Molecular Identification and Evaluation of the Genetic Diversity of Dendrobium Species Collected in Southern Vietnam
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction and Amplification
2.2. Data Analysis
3. Results
3.1. Sample Collection, Amplification, and Sequencing
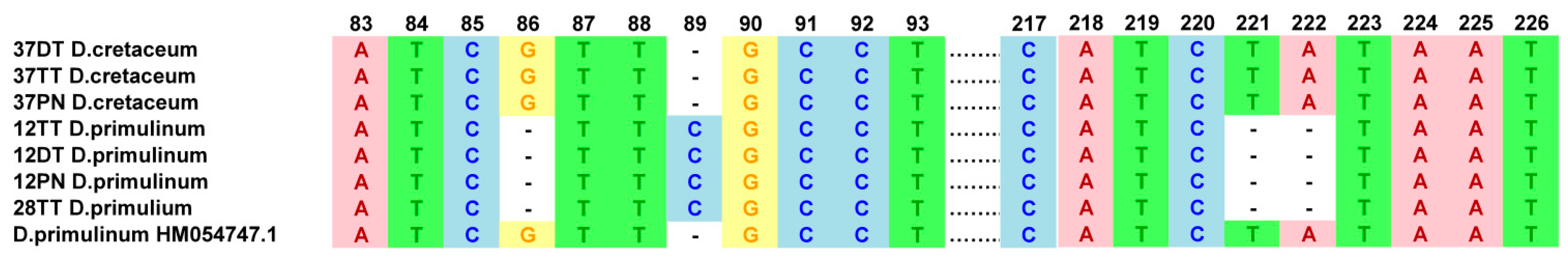
3.2. Genetic Diversity Based on Nucleotide Polymorphism and Phylogenetic Analyses
3.3. Potential Sequences for Identification of Dendrobium Species in Southern Vietnam
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Scientific Name | IUCN 2019 | Herbal | Sample Voucher | Collect Location | |
|---|---|---|---|---|---|
| 1 | D. aloifolium (Bl.) Rchb. f. | LC | 18TT | the collection of Biotechnology Center Ho Chi Minh | |
| 18DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 18PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 2 | D. amabile (Lour.) O’ Brien | 1DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||
| 1DT2 | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 1PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 3a | D. anosmum Lindl. | 27TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 27DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 6TT | the collection of Biotechnology Center Ho Chi Minh | ||||
| 6DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 3b | 15TT | the collection of Biotechnology Center Ho Chi Minh | |||
| 15DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 15PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 4 | D. aphyllum (Roxb.) C. Fisch. 1928 | LC | X | 6PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam |
| 5 | D. capillipes Rchb.f. | X | 28DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | |
| 28PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 6 | D. chrysotoxum Rchb.f; | X | 13TT | the collection of Biotechnology Center Ho Chi Minh | |
| 13DT2 | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 13PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 7 | D. cretaceum Lindl. 1847. | 37TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 37DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 37PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 8 | D. crumenatum Sw. | 34TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 34DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 9 | D. crystallinum Rchb. f. (1868) | X | 35TT | the collection of Biotechnology Center Ho Chi Minh | |
| 35DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 35PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 10 | D. densiflorum Wall. ex Lindl | 11TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 11DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 11DT2 | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 11 | D. devonianum Paxton (1840) | X | 20TT | the collection of Biotechnology Center Ho Chi Minh | |
| 20DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 20DT2 | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 12 | D. farmeri Paxton Lindl.f.Rchb.f. | 14DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||
| 14DT2 | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 14PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 13 | D. fimbriatum Hook (1823) | X | 22TT | the collection of Biotechnology Center Ho Chi Minh | |
| 22DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 22DT2 | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 14 | D. hercoglossum Rchb. f. 1886 | X | 21TT | the collection of Biotechnology Center Ho Chi Minh | |
| 21DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 21PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 15 | D. intricatum Gagnep (1930) | 36TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 36DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 16 | D. linguella Rchb. f. 1882 | 33TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 17 | D. nobile Lindl. | X | 30TT | the collection of Biotechnology Center Ho Chi Minh | |
| 30DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 30PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 18 | D. parishii Rchb. f 1863 | 38R-DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||
| 19 | D. primulinum Lindl | X | 28TT | the collection of Biotechnology Center Ho Chi Minh | |
| 12TT | the collection of Biotechnology Center Ho Chi Minh | ||||
| 12DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 12PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 20 | D. pulchellum Roxb. ex Lindl. | 10TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 10DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 10DT2 | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 10PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 21 | D. salaccense (Bl.) Lindl. | X | 24DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | |
| 22 | D. secundum (Bl.) Lindl. | 17TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 17DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 23 | D. signatum Rchb. f. 1884 | 2DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||
| 2PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 2TT | the collection of Biotechnology Center Ho Chi Minh | ||||
| 24 | D. sulcatum Lindl. (1838) | 5DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||
| 25 | D. superbum Rchb.f. | 3TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 3DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 26 | D. tortile Lindl | 32TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 27 | D. venustum Teijsm. & Binn. 1864 | 26TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 26DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 26L | the collection in Long An Province, Vietnam | ||||
| 29TT | the collection of Biotechnology Center Ho Chi Minh | ||||
| 28 | D. anosmum × D. parishi | 38TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 38DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 38DT2 | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 29 | D. anosmum × D. aphyllum | CN | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||
| 30 | D. Gatton Sunray | 31TT | the collection of Biotechnology Center Ho Chi Minh |
Appendix B
| ITS | matK | rbcL | trnH-psbA | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SPECIES | VOUCHER | Obtained from This Study | Obtained from Genbank | Obtained from This Study | Obtained from Genbank | Obtained from This Study | Obtained from Genbank | Obtained from This Study | Obtained from Genbank |
| D. aloifolium | 18DT | MT004837 | AY239951.1 | MT019381 | AB847694.1 | Not available | KC660972.1 | Not available | |
| 18TT | MT004836 | MT019380 | MT019343 | MT019476 | |||||
| 18PN | MT004838 | MT019379 | Not available | Not available | |||||
| D. anosmum | 3TT | MT004839 | JN388570.1 | MT019385 | KY966807.1 | MT019346 | KJ944591.1 | MT019457 | |
| 3DT | MT004840 | KP743544.1 | MT019386 | AB972311.1 | Not available | MT019456 | |||
| 15DT | MT004841 | MK522219.1 | MT019387 | MG490279.1 | Not available | MT019474 | |||
| 15TT | MT004842 | KJ944630.1 | Not available | MT019345 | MT019472 | ||||
| 15PN | MT004843 | AB593499.1 | MT019388 | Not available | MT019473 | ||||
| 27TT | MT004844 | MT019389 | MT019344 | MT019489 | |||||
| 27DT | MT004845 | MT019390 | Not available | MT019490 | |||||
| 6TT | MT004846 | MT019391 | MT019347 | MT019459 | |||||
| 6DT | MT004847 | MT019392 | MT019460 | ||||||
| D. aphyllum | 6PN | MT004848 | KJ210415.1 | MT019393 | AB847736.1 | Not available | MT019461 | ||
| KJ210414.1 | GU565188.1 | ||||||||
| KJ210413.1 | KF143640.1 | ||||||||
| KF143430.1 | |||||||||
| HM054561.1 | |||||||||
| D. capillipes | 28DT | MT004849 | KY966519.1 | MT019395 | KF143643.1 | Not available | FJ216545.1 | MT019493 | MF437027.1 |
| 28PN | MT004850 | AF362035.1 | MT019396 | MG490258.1 | Not available | KF177576.1 | MT019492 | ||
| KF143433.1 | MG490256.1 | ||||||||
| HQ114224.1 | MF409028.1 | ||||||||
| MK522242.1 | |||||||||
| AB593515.1 | |||||||||
| D. chrysotoxum | 13PN | MT004851 | HQ114223.1 | MT019398 | KF143654.1 | Not available | FJ216582.1 | Not available | MF437024.1 |
| 13TT | MT004852 | HQ114222.1 | MT019397 | FJ794062.1 | MT019349 | FJ216544.1 | MT019468 | MF437025.1 | |
| 13DT2 | MT004853 | HQ114221.1 | MT019399 | MG490221.1 | Not available | FJ216576.1 | MT019469 | ||
| HM590383.1 | MG490220.1 | HM055094.1 | |||||||
| MK522232.1 | KY966816.1 | JF713157.1 | |||||||
| MK483291.1 | KT778725.1 | ||||||||
| MK483283.1 | HM055093.1 | ||||||||
| MK483272.1 | |||||||||
| MK483266.1 | |||||||||
| KX440955.1 | |||||||||
| KX440953.1 | |||||||||
| AB593533.1 | |||||||||
| D. cretaceum | 37TT | MT004854 | KJ944626.1 | MT019400 | KF957845.1 | MT019359 | KJ944587.1 | MT019512 | |
| 37DT | MT004855 | KY966528.1 | MT019401 | KY966818.1 | Not available | MT019510 | |||
| 37PN | MT004856 | MT019402 | Not available | MT019511 | |||||
| D. crumenatum | 34TT | MT004864 | AY239963.1 | MT019403 | AB847734.1 | MT019350 | JF713166.1 | MT019500 | |
| 34PN | MT004865 | AY273708.1 | MT019404 | AB972308.1 | JF713165.1 | MT019501 | |||
| JN388587.1 | JF713164.1 | ||||||||
| HM590370.1 | |||||||||
| MK522246.1 | |||||||||
| AB972336.1 | |||||||||
| MH763846.1 | |||||||||
| AB593537.1 | |||||||||
| D. primulinum | 12TT | MT004857 | HM054747.1 | MT019427 | AB847845.1 | MT019368 | KF177640.1 | MT019466 | |
| 12DT | MT004858 | HQ114242.1 | MT019428 | GU565190.1 | Not available | FJ216563.1 | Not available | ||
| 12PN | MT004859 | MK522184.1 | MT019429 | KF143708.1 | Not available | JF713206.1 | MT019467 | ||
| 28TT | MT004860 | KP265001.1 | MT019394 | FJ794064.1 | MT019348 | JF713205.1 | MT019491 | ||
| MK483269.1 | KF957844.1 | JF713204.1 | |||||||
| KT778755.1 | AF445450.1 | HM055143.1 | |||||||
| KJ944625.1 | MK603116.1 | HM055142.1 | |||||||
| AB593641.1 | MG490265.1 | HM055141.1 | |||||||
| MG490264.1 | HM055140.1 | ||||||||
| MG490242.1 | HM055139.1 | ||||||||
| KT778724.1 | |||||||||
| KJ944586.1 | |||||||||
| D. devonianum | 20TT | MT004861 | KJ210443.1 | MT019411 | AB847744.1 | MT019377 | KJ187367.1 | MT019477 | |
| 20DT | MT004862 | KJ210441.1 | MT019412 | MG490252.1 | Not available | FJ216566.1 | MT019478 | ||
| 20DT2 | MT004863 | KF143453.1 | MT019413 | Not available | KJ187368.1 | Not available | |||
| KP743545.1 | JF713174.1 | ||||||||
| KC205194.1 | JF713173.1 | ||||||||
| HQ114244.1 | JF713172.1 | ||||||||
| KT778760.1 | KJ944584.1 | ||||||||
| AB593548.1 | |||||||||
| D. fimbriatum | 22DT | MT004869 | JN388588.1 | MT019418 | AB519776.1 | Not available | AB519784.1 | MT019484 | KT792701.1 |
| 22TT | MT004870 | KF143461.1 | MT019417 | AB847758.1 | MT019356 | KF177603.1 | MT019482 | ||
| 22DT2 | MT004871 | HM054637.1 | MT019419 | GU565189.1 | Not available | FJ216550.1 | MT019483 | ||
| HM054636.1 | KF143671.1 | JF713178.1 | |||||||
| HM054632.1 | AF448863.1 | JF713177.1 | |||||||
| HQ114229.1 | MK616656.1 | HM055105.1 | |||||||
| HM590392.1 | MG490240.1 | HM055104.1 | |||||||
| MK522230.1 | HM055103.1 | ||||||||
| MK483290.1 | HM055102.1 | ||||||||
| MK483275.1 | HM055101.1 | ||||||||
| MK483271.1 | KT778732.1 | ||||||||
| D. hercoglossum | 33TT | MT004874 | KJ210457.1 | MT019423 | AB847777.1 | MT019362 | KJ187382.1 | MT019499 | |
| 21TT | MT004909 | KF143472.1 | MT019420 | KF143682.1 | MT019366 | MT019479 | |||
| 21PN | MT004910 | KF143471.1 | MT019422 | KF143681.1 | Not available | MT019480 | |||
| 21DT | MT004911 | KC205188.1 | MT019421 | KP159292.1 | Not available | MT019481 | |||
| HM590381.1 | AB972305.1 | ||||||||
| MK522187.1 | MG490274.1 | ||||||||
| KP265004.1 | |||||||||
| AB593580.1 | |||||||||
| D. intricatum | 36TT | MT004872 | AB593586.1 | MT019446 | MT019360 | MT019504 | |||
| 36DT | MT004873 | MT019447 | Not available | Not available | |||||
| D. nobile | 30TT | MT004875 | JN388579.1 | MT019424 | AB847821.1 | MT019363 | EF590519.1 | MT019495 | KT792690.1 |
| 30DT | MT004876 | MH120176.1 | MT019425 | KP159296.1 | Not available | AB519785.1 | MT019497 | ||
| 30PN | MT004877 | MH120175.1 | MT019426 | KY966854.1 | Not available | KF177635.1 | MT019496 | ||
| MH120174.1 | KF177634.1 | ||||||||
| MH120173.1 | MK159250.1 | ||||||||
| MH120172.1 | MK159249.1 | ||||||||
| MH120171.1 | FJ216583.1 | ||||||||
| HM054717.1 | FJ216577.1 | ||||||||
| MK522225.1 | FJ216570.1 | ||||||||
| HM590382.1 | GQ248590.1 | ||||||||
| HM055130.1 | |||||||||
| HM055129.1 | |||||||||
| HM055128.1 | |||||||||
| HM055127.1 | |||||||||
| KT778720.1 | |||||||||
| D. amabile | 1DT | MT004878 | MK522209.1 | MT019382 | AB847690.1 | MT019376 | MT019451 | MF437029.1 | |
| 1DT2 | MT004879 | AB593495.1 | MT019384 | MT019375 | Not available | ||||
| 1PN | MT004880 | MT019383 | Not available | MT019452 | |||||
| D. farmeri | 14DT | MT004881 | KX600516.1 | MT019414 | AB847757.1 | Not available | HM055100.1 | MT019471 | MF437022.1 |
| 14PN | MT004882 | KJ672671.1 | MT019415 | KY966830.1 | Not available | HM055099.1 | MT019470 | ||
| 14DT2 | MT004883 | HM054631.1 | MT019416 | MF409019.1 | MT019355 | HM055098.1 | Not available | ||
| HM054630.1 | |||||||||
| KY966540.1 | |||||||||
| AB593561.1 | |||||||||
| D. densiflorum | 11DT2 | MT004884 | KJ210438.1 | MT019410 | AB847742.1 | Not available | MG025946.1 | Not available | MF579382.1 |
| 11TT | MT004885 | KJ210436.1 | MT019408 | KF143661.1 | MT019354 | FJ216580.1 | MT019464 | KT792697.1 | |
| 11DT | MT004886 | KJ210435.1 | MT019409 | MG490231.1 | Not available | JF713171.1 | MT019465 | ||
| HQ114255.1 | KY966823.1 | JF713170.1 | |||||||
| HQ114254.1 | MF409022.1 | JF713169.1 | |||||||
| MK522257.1 | JF713168.1 | ||||||||
| JF713167.1 | |||||||||
| HM055096.1 | |||||||||
| KT778728.1 | |||||||||
| D. pulchellum | 10TT | MT004887 | KY966577.1 | Not available | AB519778.1 | Not available | KF177644.1 | Not available | |
| 10DT | MT004888 | KJ210492.1 | MT019430 | AB519777.1 | MT019369 | AB519789.1 | MT019463 | ||
| 10DT2 | MT004889 | KF143503.1 | MT019432 | KF143712.1 | MT019370 | AB519790.1 | Not available | ||
| 10PN | MT004890 | AB593643.1 | MT019431 | KY966867.1 | MT019371 | MT019462 | |||
| D. salaccense | JN388577.1 | AF445451.1 | KF177648.1 | ||||||
| KJ210494.1 | KF177647.1 | ||||||||
| KF143506.1 | |||||||||
| HQ114260.1 | |||||||||
| MK522259.1 | |||||||||
| KJ210493.1 | |||||||||
| D. secundum | 17DT | MT004892 | AY239993.1 | MT019435 | AB847862.1 | Not available | Not available | ||
| 17TT | MT004893 | MK522237.1 | MT019434 | KY966870.1 | MT019367 | MT019475 | |||
| AB972355.1 | AB972327.1 | ||||||||
| AB593660.1 | |||||||||
| D. signatum | 2TT | MT004894 | AB972330.1 | MT019436 | AB972302.1 | MT019374 | MG324300.1 | MT019453 | |
| 2DT | MT004895 | AB593662.1 | MT019437 | AB847864.1 | MT019373 | MT019454 | |||
| 2PN | MT004896 | MT019438 | MT019372 | MT019455 | |||||
| D. tortile | 32TT | MT004897 | MK522211.1 | MT019445 | AB847878.1 | MT019361 | MT019498 | ||
| KY966585.1 | KY966874.1 | ||||||||
| EU477511.1 | |||||||||
| AB593678.1 | |||||||||
| D. venustum | 26TT | MT004898 | AB847676.1 | MT019440 | AB847886.1 | MT019365 | MT019486 | ||
| 26DT | MT004899 | MT019441 | Not available | MT019487 | |||||
| 26L | MT004900 | MT019442 | Not available | MT019488 | |||||
| 29TT | MT004901 | MT019443 | MT019364 | MT019494 | |||||
| D. parishii | 38RDT | MT004902 | KC568303.1 | Not available | Not available | MT019508 | |||
| EU121417.1 | |||||||||
| KY966570.1 | |||||||||
| KX522639.1 | |||||||||
| KC205202.1 | |||||||||
| HM054736.1 | |||||||||
| HM054735.1 | |||||||||
| HM590378.1 | |||||||||
| KJ944629.1 | |||||||||
| MK522227.1 | |||||||||
| MK483284.1 | |||||||||
| AB972344.1 | |||||||||
| AB593630.1 | |||||||||
| D. sulcatum | 5DT | MT004903 | KF143517.1 | MT019439 | KF143726.1 | MT019358 | KF177658.1 | MT019458 | MF579383.1 |
| MK522262.1 | KY966873.1 | KY440172.1 | |||||||
| EU477510.1 | |||||||||
| AB593670.1 | |||||||||
| D. hancockii | 24DT | MT004891 | JN388591.1 | MT019433 | AB847771.1 | MT019357 | MT019485 | ||
| DQ058787.1 | GU565195.1 | ||||||||
| AF362025.1 | KF143677.1 | ||||||||
| KF143467.1 | FJ794051.1 | ||||||||
| HQ114259.1 | |||||||||
| KP159297.1 | |||||||||
| AB593575.1 | |||||||||
| D. crystallinum | 35TT | MT004866 | AB593538.1 | MT019405 | AB847735.1 | MT019351 | FJ216564.1 | Not available | |
| 35DT | MT004867 | KC205205.1 | MT019406 | GU565192.1 | MT019352 | KF177590.1 | MT019503 | ||
| 35PN | MT004868 | HQ114243.1 | MT019407 | KF143657.1 | MT019353 | KJ944594.1 | MT019502 | ||
| KJ944633.1 | KF957852.1 | KT778733.1 | |||||||
| KT778764.1 | MG490248.1 | ||||||||
| KY966693.1 | |||||||||
| AF445447.1 | |||||||||
| D. Gatton sunray | 31TT | MT004904 | MT019444 | ||||||
| D. anosmum × D. parishii | 38TT | MT004905 | MT019448 | MT019378 | MT019505 | ||||
| 38DT | MT004906 | MT019449 | MT019507 | ||||||
| 38DT2 | MT004907 | MT019450 | MT019506 | ||||||
| D. anosmum × D. aphyllum | CN | MT004908 | MT019509 | ||||||
Appendix C
| No | Species | ITS | ITS2 | matK | rbcL | trnH-psbA | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tree-Based | Indel-Based | Tree-Based | Indel-Based | Tree-Based | Indel-Based | Tree-Based | Indel-Based | Tree-Based | Indel-Based | ||
| 1 | D. aloifolium | + | + | + | + | + | |||||
| 2 | D. amabile | + | + | + | ― | ― | |||||
| 3 | D. anosmum (synonym name D. superbum) | ― | ― | ― | ― | ― | |||||
| 4 | D. aphyllum | + | + | + | ― | ― | |||||
| 5 | D. capillipes | + | + | + | ― | ― | |||||
| 6 | D. chrysotoxum | + | + | + | ― | ― | |||||
| 7 | D. cretaceum | ― | + | ― | ― | ― | ― | ||||
| 8 | D. crumenatum | + | + | + | + | + | |||||
| 9 | D. crystallinum | + | + | + | ― | ― | |||||
| 10 | D. densiflorum | + | + | ― | ― | ― | |||||
| 11 | D. farmeri | + | + | ― | ― | ― | |||||
| 12 | D. fimbriatum | + | + | ― | ― | ― | |||||
| 13 | D. hercoglossum (synonym name D. linguella) | ― | ― | ― | ― | ― | |||||
| 14 | D. intricatum | + | + | + | ― | ― | |||||
| 15 | D. nobile | ― | ― | ― | ― | ― | |||||
| 16 | D. parishii | + | + | not available | not available | ― | |||||
| 17 | D. primulinum | ― | + | ― | ― | ― | ― | ||||
| 18 | D. pulchellum | + | + | + | ― | + | |||||
| 19 | D. hancockii (previously named D. salaccense) | + | + | + | + | + | |||||
| 20 | D. secundum | + | + | ― | + | ― | |||||
| 21 | D. signatum | ― | ― | ― | ― | ― | |||||
| 22 | D. sulcatum | + | + | + | ― | ― | |||||
| 23 | D. tortile | ― | ― | ― | ― | ― | |||||
| 24 | D. venustum | + | + | + | + | + | |||||
| Identified species | 17/24 | 2 | 17/24 | 0 | 12/23 | 0 | 5/23 | 0 | 5/24 | 0 | |
| 19/24 | 17/24 | ||||||||||
Appendix D
| Species | Voucher | ITS | ITS2 | matK | rbcL | trnH-psbA | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Best Match (%) | Best Close Match (%) | Best Match (%) | Best Close Match (%) | Best Match (%) | Best Close Match (%) | Best Match (%) | Best Close Match (%) | Best Match (%) | Best Close Match (%) | |||||||||||||||||||||||||||
| Correct | Ambiguous | Incorrect | Correct | Ambiguous | Incorrect | No Match | Correct | Ambiguous | Incorrect | Correct | Ambiguous | Incorrect | No Match | Correct | Ambiguous | Incorrect | Correct | Ambiguous | Incorrect | No Match | Correct | Ambiguous | Incorrect | Correct | Ambiguous | Incorrect | No Match | Correct | Ambiguous | Incorrect | Correct | Ambiguous | Incorrect | No Match | ||
| D. aloifolium | 18TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 18DT | X | X | X | X | X | X | ||||||||||||||||||||||||||||||
| 18PN | X | X | X | X | X | X | ||||||||||||||||||||||||||||||
| D. amabile | 1DT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 1DT2 | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 1PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. anosmum | 27TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 27DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 6TT | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| 6DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 15TT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 15DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 15PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. superbum | 3TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 3DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. aphyllum | 6PN | x | x | X | X | X | X | X | X | |||||||||||||||||||||||||||
| D. capillipes | 28DT | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||||
| 28PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. chrysotoxum | 13TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 13DT2 | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 13PN | X | X | X | X | X | X | ||||||||||||||||||||||||||||||
| D. cretaceum | 37TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 37DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 37PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. crumenatum | 34TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 34PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. crystallinum | 35TT | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||||
| 35DT | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| 35PN | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| D. densiflorum | 11TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 11DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 11DT2 | X | X | X | X | X | X | ||||||||||||||||||||||||||||||
| D. farmeri | 14DT | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||||
| 14DT2 | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 14PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. fimbriatum | 22TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 22DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 22DT2 | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. hercoglossum | 21TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 21DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 21PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. linguella | 33TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| D. intricatum | 36TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 36DT | X | X | X | X | X | X | ||||||||||||||||||||||||||||||
| D. nobile | 30TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 30DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 30PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. parishii | 38R-DT | X | X | X | X | X | X | |||||||||||||||||||||||||||||
| D. primulinum | 28TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 12TT | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| 12DT | X | X | X | X | X | X | ||||||||||||||||||||||||||||||
| 12PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. pulchellum | 10TT | X | X | X | X | |||||||||||||||||||||||||||||||
| 10DT | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| 10DT2 | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| 10PN | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| D. salaccense | 24DT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| D. secundum | 17TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 17DT | X | X | X | X | X | X | ||||||||||||||||||||||||||||||
| D. signatum | 2DT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 2PN | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| 2TT | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| D. sulcatum | 5DT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| D. tortile | 32TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| D. venustum | 26TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 26DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 26L | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 29TT | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
Appendix E
| No | Species | Identified Species Uisng ITS | |
|---|---|---|---|
| Our Study | Tran et al. (2018) | ||
| 1 | D. aloifolium | + | not included |
| 2 | D. amabile | + | ― |
| 3 | D. anosmum (synonym name D. superbum) | ― | + |
| 4 | D. aphyllum | + | + |
| 5 | D. capillipes | + | + |
| 6 | D. chrysotoxum | + | + |
| 7 | D. cretaceum | + | not included |
| 8 | D. crumenatum | + | not included |
| 9 | D. crystallinum | + | not included |
| 10 | D. densiflorum | + | not included |
| D.devonianum | ― | not included | |
| 11 | D. farmeri | + | ― |
| 12 | D. fimbriatum | + | + |
| 13 | D. hercoglossum (synonym name D. linguella) | ― | not included |
| 14 | D. intricatum | + | not included |
| 15 | D. nobile | ― | + |
| 16 | D. parishii | + | + |
| 17 | D. primulinum | + | + |
| 18 | D. pulchellum | + | not included |
| 19 | D. hancockii (previously named D. salaccense) | + | + |
| 20 | D. secundum | + | not included |
| 21 | D. signatum | ― | not included |
| 22 | D. sulcatum | + | not included |
| 23 | D. tortile | ― | ― |
| 24 | D. venustum | + | not included |
| 25 | D. anosmum × D. parishi | ― | not included |
| 26 | D. anosmum × D. aphyllum | ― | not included |
| 27 | D. Gatton Sunray | ― | not included |
| 28 | D. findlayanum | not included | + |
| 29 | D. moschatum | not included | + |
| 30 | D. chrysanthum | not included | + |
| 31 | D. thyrsiflorum | not included | + |
| 32 | D. wattii | not included | + |
| 33 | D. jenkinsii | not included | + |
| 34 | D. haveyanum | not included | ― |
| 35 | D. aduncum | not included | + |
| 36 | D.brymerianum | not included | + |
| 37 | D. draconis | not included | + |
| 38 | D. christyanum | not included | + |
| 28 species | 23 species | ||
| 19 identified species | 19 identified species | ||
References
- Leitch, I.J.; Kahandawala, I.; Suda, J.; Hanson, L.; Ingrouille, M.J.; Chase, M.W.; Fay, M.F. Genome size diversity in orchids: Consequences and evolution. Ann. Bot. 2009, 104, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Takamiya, T.; Wongsawad, P.; Tajima, N.; Shioda, N.; Lu, J.F.; Wen, C.L.; Wu, J.B.; Handa, T.; Iijima, H.; Kitanaka, S.; et al. Identification of Dendrobium species used for herbal medicines based on ribosomal DNA internal transcribed spacer sequence. Biol. Pharm. Bull. 2011, 34, 779–782. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Burke, J.M.; Bayly, M.J.; Adams, P.B.; Ladiges, P.Y. Molecular phylogenetic analysis of Dendrobium (Orchidaceae), with emphasis on the Australian section Dendrocoryne, and implications for generic classification. Aust. Sys. Bot. 2008, 21, 1–14. [Google Scholar] [CrossRef]
- Yukawa, T.; Kita, K.; Handa, T. DNA phylogeny and morphological diversification of Australian Dendrobium (Orchidaceae). In Monocots: Systematics and Evolution; Wilson, K.L., Morrison, D.A., Eds.; CSIRO Publishing: Collingwood, Australia, 2000; pp. 465–471. [Google Scholar]
- Xiang, X.G.; Schuiteman, A.; Li, D.Z.; Huang, W.C.; Chung, S.W.; Li, J.W.; Zhou, H.L.; Jin, W.T.; Lai, Y.J.; Li, Z.Y.; et al. Molecular systematics of Dendrobium (Orchidaceae, Dendrobieae) from mainland Asia based on plastid and nuclear sequences. Mol. Phylogenet. Evol. 2013, 69, 950–960. [Google Scholar] [CrossRef]
- Xu, S.; Li, D.; Li, J.; Xiang, X.; Jin, W.; Huang, W.; Jin, X.; Huang, L. Evaluation of the DNA barcodes in Dendrobium (Orchidaceae) from mainland Asia. PLoS ONE 2015, 10, e0115168. [Google Scholar] [CrossRef]
- Feng, S.-G.; Lu, J.-J.; Gao, L.; Liu, J.-J.; Wang, H.-Z. Molecular phylogeny analysis and species identification of Dendrobium (Orchidaceae) in China. Biochem. Genet. 2014, 52, 127–136. [Google Scholar] [CrossRef]
- Peyachoknagul, S.; Mongkolsiriwatana, C.; Srikulnath, S.; Huehne, P.; Srikulnath, K. Identification of native Dendrobium species in Thailand by PCR-RFLP of rDNA-ITS and chloroplast DNA. ScienceAsia 2014, 40, 113–120. [Google Scholar] [CrossRef]
- Srikulnath, K.; Sawasdichai, S.; Jantapanon, T.K.; Pongtongkam, P.; Peyachoknagul, S. Phylogenetic relationship of Dendrobium species in Thailand inferred from chloroplast matK gene and nuclear rDNA ITS region. Hort J. 2015, 84, 243–252. [Google Scholar] [CrossRef]
- Asahina, H.; Shinozaki, J.; Masuda, K.; Morimitsu, Y.; Satake, M. Identification of medicinal Dendrobium species by phylogenetic analyses using matK and rbcL sequences. J. Nat. Med. 2010, 64, 133–138. [Google Scholar] [CrossRef]
- Hollingsworth, P.; Forrest, L.; Spouge, J.; Hajibabaei, M.; Ratnasingham, S.; Bank, M.; Chase, M.; Cowan, R.; Erickson, D.; Fazekas, A. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar]
- Chattopadhyay, P.; Banerjee, G.; Banerjee, N. Distinguishing orchid species by DNA barcoding: Increasing the resolution of population studies in plant biology. Omics 2017, 21, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shi, L.L.; Zhou, J.; Zhu, G.P. DNA barcoding identification of Dendrobium huoshanense and its adulterants. Zhongguo Zhong Yao Za Zhi 2018, 43, 4055–4061. [Google Scholar] [PubMed]
- Liu, H.; Fang, C.; Zhang, T.; Guo, L.; Ye, Q. Molecular authentication and differentiation of Dendrobium species by rDNA ITS region sequence analysis. AMB Express 2019, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.D.; Khuat, H.T.; La, T.N.; Nguyen, T.T.T.; Pham, B.H.; Nguyen, T.K.; Tran, H.D.; Do, M.T. Identification of Vietnamese native Dendrobium species based on ribosomal DNA internal transcribed spacer sequence. Adv. Stud. Biol. 2018, 10, 1–12. [Google Scholar]
- Nguyen, N.H.; Tran, H.D.; Duong, H.X.; Huynh, H.D. Phylogenetic relationship between several Dendrobium strains based on ITS sequence. Vietnam J. Sci. Technol. Agri. 2017, 12, 41–45. (In Vietnamese) [Google Scholar]
- Nguyen, N.H. ITS sequence of several strains of Dendrobium thyrsiflorum. J. Sci. Univ. Educ. 2018, 15, 149–153. (In Vietnamese) [Google Scholar]
- Waud, M.; Busschaert, P.; Ruyters, S.; Jacquemyn, H.; Lievens, B. Impact of primer choice on characterization of orchid mycorrhizal communities using 454 pyrosequencing. Mol. Ecol. Resour. 2014, 14, 679–699. [Google Scholar] [CrossRef]
- Kyndt, T.; Van, B.; Romeijn, E.; Romero, P.; Scheldeman, X.; Goetghebeur, P.; Van Damme, P.; Gheysen, G. Molecular phylogeny and evolution of Caricaceae based on rDNA internal transcribed spacers and chloroplast sequence data. Mol. Phylogenet. Evol. 2005, 37, 442–459. [Google Scholar] [CrossRef]
- Yao, P.-C.; Gao, H.-Y.; Wei, Y.-N.; Zhang, J.-H.; Chen, X.-Y.; Li, H.-Q. Evaluating sampling strategy for DNA barcoding study of coastal and inland halo-tolerant Poaceae and Chenopodiaceae: A case study for increased sample size. PLoS ONE 2017, 12, e0185311. [Google Scholar] [CrossRef]
- Cabelin, V.L.; Alejandro, G.J. Efficiency of matK, rbcL, trnH-psbA, and trnL-F (cpDNA) to molecularly authenticate philippine ethnomedicinal apocynaceae through DNA barcoding. Pharmacogn. Mag. 2016, 12, S384–S388. [Google Scholar] [CrossRef]
- Geospiza Inc. FinchTV. Available online: http://www.geospiza.com/Products/finchtv.shtml (accessed on 10 January 2020).
- Manolo, G.; Stéphane, G.; Olivier, G. SeaView Version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Jiang, Y.; Wang, S.; Jiang, M.; Chen, Z.; Ying, Q.; Wang, H. Molecular identification of Dendrobium species (Orchidaceae) based on the DNA barcode ITS2 region and its application for phylogenetic study. Int. J. Mol. Sci. 2015, 16, 21975–21988. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Song, J.; Liu, C.; Luo, K.; Han, J.; Li, Y.; Pang, X.; Xu, H.; Zhu, Y.; Xiao, P.; et al. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS ONE 2010, 5, e13102. [Google Scholar] [CrossRef]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef]
- Vu, H.-T.; Huynh, P.; Tran, H.-D.; Le, L. In silico study on molecular sequences for identification of paphiopedilum species. Evol. Bioinform. 2018, 14. [Google Scholar] [CrossRef]
- Vu, H.-T.; Vu, Q.-L.; Nguyen, T.-D.; Tran, N.; Nguyen, T.-C.; Duong, T.; Nguyen, T.-K.; Le, L. Genetic Diversity and Identification of Vietnamese Paphiopedilum Species Using DNA Sequences. Biology 2019, 9, 9. [Google Scholar] [CrossRef]
- Guo, Y.-Y.; Huang, L.-Q.; Liu, Z.-J.; Wang, X.-Q. Promise and challenge of DNA barcoding in venus slipper (Paphiopedilum). PLoS ONE 2016, 11, e0146880. [Google Scholar] [CrossRef]
- Wattoo, J.; Saleem, Z.; Shahzad, M.S.; Arif, A.; Hameed, A.; Saleem, A. DNA Barcoding: Amplification and sequence analysis of rbcl and matK genome regions in three divergent plant species. Adv. Life Sci. 2016, 4, 3–7. [Google Scholar]
- Yao, H.; Song, J.Y.; Ma, X.Y.; Liu, C.; Li, Y.; Xu, H.X.; Han, J.P.; Duan, L.S.; Chen, S.L. Identification of Dendrobium species by a candidate DNA barcode sequence: The chloroplast psbA-trnH intergenic region. Planta Med. 2009, 75, 667–669. [Google Scholar] [CrossRef]
- Gigot, G.; Van Alphen-Stahl, J.; Bogarin, D.; Warner, J.; Chase, M.W.; Savolainen, V. Finding a suitable DNA barcode for Mesoamerican orchids. Lankesteriana Int. J. Orchid. 2007, 7, 200–203. [Google Scholar] [CrossRef][Green Version]
- Moudi, M. Phylogenetic analysis among four sections of Genus Dendrobium Sw. (Orchidaceae) in Peninsular Malaysia using rbcL sequence data. Int. J. Bioassays 2013, 2, 932–937. [Google Scholar]
- Singh, H.K.; Parveen, I.; Raghuvanshi, S.; Babbar, S.B. The loci recommended as universal barcodes for plants on the basis of floristic studies may not work with congeneric species as exemplified by DNA barcoding of Dendrobium species. BMC Res. Notes 2012, 5, 42. [Google Scholar] [CrossRef]
- Li, D.; Li, Z.; Mao, P.; Yan, X.; Chun, Z.; Ma, X. Phylogenetic analysis and identification of Dendrobium species based on ribosomal DNA internal transcribed spacer (ITS) sequence. Acta Hortic. Sin. 2012, 39, 1539–1550. [Google Scholar]
- Liu, Y.; Chen, R.; Lin, S.; Chen, Y.; Chin, S.; Chen, F.; Lee, C. Analysis of sequence diversity through internal transcribed spacers and simple sequence repeats to identify Dendrobium species. Genet. Mol. Res. 2014, 13, 2709–2717. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, X. Barcoding the Dendrobium (Orchidaceae) species and analysis of the intragenomic variation based on the internal transcribed spacer 2. BioMed Res. Int. 2017, 2017, 2734960. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Huang, J.; Li, P.-X.; Yang, H.-J.; Zhang, Y.-Q.; Zhang, G.-Q.; Chen, L.-J.; Niu, Y.-X.; Luo, Y.-B. Morphological type identification of self-incompatibility in dendrobium and its phylogenetic evolution pattern. Int. J. Mol. Sci. 2018, 19, 2595. [Google Scholar] [CrossRef]

| Barcode | Primer Name | Primer Sequence | Thermal Cycle | Source |
|---|---|---|---|---|
| ITS | ITS1F | 5′CTTGGTCATTTAGAGGAAGTAA3′ | Denaturing: 94 °C/30 sec Annealing: 55 °C/40 sec Extending: 72 °C/1 min | [17,18] |
| ITS4R | 5′TCCTCCGCTTATTGATATGC3′ | |||
| matK | 390F | 5′CGATCTATTCATTCAATATTTC3′ | Denaturing: 94 °C/1 min Annealing: 48 °C/30 sec Extending: 72 °C/1 min | [6,19] |
| 1326R | 5′TCTAGCACACGAAAGTCGAAGT3′ | |||
| rbcL | aF | 5′ATGTCACCACAAACAGAGACTAAAGC3′ | Denaturing: 94 °C/30 sec Annealing: 55 °C/1 min Extending: 70 °C/1 min | [20] |
| aR | 5′CTTCTGCTACAAATAAGAATCGATCTCTCCA3′ | |||
| trnH-psbA | trnHF_05 | 5′CGCGCATGGTGGATTCACAATCC3′ | Denaturing: 95 °C/30 sec Annealing: 5 °C/20 sec Extending: 72 °C/20 sec | [21] |
| psbA3′f | 5′GTTATGCATGAACGTAATGCTC3′ |
| Region | Length | Number of Samples | Number of Species | Variable Site (%) | Parsimony (%) | Single-ton (%) | Indel | Identified Species Based on the Phylogenetic Tree | Identified Species Based on the Phylogenetic Tree and Indel Information |
|---|---|---|---|---|---|---|---|---|---|
| ITS | 639 | 68 | 24 | 362 (56.65) | 338 (52.89) | 24 (3.75) | 15 | 17/24 | 19/24 |
| ITS2 | 253 | 68 | 24 | 167 (66.00) | 152 (60.07) | 15 (5.92) | 12 | 17/24 | 17/24 |
| matK | 822 | 65 | 23 | 84 (10.21) | 53 (6.44) | 31 (3.77) | 3 | 12/23 | 12/23 |
| rbcL | 501 | 34 | 21 | 26 (6.58) | 16 (4.59) | 10 (1.99) | 0 | 5/23 | 5/23 |
| trnH-psbA | 782 | 56 | 24 | 65 (8.31) | 46 (5.88) | 17 (2.17) | 13 | 5/24 | 5/24 |
| Barcode | No Sequences | Best Match (%) | Best Close Match (%) | |||||
|---|---|---|---|---|---|---|---|---|
| Correct | Ambiguous | Incorrect | Correct | Ambiguous | Incorrect | No Match | ||
| ITS | 68 | 55 (80.88) | 2 (2.94) | 11 (16.17) | 51 (75.00) | 2 (2.94) | 5 (7.35) | 10 (14.70) |
| ITS2 | 68 | 57 (83.82) | 4 (5.88) | 7 (10.29) | 52 (76.47) | 3 (4.41) | 4 (5.88) | 9 (13.23) |
| matK | 65 | 44 (67.69) | 16 (24.61) | 5 (7.69) | 44 (67.69) | 15 (23.07) | 5 (7.69) | 1 (1.53) |
| rbcL | 34 | 7 (20.58) | 24 (70.58) | 3 (8.82) | 7 (20.58) | 24 (70.58) | 3 (8.82) | 0 (0.00) |
| trnH-psbA | 56 | 38 (67.85) | 6 (10.71) | 12 (21.42) | 38 (67.85) | 6 (10.71) | 12 (21.42) | 0 (0.00) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, N.-H.; Vu, H.-T.; Le, N.-D.; Nguyen, T.-D.; Duong, H.-X.; Tran, H.-D. Molecular Identification and Evaluation of the Genetic Diversity of Dendrobium Species Collected in Southern Vietnam. Biology 2020, 9, 76. https://doi.org/10.3390/biology9040076
Nguyen N-H, Vu H-T, Le N-D, Nguyen T-D, Duong H-X, Tran H-D. Molecular Identification and Evaluation of the Genetic Diversity of Dendrobium Species Collected in Southern Vietnam. Biology. 2020; 9(4):76. https://doi.org/10.3390/biology9040076
Chicago/Turabian StyleNguyen, Nhu-Hoa, Huyen-Trang Vu, Ngoc-Diep Le, Thanh-Diem Nguyen, Hoa-Xo Duong, and Hoang-Dung Tran. 2020. "Molecular Identification and Evaluation of the Genetic Diversity of Dendrobium Species Collected in Southern Vietnam" Biology 9, no. 4: 76. https://doi.org/10.3390/biology9040076
APA StyleNguyen, N.-H., Vu, H.-T., Le, N.-D., Nguyen, T.-D., Duong, H.-X., & Tran, H.-D. (2020). Molecular Identification and Evaluation of the Genetic Diversity of Dendrobium Species Collected in Southern Vietnam. Biology, 9(4), 76. https://doi.org/10.3390/biology9040076





