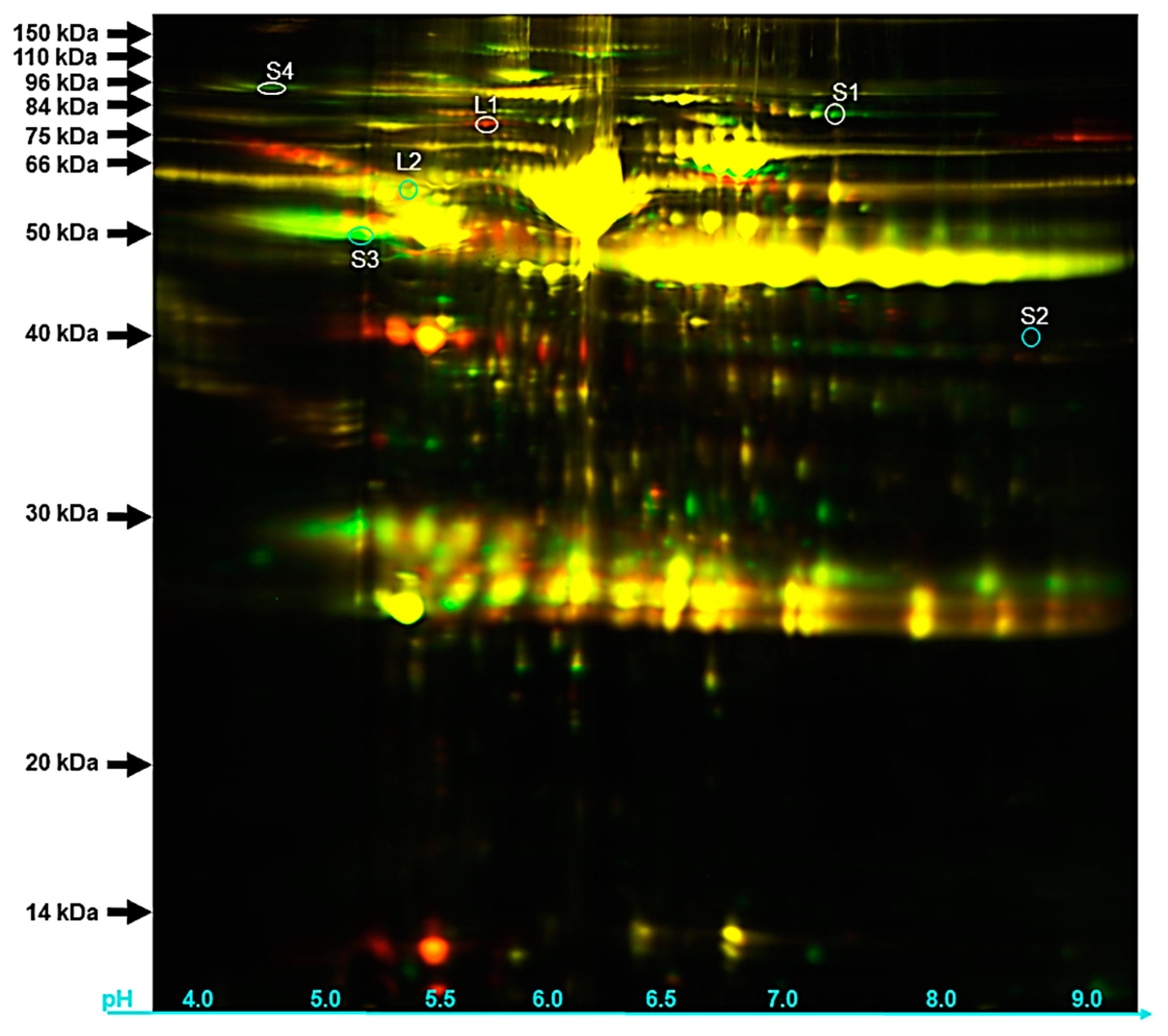
3.4. Protein Identification by Gel Based (2 Dimension-Differential in-gel Electrophoresis or 2D-DIGE)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AHSG | Alpha-2-HS-glycoprotein |
| A2M | Alpha-2-macroglobulin |
| BP | Biological process |
| CC | Cellular component |
| HRG | Histidine-rich glycoprotein |
| E2 | Estradiol |
| FF | Follicular fluid |
| GO | Gene ontology |
| HRG | Histidine-rich glycoprotein |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LFF | Large follicular fluid |
| LNA | Large non-atretic |
| MF | Molecular function |
| NA | Non-atretic |
| PLG | Plasminogen |
| PPI | Protein-to-protein interactions |
| SFF | Small follicular fluid |
| SNA | Small non-atretic |
| 2D-DIGE | Two Dimension-Differential In Gel Electrophoresis |
References
- Fortune, J. Ovarian follicular growth and development in mammals. Biol. Reprod. 1994, 50, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Algriany, O.; Bevers, M.; Schoevers, E.; Colenbrander, B.; Dieleman, S. Follicle size-dependent effects of sow follicular fluid on in vitro cumulus expansion, nuclear maturation and blastocyst formation of sow cumulus oocytes complexes. Theriogenology 2004, 62, 1483–1497. [Google Scholar] [CrossRef] [PubMed]
- Ferrazza, R.D.A.; Garcia, H.D.M.; Schmidt, E.M.D.S.; Mihm Carmichael, M.; Souza, F.F.D.; Burchmore, R.; Sartori, R.; Eckersall, P.D.; Ferreira, J.C.P. Quantitative proteomic profiling of bovine follicular fluid during follicle development. Biol. Reprod. 2017, 97, 835–849. [Google Scholar] [CrossRef] [PubMed]
- O’callaghan, D.; Yaakub, H.; Hyttel, P.; Spicer, L.; Boland, M. Effect of nutrition and superovulation on oocyte morphology, follicular fluid composition and systemic hormone concentrations in ewes. J. Reprod. Fertil. 2000, 118, 303–314. [Google Scholar] [CrossRef]
- Nandi, S.; Kumar, V.G.; Manjunatha, B.; Gupta, P. Biochemical composition of ovine follicular fluid in relation to follicle size. Dev. Growth Differ. 2007, 49, 61–66. [Google Scholar] [CrossRef]
- Fu, Q.; Huang, Y.; Wang, Z.; Chen, F.; Huang, D.; Lu, Y.; Liang, X.; Zhang, M. Proteome profile and quantitative proteomic analysis of buffalo (Bubalusbubalis) follicular fluid during follicle development. Int. J. Mol. Sci. 2016, 17, 618. [Google Scholar] [CrossRef]
- Paula, A.J.; Lobo, M.; Monteiro-Moreira, A.; Moreira, R.; Melo, C.; Souza-Fabjan, J.; Araújo, A.; Melo, L.; Teixeira, D.; Moura, A. Proteomic analysis of follicular fluid from tropically-adapted goats. Anim. Reprod. Sci. 2018, 188, 35–44. [Google Scholar] [CrossRef]
- von Wolff, M.; Kollmann, Z.; Flück, C.; Stute, P.; Marti, U.; Weiss, B.; Bersinger, N.A. Gonadotrophin stimulation for in vitro fertilization significantly alters the hormone milieu in follicular fluid: A comparative study between natural cycle IVF and conventional IVF. Hum. Reprod. 2014, 29, 1049–1057. [Google Scholar] [CrossRef]
- Otoi, T.; Yamamoto, K.; Koyama, N.; Tachikawa, S.; Suzuki, T. Bovine oocyte diameter in relation to developmental competence. Theriogenology 1997, 48, 769–774. [Google Scholar] [CrossRef]
- Bagg, M.A.; Nottle, M.B.; Armstrong, D.T.; Grupen, C.G. Relationship between follicle size and oocyte developmental competence in prepubertal and adult pigs. Reprod. Fertil. Dev. 2007, 19, 797–803. [Google Scholar] [CrossRef]
- Motlik, J.; Crozet, N.; Fulka, J. Meiotic competence in vitro of pig oocytes isolated from early antral follicles. Reproduction 1984, 72, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Eppig, J.J. Coordination of nuclear and cytoplasmic oocyte maturation in eutherian mammals. Reprod. Fertil. Dev. 1996, 8, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Fowler, K.E.; Mandawala, A.A.; Griffin, D.K.; Walling, G.A.; Harvey, S.C. The production of pig preimplantation embryos in vitro: Current progress and future prospects. Reprod. Biol. 2018, 18, 203–211. [Google Scholar] [CrossRef]
- Grupen, C.G. The evolution of porcine embryo in vitro production. Theriogenology 2014, 81, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Dumesic, D.A.; Meldrum, D.R.; Katz-Jaffe, M.G.; Krisher, R.L.; Schoolcraft, W.B. Oocyte environment: Follicular fluid and cumulus cells are critical for oocyte health. Fertil. Steril. 2015, 103, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Zuccotti, M.; Merico, V.; Cecconi, S.; Redi, C.A.; Garagna, S. What does it take to make a developmentally competent mammalian egg? Hum. Reprod. Update 2011, 17, 525–540. [Google Scholar] [CrossRef]
- Conti, M.; Franciosi, F. Acquisition of oocyte competence to develop as an embryo: Integrated nuclear and cytoplasmic events. Hum. Reprod. Update 2018, 24, 245–266. [Google Scholar] [CrossRef]
- Fair, T.; Hyttel, P.; Greve, T. Bovine oocyte diameter in relation to maturational competence and transcriptional activity. Mol. Reprod. Dev. 1995, 42, 437–442. [Google Scholar] [CrossRef]
- Crozet, N.; Dahirel, M.; Gall, L. Meiotic competence of in vitro grown goat oocytes. Reproduction 2000, 118, 367–373. [Google Scholar] [CrossRef][Green Version]
- Marchal, R.; Vigneron, C.; Perreau, C.; Bali-Papp, A.; Mermillod, P. Effect of follicular size on meiotic and developmental competence of porcine oocytes. Theriogenology 2002, 57, 1523–1532. [Google Scholar] [CrossRef]
- Motlík, J.; Fulka, J. Factors affecting meiotic competence in pig oocytes. Theriogenology 1986, 25, 87–96. [Google Scholar] [CrossRef]
- Barberi, M.; Ermini, B.; Morelli, M.B.; Ermini, M.; Cecconi, S.; Canipari, R. Follicular fluid hormonal profile and cumulus cell gene expression in controlled ovarian hyperstimulation with recombinant FSH: Effects of recombinant LH administration. J. Assist. Reprod. Genet. 2012, 29, 1381–1391. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Marchal, R.; Feugang, J.; Perreau, C.; Venturi, E.; Terqui, M.; Mermillod, P. Meiotic and developmental competence of prepubertal and adult swine oocytes. Theriogenology 2001, 56, 17–29. [Google Scholar] [CrossRef]
- Vatzias, G.; Hagen, D.R. Effects of porcine follicular fluid and oviduct-conditioned media on maturation and fertilization of porcine oocytes in vitro. Biol. Reprod. 1999, 60, 42–48. [Google Scholar] [CrossRef]
- Benkhalifa, M.; Madkour, A.; Louanjli, N.; Bouamoud, N.; Saadani, B.; Kaarouch, I.; Chahine, H.; Sefrioui, O.; Merviel, P.; Copin, H. From global proteome profiling to single targeted molecules of follicular fluid and oocyte: Contribution to embryo development and IVF outcome. Expert Rev. Proteom. 2015, 12, 407–423. [Google Scholar] [CrossRef]
- Feugang, J.M.; Liao, S.F.; Willard, S.T.; Ryan, P.L. In-depth proteomic analysis of boar spermatozoa through shotgun and gel-based methods. BMC Genom. 2018, 19, 62. [Google Scholar] [CrossRef]
- Angelucci, S.; Ciavardelli, D.; Di Giuseppe, F.; Eleuterio, E.; Sulpizio, M.; Tiboni, G.M.; Giampietro, F.; Palumbo, P.; Di Ilio, C. Proteome analysis of human follicular fluid. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2006, 1764, 1775–1785. [Google Scholar] [CrossRef]
- Bijttebier, J.; Tilleman, K.; Dhaenens, M.; Deforce, D.; Van Soom, A.; Maes, D. Comparative proteome analysis of porcine follicular fluid and serum reveals that excessive α2-macroglobulin in serum hampers successful expansion of cumulus-oocyte complexes. Proteomics 2009, 9, 4554–4565. [Google Scholar] [CrossRef]
- Sun, Y.L.; Ping, Z.G.; Li, C.J.; Sun, Y.F.; Yi, K.L.; Chen, L.; Li, X.Y.; Wang, X.L.; Zhou, X. Comparative Proteomic Analysis of Follicular Fluids from Normal and Cystic Follicles in Sows. Reprod. Domest. Anim. 2011, 46, 889–895. [Google Scholar] [CrossRef]
- Fahiminiya, S.; Labas, V.; Roche, S.; Dacheux, J.-L.; Gérard, N. Proteomic analysis of mare follicular fluid during late follicle development. Proteome Sci. 2011, 9, 54. [Google Scholar] [CrossRef]
- Dutra, G.; Ishak, G.; Pechanova, O.; Pechan, T.; Peterson, D.; Jacob, J.; Willard, S.; Ryan, P.; Gastal, E.; Feugang, J. Seasonal variation in equine follicular fluid proteome. Reprod. Biol. Endocrinol. 2019, 17, 29. [Google Scholar] [CrossRef] [PubMed]
- Nagai, T. In vitro maturation and fertilization of pig oocytes. Anim. Reprod. Sci. 1996, 42, 153–163. [Google Scholar] [CrossRef]
- Bing, Y.; Nagai, T.; Rodriguez-Martinez, H. Effects of cysteamine, FSH and estradiol-17β on in vitro maturation of porcine oocytes. Theriogenology 2001, 55, 867–876. [Google Scholar] [CrossRef]
- Paes, V.M.; Liao, S.F.; Figueiredo, J.R.; Willard, S.T.; Ryan, P.L.; Feugang, J.M. Proteome changes of porcine follicular fluid during follicle development. J. Anim. Sci. Biotechnol. 2019, 10, 94. [Google Scholar] [CrossRef]
- Murdoch, W. Inhibition by oestradiol of oxidative stress-induced apoptosis in pig ovarian tissues. Reproduction 1998, 114, 127–130. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lundgren, D.H.; Hwang, S.-I.; Wu, L.; Han, D.K. Role of spectral counting in quantitative proteomics. Expert Rev. Proteom. 2010, 7, 39–53. [Google Scholar] [CrossRef]
- Rodgers, R.J.; Irving-Rodgers, H.F. Formation of the ovarian follicular antrum and follicular fluid. Biol. Reprod. 2010, 82, 1021–1029. [Google Scholar] [CrossRef]
- Matsuda, F.; Inoue, N.; Manabe, N.; Ohkura, S. Follicular growth and atresia in mammalian ovaries: Regulation by survival and death of granulosa cells. J. Reprod. Dev. 2012, 58, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Tu, D.; Yuan, L.-Y.; Yi, J.-E.; Tian, Y. T-2 toxin regulates steroid hormone secretion of rat ovarian granulosa cells through cAMP-PKA pathway. Toxicol. Lett. 2015, 232, 573–579. [Google Scholar] [CrossRef]
- Guzel, Y.; Bildik, G.; Oktem, O. Sphingosine-1-phosphate protects human ovarian follicles from apoptosis in vitro. Eur. J. Obstet. Gynecol. Reprod. Biol. 2018, 222, 19–24. [Google Scholar] [CrossRef]
- Xu, J.; Lawson, M.S.; Xu, F.; Du, Y.; Tkachenko, O.Y.; Bishop, C.V.; Pejovic-Nezhat, L.; Seifer, D.B.; Hennebold, J.D. Vitamin D3 Regulates Follicular Development and Intrafollicular Vitamin D Biosynthesis and Signaling in the Primate Ovary. Front. Physiol. 2018, 9, 1600. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, A.W.; Anisowicz, A. Cation and protein composition of ovarian follicular fluid of the pig: Relation to follicle size. Biol. Reprod. 1974, 11, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Andersen, M.M.; Krøll, J.; Byskov, A.; Faber, M. Protein composition in the fluid of individual bovine follicles. Reproduction 1976, 48, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Shiny, J.; Deshmukh, B.; Dhaware, S.; Borkar, S. Follicular fluid protein profile in buffalo. Vet. Sci. Res. J. 2015, 6, 71–79. [Google Scholar]
- Lippolis, J.D.; Reinhardt, T.A. Utility, limitations, and promise of proteomics in animal science. Vet. Immunol. Immunopathol. 2010, 138, 241–251. [Google Scholar] [CrossRef]
- Iwase, A.; Kobayashi, H.; Goto, M.; Nakahara, T.; Nakamura, T.; Kondo, M.; Nagatomo, Y.; Kotani, T.; Kikkawa, F. A proteomic analysis of human follicular fluid: Comparison between fertilized oocytes and non-fertilized oocytes in the same patient. J. Assist. Reprod. Genet. 2013, 30, 1231–1238. [Google Scholar]
- Lu, C.-H.; Lee, R.K.-K.; Hwu, Y.-M.; Lin, M.-H.; Yeh, L.-Y.; Chen, Y.-J.; Lin, S.-P.; Li, S.-H. Involvement of the serine protease inhibitor, SERPINE2, and the urokinase plasminogen activator in cumulus expansion and oocyte maturation. PLoS ONE 2013, 8, e74602. [Google Scholar] [CrossRef]
- Chen, L.; Mao, S.; McLean, L.R.; Powers, R.W.; Larsen, W.J. Proteins of the inter-alpha-trypsin inhibitor family stabilize the cumulus extracellular matrix through their direct binding with hyaluronic acid. J. Biol. Chem. 1994, 269, 28282–28287. [Google Scholar]
- Zhang, M.; Xia, G.; Zhou, B.; Wang, C. Gonadotropin-controlled mammal oocyte meiotic resumption. Front. Biosci. 2007, 12, 282–296. [Google Scholar] [CrossRef][Green Version]
- Zhang, H.; Lin, H.-Y.; Yang, Q.; Wang, H.-X.; Chai, K.X.; Chen, L.-M.; Zhu, C. Expression of prostasin serine protease and protease nexin-1 (PN-1) in rhesus monkey ovary during menstrual cycle and early pregnancy. J. Histochem. Cytochem. 2007, 55, 1237–1244. [Google Scholar] [CrossRef]
- Fusi, F.M.; Bernocchi, N.; Ferrari, A.; Bronson, R. Integrins and adhesion molecules: Is vitronectin the velcro that binds the gametes together? MHR Basic Sci. Reprod. Med. 1996, 2, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Boissonnas, C.C.; Montjean, D.; Lesaffre, C.; Auer, J.; Vaiman, D.; Wolf, J.P.; Ziyyat, A. Role of sperm αvβ3 integrin in mouse fertilization. Dev. Dyn. An Off. Publ. Am. Assoc. Anat. 2010, 239, 773–783. [Google Scholar] [CrossRef]
- Zhang, L.; Bell, B.A.; Li, Y.; Caspi, R.R.; Lin, F. Complement component C4 regulates the development of experimental autoimmune uveitis through a T cell-intrinsic mechanism. Front. Immunol. 2017, 8, 1116. [Google Scholar] [CrossRef] [PubMed]
- Wissing, M.; Kristensen, S.; Andersen, C.; Mikkelsen, A.; Høst, T.; Borup, R.; Grøndahl, M. Identification of new ovulation-related genes in humans by comparing the transcriptome of granulosa cells before and after ovulation triggering in the same controlled ovarian stimulation cycle. Hum. Reprod. 2014, 29, 997–1010. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, A.J.; Kawamura, K.; Cheng, Y.; Fauser, B.C. Intraovarian control of early folliculogenesis. Endocr. Rev. 2015, 36, 1–24. [Google Scholar] [CrossRef] [PubMed]
- De Agostini, A.I.; Dong, J.-C.; de Vantéry Arrighi, C.; Ramus, M.-A.; Dentand-Quadri, I.; Thalmann, S.; Ventura, P.; Ibecheole, V.; Monge, F.; Fischer, A.-M. Human follicular fluid heparan sulfate contains abundant 3-O-sulfated chains with anticoagulant activity. J. Biol. Chem. 2008, 283, 28115–28124. [Google Scholar] [CrossRef]
- Jessen, T.; Odum, L. Role of tumour necrosis factor stimulated gene 6 (TSG-6) in the coupling of inter-alpha-trypsin inhibitor to hyaluronan in human follicular fluid. Reproduction 2003, 125, 27–31. [Google Scholar] [CrossRef]
- Appeltant, R.; Beek, J.; Maes, D.; Bijttebier, J.; Van Steendam, K.; Nauwynck, H.; Van Soom, A. Hampered cumulus expansion of porcine cumulus-oocyte complexes by excessive presence of alpha2-macroglobulin is likely mediated via inhibition of zinc-dependent metalloproteases. Anim. Sci. J. 2017, 88, 1279–1290. [Google Scholar] [CrossRef]
- Kalab, P.; Schultz, R.M.; Kopf, G.S. Modifications of the mouse zona pellucida during oocyte maturation: Inhibitory effects of follicular fluid, fetuin, and α2HS-glycoprotein. Biol. Reprod. 1993, 49, 561–567. [Google Scholar] [CrossRef]
- Tsuchida-Straeten, N.; Ensslen, S.; Schäfer, C.; Wöltje, M.; Denecke, B.; Moser, M.; Gräber, S.; Wakabayashi, S.; Koide, T.; Jahnen-Dechent, W. Enhanced blood coagulation and fibrinolysis in mice lacking histidine-rich glycoprotein (HRG). J. Thromb. Haemost. 2005, 3, 865–872. [Google Scholar] [CrossRef]
- Dixelius, J.; Olsson, A.-K.; Thulin, Å.; Lee, C.; Johansson, I.; Claesson-Welsh, L. Minimal active domain and mechanism of action of the angiogenesis inhibitor histidine-rich glycoprotein. Cancer Res. 2006, 66, 2089–2097. [Google Scholar] [CrossRef] [PubMed]
- Robker, R.L.; Richards, J.S. Hormone-induced proliferation and differentiation of granulosa cells: A coordinated balance of the cell cycle regulators cyclin D2 and p27Kip1. Mol. Endocrinol. 1998, 12, 924–940. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Ding, Y.; Liu, J.; Heng, D.; Xu, K.; Liu, W.; Zhang, C. Nitric oxide–mediated regulation of GLUT by T3 and follicle-stimulating hormone in rat granulosa cells. Endocrinology 2017, 158, 1898–1915. [Google Scholar] [CrossRef] [PubMed]
- Oikonomopoulou, K.; Ricklin, D.; Ward, P.A.; Lambris, J.D. Interactions between coagulation and complement—their role in inflammation. In Seminars in Immunopathology; Springer: Berlin, Germany, 2012; pp. 151–165. [Google Scholar]
- Jesam, C.; Salvatierra, A.M.; Schwartz, J.L.; Croxatto, H.B. Suppression of follicular rupture with meloxicam, a cyclooxygenase-2 inhibitor: Potential for emergency contraception. Hum. Reprod. 2009, 25, 368–373. [Google Scholar] [CrossRef][Green Version]
- Zamah, A.M.; Hassis, M.E.; Albertolle, M.E.; Williams, K.E. Proteomic analysis of human follicular fluid from fertile women. Clin. Proteom. 2015, 12, 5. [Google Scholar] [CrossRef]
- La Salle, S.; Palmer, K.; O’Brien, M.; Schimenti, J.C.; Eppig, J.; Handel, M.A. Spata22, a novel vertebrate-specific gene, is required for meiotic progress in mouse germ cells. Biol. Reprod. 2012, 86, 12–41. [Google Scholar] [CrossRef]
- Kato, K.; Satouh, Y.; Nishimasu, H.; Kurabayashi, A.; Morita, J.; Fujihara, Y.; Oji, A.; Ishitani, R.; Ikawa, M.; Nureki, O. Structural and functional insights into IZUMO1 recognition by JUNO in mammalian fertilization. Nat. Commun. 2016, 7, 1–9. [Google Scholar] [CrossRef]
- Gaikwad, A.S.; Anderson, A.L.; Merriner, D.J.; O’Connor, A.E.; Houston, B.J.; Aitken, R.J.; O’Bryan, M.K.; Nixon, B. GLIPR1L1 is an IZUMO-binding protein required for optimal fertilization in the mouse. BMC Biol. 2019, 17, 86. [Google Scholar] [CrossRef]
| Classification | Gene Ontology (GO) Term | Observed | Background | False Discovery |
|---|---|---|---|---|
| Category | Identification/Description | Gene (n) | Gene (n) | Rate |
| Cellular component | GO:0005576/Extracellular region | 11 | 352 | 9.20 × 10−9 |
| GO:0005615/Extracellular space | 5 | 185 | 0.00076 | |
| GO:00725615/Blood microparticle | 2 | 5 | 0.00076 | |
| Biological process | GO:0010951/Negative regulation of endopeptidase activity | 5 | 31 | 6.01 × 10−6 |
| Biological process | GO:0043086/Negative regulation of catalytic activity | 6 | 68 | 6.01 × 10−6 |
| GO:0050790/Regulation of catalytic activity | 7 | 161 | 6.01 × 10−6 | |
| GO:0051336/Regulation of hydrolase activity | 6 | 93 | 6.01 × 10−6 | |
| GO:0065007/biological regulation | 11 | 716 | 1.75 × 10−5 | |
| GO:0048519/negative regulation of biological process | 7 | 280 | 8.86 × 10−5 | |
| GO:0006950/response to stress | 6 | 224 | 0.00026 | |
| GO:0050789/regulation of biological process | 9 | 648 | 0.00026 | |
| GO:1901564/organonitrogen compound metabolic process | 7 | 375 | 0.00037 | |
| GO:0043170/macromolecule metabolic process | 7 | 388 | 0.00044 | |
| Molecular function | GO:0030234/Enzyme regulator activity | 7 | 90 | 1.72 × 10−7 |
| Molecular function | GO:0004857/Enzyme inhibitor activity | 6 | 55 | 1.98 × 10−7 |
| Molecular function | GO:0004866/Endopeptidase inhibitor activity | 5 | 30 | 3.96 × 10−7 |
| GO:0004867/Serine-type endopeptidase inhibitor activity | 3 | 16 | 0.0001 | |
| GO:0004252/Serine-type endopeptidase activity | 2 | 21 | 0.0109 | |
| GO:0005215/Transporter activity | 3 | 90 | 0.0109 |
| Reactome Pathway Term | Observed | Background | False Discovery | |
|---|---|---|---|---|
| Identification | Description | Gene (n) | Gene (n) | Rate |
| Up-Regulated Proteins in SNA | ||||
| ssc392499 | Metabolism of proteins | 11 | 1310 | 0.0107 |
| Ssc163685 | Integration of energy metabolism | 3 | 66 | 0.0301 |
| ssc597592 | Post-translational protein modification | 9 | 1076 | 0.0301 |
| ssc8957275 | Post-translational protein phosphorylation | 3 | 74 | 0.0301 |
| ssc381426 | Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) | 3 | 83 | 0.0307 |
| ssc109582 | Hemostasis | 5 | 412 | 0.0356 |
| ssc1474244 | Extracellular matrix organization | 4 | 219 | 0.0356 |
| ssc163359 | Glucagon signaling in metabolic regulation | 2 | 21 | 0.0356 |
| ssc418597 | G alpha (z) signaling events | 2 | 22 | 0.0356 |
| ssc432040 | Vasopressin regulates renal water homeostasis via Aquaporins | 2 | 26 | 0.0356 |
| Specific Proteins to SNA | ||||
| ssc0392499 | Metabolism of proteins | 78 | 1310 | 0.0188 |
| ssc1640170 | Cell cycle | 35 | 461 | 0.0305 |
| ssc5610787 | Hedgehog’off state | 12 | 87 | 0.0305 |
| ssc597592 | Post-translational protein modification | 65 | 1076 | 0.0305 |
| ssc69278 | Cell cycle, mitotic | 31 | 398 | 0.0305 |
| Up-Regulated Proteins in LNA | ||||
| SSC-114608 | Platelet degranulation | 5 | 78 | 0.0002 |
| SSC-140837 | Intrinsic Pathway of Fibrin Clot Formation | 3 | 13 | 0.00032 |
| SSC-109582 | Hemostasis | 7 | 412 | 0.0014 |
| SSC-75205 | Dissolution of Fibrin Clot | 2 | 6 | 0.0032 |
| SSC-140875 | Common Pathway of Fibrin Clot Formation | 2 | 14 | 0.0119 |
| SSC-977606 | Regulation of Complement cascade | 2 | 32 | 0.0481 |
| Sample Type | KEGG Pathway Identification | Observed | Background | False |
|---|---|---|---|---|
| (Dysregulation) | Identification/Description | Gene Count | Gene Count | Discovery Rate |
| SNA-specific | ssc04540/Gap junction | 11 | 73 | 0.0418 |
| ssc04915/Estrogen signaling pathway | 13 | 109 | 0.0418 | |
| Ssc04971/Gastric acid secretion | 10 | 60 | 0.0418 | |
| LNA-specific | ssc04974/Protein digestion and absorption | 13 | 70 | 0.0048 |
| LNA-specific | ssc04510/Focal adhesion | 18 | 163 | 0.0341 |
| ssc05165/Human papillomavirus infection | 24 | 259 | 0.041 | |
| Up-regulated SNA (↑SNA ↓LNA) | ssc04512/ECM-receptor interaction | 3 | 66 | 0.0184 |
| Up-regulated SNA (↑SNA ↓LNA) | ssc04713/Circadian entrainment | 3 | 72 | 0.0184 |
| ssc05165/Human papillomavirus infection | 5 | 259 | 0.0184 | |
| ssc04371/Apelin signaling pathway | 3 | 108 | 0.0358 | |
| ssc05200/Pathways in cancer | 5 | 428 | 0.0358 | |
| ssc04151/PI3K-Akt signaling pathway | 4 | 284 | 0.0439 | |
| Up-regulated LNA (↑LNA ↓SNA) | ssc04610/Complement and coagulation cascades | 7 | 73 | 3.33 × 10−8 |
| Up-regulated LNA (↑LNA ↓SNA) | ssc05150/Staphylococcus aureus infection | 4 | 39 | 8.17 × 10−5 |
| Protein | SNA | LNA | |||
|---|---|---|---|---|---|
| Name | GI | Shotgun | Gel-Based | Shotgun | Gel-Based |
| Complement C4 precursor | 178056221 | ↓ | ↓ | ↑ | ↑ |
| Alpha-1-antichymotrypsin 2 precursor | 47523270 | ↓ | ↓ | ↑ | ↑ |
| Plasminogen precursor | 113205806 | ↑ | ↑ | ↓ | ↓ |
| Nexin-1 precursor | 47523638 | ↑ | ↑ | ↓ | ↓ |
| Vitronectin precursor | 55741847 | ↑ | ↑ | ↓ | ↓ |
| Inter/alpha-trypsin inhibitor heavy chain H2 precursor | 47522678 | ↑ | ↑ | ↓ | ↓ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paes, V.M.; de Figueiredo, J.R.; Ryan, P.L.; Willard, S.T.; Feugang, J.M. Comparative Analysis of Porcine Follicular Fluid Proteomes of Small and Large Ovarian Follicles. Biology 2020, 9, 101. https://doi.org/10.3390/biology9050101
Paes VM, de Figueiredo JR, Ryan PL, Willard ST, Feugang JM. Comparative Analysis of Porcine Follicular Fluid Proteomes of Small and Large Ovarian Follicles. Biology. 2020; 9(5):101. https://doi.org/10.3390/biology9050101
Chicago/Turabian StylePaes, Victor M., José R. de Figueiredo, Peter L. Ryan, Scott T. Willard, and Jean M. Feugang. 2020. "Comparative Analysis of Porcine Follicular Fluid Proteomes of Small and Large Ovarian Follicles" Biology 9, no. 5: 101. https://doi.org/10.3390/biology9050101
APA StylePaes, V. M., de Figueiredo, J. R., Ryan, P. L., Willard, S. T., & Feugang, J. M. (2020). Comparative Analysis of Porcine Follicular Fluid Proteomes of Small and Large Ovarian Follicles. Biology, 9(5), 101. https://doi.org/10.3390/biology9050101






