Putative Protein Biomarkers of Escherichia coli Antibiotic Multiresistance Identified by MALDI Mass Spectrometry
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Bacterial Strains
2.2. Antimicrobial Susceptibility Test and Characterization of Resistance Genes
2.3. Proteomics Study
2.3.1. Bacterial Culture with and without Antibiotics
2.3.2. MALDI-TOF Mass Spectrometry
2.3.3. Statistics and Bioinformatics
3. Results and Discussion
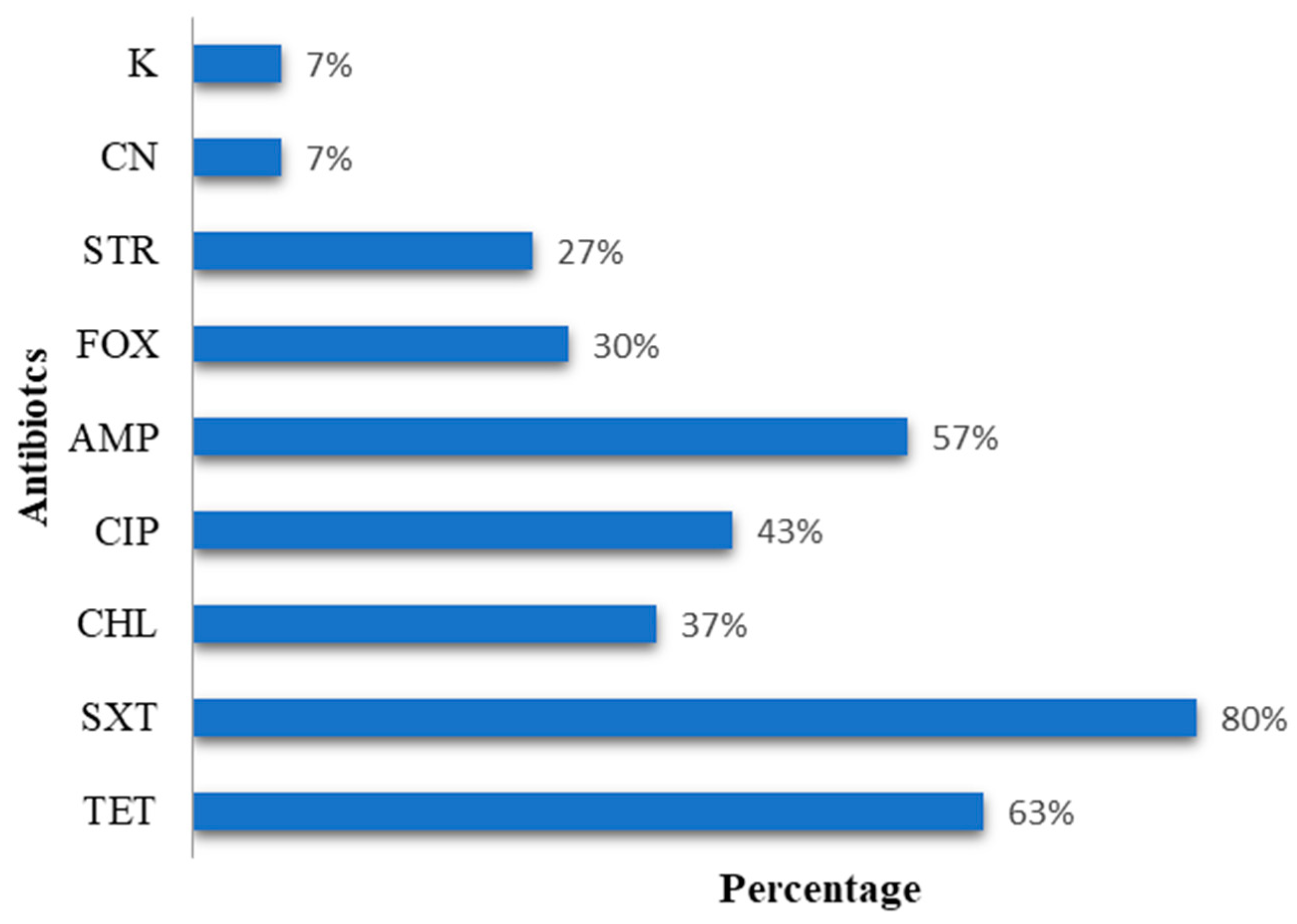
3.1. Phenotypic and Genetic Characterization
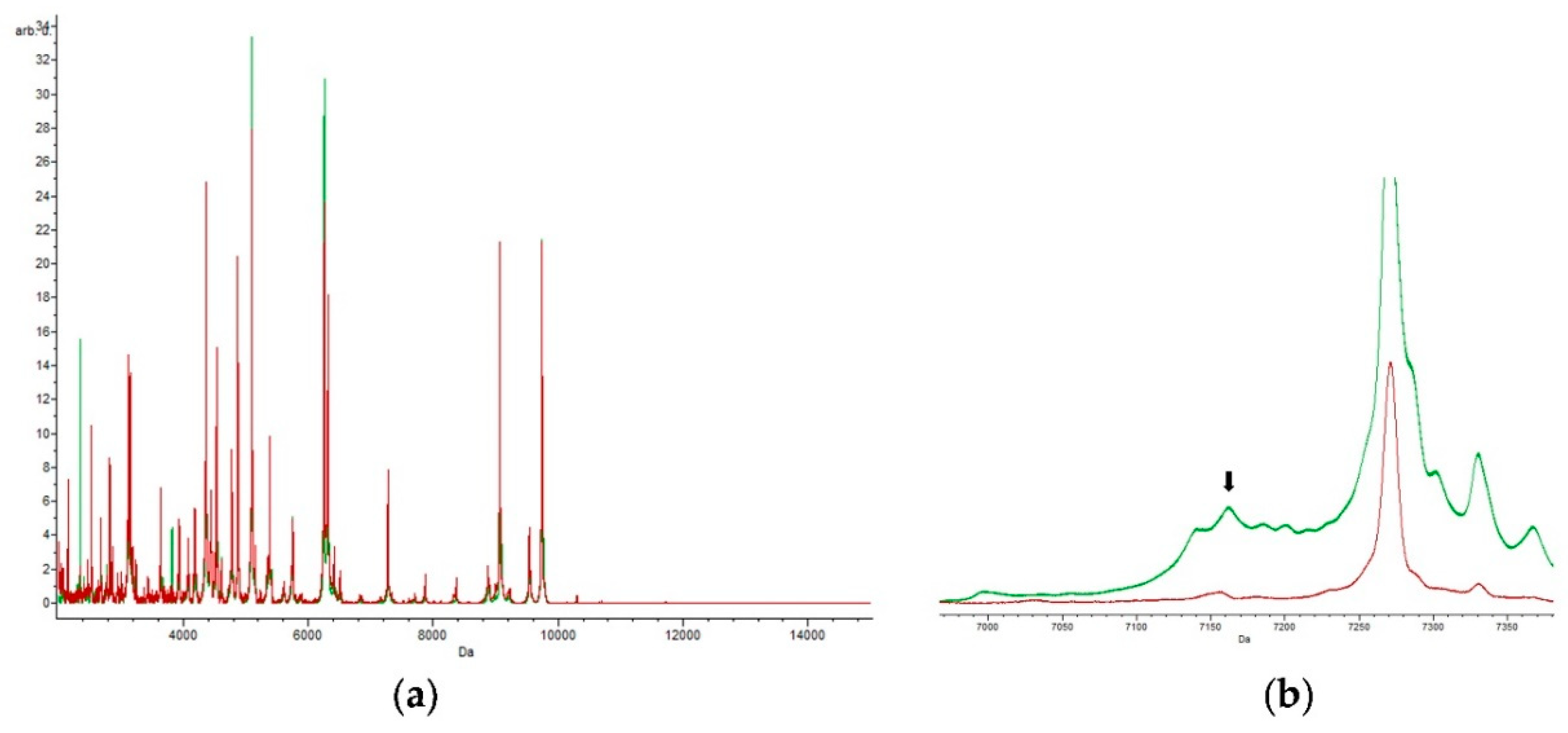
3.2. Specific Peak for E. coli
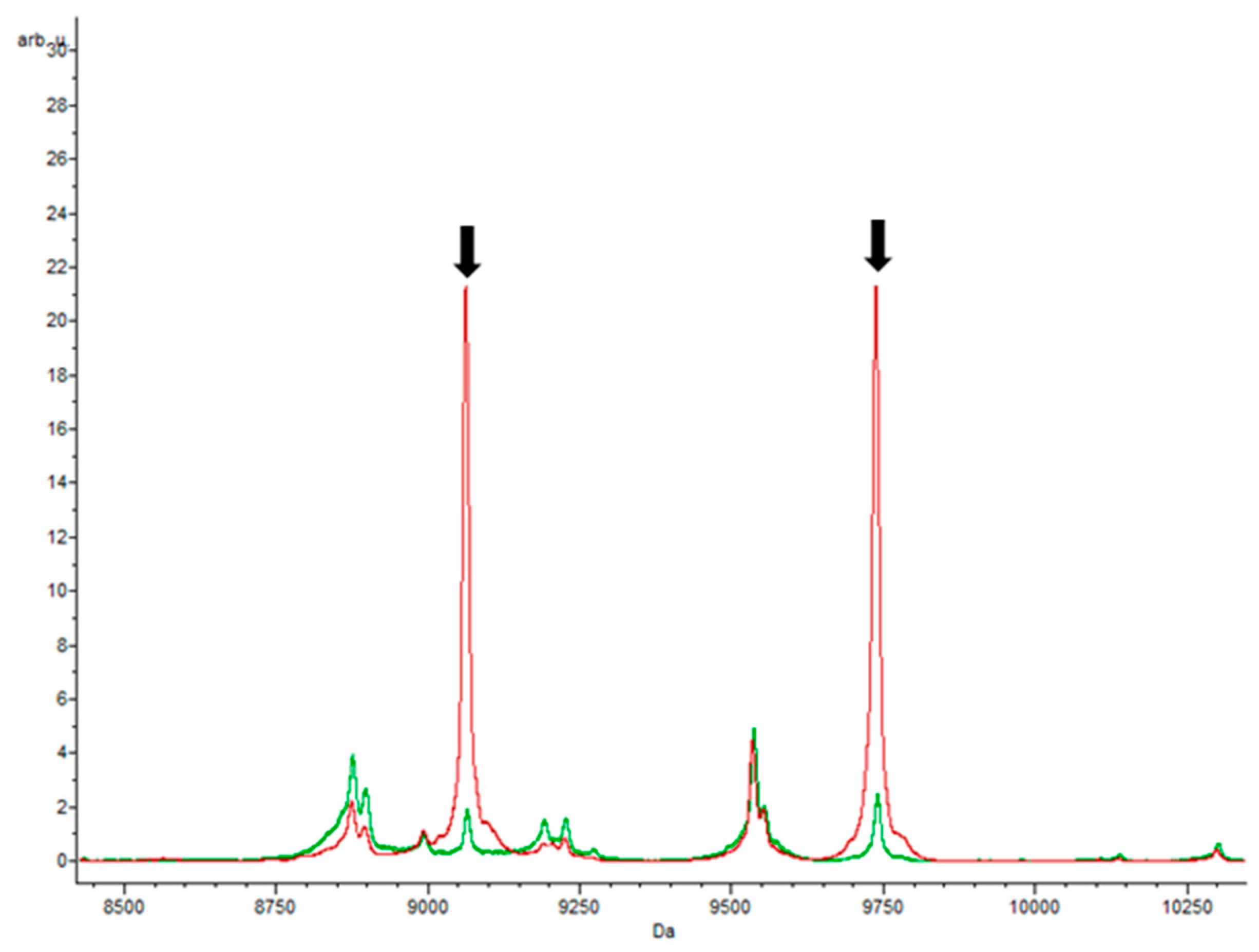
3.3. Specific Peaks for E. coli Strains Grown in Each Antibiotic
3.3.1. Trimethoprim–Sulfamethoxazole and Ampicillin
3.3.2. Ampicilin
3.3.3. Chloramphenicol
3.3.4. Tetracycline
3.3.5. Ciprofloxacin
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alberts, B.; Johnson, A.; Lewis, J.; Morgan, D.; Raff, M.; Roberts, K.; Walter, P.; Wilson, J.; Hunt, T. Biologia Molecular da Célula; Artmed Editora: Porto Alegre, Brazil, 2010. [Google Scholar]
- Costa, D.; Poeta, P.; Saenz, Y.; Vinue, L.; Rojo-Bezares, B.; Jouini, A.; Zarazaga, M.; Rodrigues, J.; Torres, C. Detection of Escherichia coli harbouring extended-spectrum beta-lactamases of the CTX-M, TEM and SHV classes in faecal samples of wild animals in Portugal. J. Antimicrob. Chemother. 2006, 58, 1311–1312. [Google Scholar] [CrossRef] [PubMed]
- Martel, A.; Devriese, L.A.; Decostere, A.; Haesebrouck, F. Presence of macrolide resistance genes in streptococci and enterococci isolated from pigs and pork carcasses. Int. J. Food Microbiol. 2003, 84, 27–32. [Google Scholar] [CrossRef]
- Radhouani, H.; Silva, N.; Poeta, P.; Torres, C.; Correia, S.; Igrejas, G. Potential impact of antimicrobial resistance in wildlife, environment and human health. Front. Microbiol 2014, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef]
- Soares, G.M.; Figueiredo, L.C.; Faveri, M.; Cortelli, S.C.; Duarte, P.M.; Feres, M. Mechanisms of action of systemic antibiotics used in periodontal treatment and mechanisms of bacterial resistance to these drugs. J. Appl. Oral Sci. 2012, 20, 295–309. [Google Scholar] [CrossRef]
- Tortora, G.J.; Funke, B.R.; Case, C.L. Introducción a la Microbiología; Ed. Médica Panamericana: Ciudad de México, México, 2007. [Google Scholar]
- Geyer, P.E.; Holdt, L.M.; Teupser, D.; Mann, M. Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol. 2017, 13, 942. [Google Scholar] [CrossRef]
- Sigdel, T.K.; Sarwal, M.M. The proteogenomic path towards biomarker discovery. Pediatric Transpl. 2008, 12, 737–747. [Google Scholar] [CrossRef]
- Veenstra, T.D. Global and targeted quantitative proteomics for biomarker discovery. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2007, 847, 3–11. [Google Scholar] [CrossRef]
- Radhouani, H.; Pinto, L.; Poeta, P.; Igrejas, G. After genomics, what proteomics tools could help us understand the antimicrobial resistance of Escherichia coli? J. Proteom. 2012, 75, 2773–2789. [Google Scholar] [CrossRef]
- Wang, H.Y.; Lien, F.; Liu, T.P.; Chen, C.H.; Chen, C.J.; Lu, J.J. Application of a MALDI-TOF analysis platform (ClinProTools) for rapid and preliminary report of MRSA sequence types in Taiwan. PeerJ 2018, 6, e5784. [Google Scholar] [CrossRef]
- Krause, E.; Wenschuh, H.; Jungblut, P.R. The dominance of arginine-containing peptides in MALDI-derived tryptic mass fingerprints of proteins. Anal. Chem. 1999, 71, 4160–4165. [Google Scholar] [CrossRef] [PubMed]
- Ryzhov, V.; Fenselau, C. Characterization of the protein subset desorbed by MALDI from whole bacterial cells. Anal. Chem. 2001, 73, 746–750. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Han, J.J.; Altwerger, G.; Kohn, E.C. Proteomics and biomarkers in clinical trials for drug development. J. Proteom. 2011, 74, 2632–2641. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Standards for Antimicrobial Susceptibility Testing; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- Schnellmann, C.; Gerber, V.; Rossano, A.; Jaquier, V.; Panchaud, Y.; Doherr, M.G.; Thomann, A.; Straub, R.; Perreten, V. Presence of new mecA and mph(C) variants conferring antibiotic resistance in Staphylococcus spp. isolated from the skin of horses before and after clinic admission. J. Clin. Microbiol. 2006, 44, 4444–4454. [Google Scholar] [CrossRef]
- Pagani, L.; Dell’Amico, E.; Migliavacca, R.; D’Andrea, M.M.; Giacobone, E.; Amicosante, G.; Romero, E.; Rossolini, G.M. Multiple CTX-M-type extended-spectrum beta-lactamases in nosocomial isolates of Enterobacteriaceae from a hospital in northern Italy. J. Clin. Microbiol. 2003, 41, 4264–4269. [Google Scholar] [CrossRef]
- Caroff, N.; Espaze, E.; Berard, I.; Richet, H.; Reynaud, A. Mutations in the ampC promoter of Escherichia coli isolates resistant to oxyiminocephalosporins without extended spectrum beta-lactamase production. FEMS Microbiol. Lett. 1999, 173, 459–465. [Google Scholar] [CrossRef][Green Version]
- Saenz, Y.; Brinas, L.; Dominguez, E.; Ruiz, J.; Zarazaga, M.; Vila, J.; Torres, C. Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob. Agents Chemother. 2004, 48, 3996–4001. [Google Scholar] [CrossRef]
- Freiwald, A.; Sauer, S. Phylogenetic classification and identification of bacteria by mass spectrometry. Nat. Protoc. 2009, 4, 732–742. [Google Scholar] [CrossRef]
- Irenge, L.M.; Ambroise, J.; Bearzatto, B.; Durant, J.F.; Chirimwami, R.B.; Gala, J.L. Whole-genome sequences of multidrug-resistant Escherichia coli in South-Kivu Province, Democratic Republic of Congo: Characterization of phylogenomic changes, virulence and resistance genes. BMC Infect. Dis. 2019, 19, 137. [Google Scholar] [CrossRef]
- Santos, T.; Capelo, J.L.; Santos, H.M.; Oliveira, I.; Marinho, C.; Goncalves, A.; Araujo, J.E.; Poeta, P.; Igrejas, G. Use of MALDI-TOF mass spectrometry fingerprinting to characterize Enterococcus spp. and Escherichia coli isolates. J. Proteom. 2015, 127, 321–331. [Google Scholar] [CrossRef]
- Taghadosi, R.; Shakibaie, M.R.; Hosseini-Nave, H. Antibiotic resistance, ESBL genes, integrons, phylogenetic groups and MLVA profiles of Escherichia coli pathotypes isolated from patients with diarrhea and farm animals in south-east of Iran. Comp. Immunol. Microbiol. Infect. Dis. 2019, 63, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Enne, V.I.; Cassar, C.; Sprigings, K.; Woodward, M.J.; Bennett, P.M. A high prevalence of antimicrobial resistant Escherichia coli isolated from pigs and a low prevalence of antimicrobial resistant E. coli from cattle and sheep in Great Britain at slaughter. FEMS Microbiol. Lett. 2008, 278, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.O.; Clegg, P.D.; Williams, N.J.; Baptiste, K.E.; Bennett, M. Antimicrobial resistance in equine faecal Escherichia coli isolates from North West England. Ann. Clin. Microbiol. Antimicrob. 2010, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Memariani, M.; Najar Peerayeh, S.; Zahraei Salehi, T.; Shokouhi Mostafavi, S.K. Occurrence of SHV, TEM and CTX-M beta-lactamase genes among enteropathogenic Escherichia coli strains isolated from children with diarrhea. Jundishapur J. Microbiol. 2015, 8, e15620. [Google Scholar] [CrossRef]
- Pitout, J.D.; Nordmann, P.; Laupland, K.B.; Poirel, L. Emergence of Enterobacteriaceae producing extended-spectrum beta-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 2005, 56, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Christner, M.; Trusch, M.; Rohde, H.; Kwiatkowski, M.; Schluter, H.; Wolters, M.; Aepfelbacher, M.; Hentschke, M. Rapid MALDI-TOF mass spectrometry strain typing during a large outbreak of Shiga-toxigenic Escherichia coli. PLoS ONE 2014, 9, e101924. [Google Scholar] [CrossRef] [PubMed]
- Bittar, F.; Ouchenane, Z.; Smati, F.; Raoult, D.; Rolain, J.M. MALDI-TOF-MS for rapid detection of staphylococcal Panton-Valentine leukocidin. Int. J. Antimicrob. Agents 2009, 34, 467–470. [Google Scholar] [CrossRef]
- Li, Y.; Shan, M.; Zhu, Z.; Mao, X.; Yan, M.; Chen, Y.; Zhu, Q.; Li, H.; Gu, B. Application of MALDI-TOF MS to rapid identification of anaerobic bacteria. BMC Infect. Dis. 2019, 19, 941. [Google Scholar] [CrossRef]
- Camara, J.E.; Hays, F.A. Discrimination between wild-type and ampicillin-resistant Escherichia coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Bioanal. Chem. 2007, 389, 1633–1638. [Google Scholar] [CrossRef]
- Bonardi, S.; Cabassi, C.S.; Longhi, S.; Pia, F.; Corradi, M.; Gilioli, S.; Scaltriti, E. Detection of extended-spectrum beta-lactamase producing Escherichia coli from mesenteric lymph nodes of wild boars (Sus scrofa). Ital. J. Food Saf. 2018, 7, 7707. [Google Scholar] [CrossRef]
- Sturenburg, E.; Storm, N.; Sobottka, I.; Horstkotte, M.A.; Scherpe, S.; Aepfelbacher, M.; Muller, S. Detection and genotyping of SHV beta-lactamase variants by mass spectrometry after base-specific cleavage of in vitro-generated RNA transcripts. J. Clin. Microbiol. 2006, 44, 909–915. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oviano, M.; Gomara, M.; Barba, M.J.; Revillo, M.J.; Barbeyto, L.P.; Bou, G. Towards the early detection of beta-lactamase-producing Enterobacteriaceae by MALDI-TOF MS analysis. J. Antimicrob. Chemother. 2017, 72, 2259–2262. [Google Scholar] [CrossRef] [PubMed]
- Sparbier, K.; Schubert, S.; Weller, U.; Boogen, C.; Kostrzewa, M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based functional assay for rapid detection of resistance against beta-lactam antibiotics. J. Clin. Microbiol. 2012, 50, 927–937. [Google Scholar] [CrossRef]
- Holland, R.D.; Duffy, C.R.; Rafii, F.; Sutherland, J.B.; Heinze, T.M.; Holder, C.L.; Voorhees, K.J.; Lay, J.O., Jr. Identification of bacterial proteins observed in MALDI TOF mass spectra from whole cells. Anal. Chem. 1999, 71, 3226–3230. [Google Scholar] [CrossRef] [PubMed]
- Siegrist, T.J.; Anderson, P.D.; Huen, W.H.; Kleinheinz, G.T.; McDermott, C.M.; Sandrin, T.R. Discrimination and characterization of environmental strains of Escherichia coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS). J. Microbiol. Methods 2007, 68, 554–562. [Google Scholar] [CrossRef]
- Hleba, L.; Charousova, I.; Cisarova, M.; Kovacik, A.; Kormanec, J.; Medo, J.; Bozik, M.; Javorekova, S. Rapid identification of Streptomyces tetracycline producers by MALDI-TOF mass spectrometry. J. Environ. Sci. Health. Part A Toxic Hazard. Subst. Environ. Eng. 2018, 53, 1083–1093. [Google Scholar] [CrossRef]
- Dortet, L.; Bonnin, R.A.; Pennisi, I.; Gauthier, L.; Jousset, A.B.; Dabos, L.; Furniss, R.C.D.; Mavridou, D.A.I.; Bogaerts, P.; Glupczynski, Y.; et al. Rapid detection and discrimination of chromosome-and MCR-plasmid-mediated resistance to polymyxins by MALDI-TOF MS in Escherichia coli: The MALDIxin test. J. Antimicrob. Chemother. 2018, 73, 3359–3367. [Google Scholar] [CrossRef]



| Animal | Number of Animal Samples Collected | Type of Samples | Period of Collected | Geographic Location of Collection |
|---|---|---|---|---|
| Pigs | 71 | Faecal samples | September 2008 to March 2009 | Slaughterhouse located in central Portugal |
| Iberian Lynx | 128 | Faecal samples | 2008 to 2010 | Doniga National Park, Sierra Morena, and Southern Spain |
| Buteo buteo | 33 | Faecal samples | September 2007 to February 2008 | Pêneda do Gerês Natural Park or others conservation areas of Portugal |
| Birds of prey | 119 | Faecal samples | April to July de 2008 | Center for Ecology, Recovery and Wildlife Surveillance |
| Boar | 77 | Faecal samples | December 2005 to February 2007 | North of Portugal |
| Chicken | 22 | Different parts of a chicken | September to December 2007 | Supermarket in Vila Real |
| Animals | E. coli Isolates | Resistance Phenotype | Resistance Genes |
|---|---|---|---|
| Chicken breast | P6A | STR CN SXT CIP | blaTEM-52aadA aac(3)-II sul2 |
| Chicken wings | A3A | TET SXT | blaCTX-M-1 tet(B) sul2 |
| A4A | AMP | blaTEM-52 tet(B) | |
| Chicken gizzard | M1A | CN SXT CHL | blaCTX-M-14cmlA aac(3)-II sul3 |
| Chicken skin | Pe4A | TET SXT | blaTEM-52 tet(A) sul sul2 |
| Birds of prey | 13 103 | AMP CIP TET K SXT | blaTEM blaCTX-M-3 tet(A) sul2 |
| 1 102 | AMP CIP TET K STR SXT | blaCTX-M-3tet(A) sul2 | |
| 2 101 | AMP CIP TET K SXT | blaTEMblaCTX-M-3tet(A) sul2 | |
| Boar | J31 | AMP CIP STR SXT | blaTEMblaCTX-M-3sul2 sul3 |
| Pigs | SU50 | TET CHL | tet(A) cmlAblaCTX-M-1 |
| SU54B | TET SXT CHL | sul3 tet(A) tet(B) cmlAblaCTX-M-1 | |
| SU54C | TET SXT CHL | sul3 tet(A) tet(B) cmlA blaCTX-M-1 | |
| SU62 | TET | tet(A) blaSHV-12 | |
| SU65 | TET | tet(B) blaCTX-M-1 | |
| SU80 | TET SXT | sul1 sul2 sul3 tet(A) blaCTX-M-1 | |
| Lynx | L16 | TET STR | tet(A) aadA blaCTX-M-14 |
| L98 | STR SXT | sul3 aadA blaSHV-12 | |
| Buteo buteo | BU10A | AMP TET SXT | sul1 sul2 sul3 tet(A) blaTEM-1 blaCTX-M-32 |
| BU10B | AMP FOX STR TET SXT | aadA ampC sul1 sul2 sul3 tet(A) blaTEM-1blaCTX-M-1 | |
| BU22A | TET STR SXT AMP | aadA sul1 sul2 sul3 tet(A)blaTEM-1 blaCTX-M-1 | |
| BU32 | STR | aadA blaTEM-1 blaCTX-M-1 | |
| BU41A | TET SXT AMP | sul1 sul2 aadA blaTEM-1blaCTX-M-1 | |
| Human | CNR695 | FOX CIP CHL SXT AMP | blaCTX-M-14 |
| CNR2630 | FOX CIP CHL SXT AMP | blaSHV-12 | |
| CNR1890 | FOX CIP CHL SXT AMP | blaCTX-M-1 | |
| CNR681 | FOX CIP CHL SXT AMP | blaCTX-M-9 | |
| CNR742 | FOX CIP CHL SXT AMP TET | blaCTX-M-14 | |
| CNR477 | FOX CIP CHL SXT AMP TET | blaCTX-M-14 | |
| BLSE176 | FOX CIP CHL SXT AMP | blaCTX-M-32 | |
| BLSE119 | FOX CIP CHL SXT AMP TET | blaCTX-M-32 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sousa, T.d.; Viala, D.; Théron, L.; Chambon, C.; Hébraud, M.; Poeta, P.; Igrejas, G. Putative Protein Biomarkers of Escherichia coli Antibiotic Multiresistance Identified by MALDI Mass Spectrometry. Biology 2020, 9, 56. https://doi.org/10.3390/biology9030056
Sousa Td, Viala D, Théron L, Chambon C, Hébraud M, Poeta P, Igrejas G. Putative Protein Biomarkers of Escherichia coli Antibiotic Multiresistance Identified by MALDI Mass Spectrometry. Biology. 2020; 9(3):56. https://doi.org/10.3390/biology9030056
Chicago/Turabian StyleSousa, Telma de, Didier Viala, Laetitia Théron, Christophe Chambon, Michel Hébraud, Patricia Poeta, and Gilberto Igrejas. 2020. "Putative Protein Biomarkers of Escherichia coli Antibiotic Multiresistance Identified by MALDI Mass Spectrometry" Biology 9, no. 3: 56. https://doi.org/10.3390/biology9030056
APA StyleSousa, T. d., Viala, D., Théron, L., Chambon, C., Hébraud, M., Poeta, P., & Igrejas, G. (2020). Putative Protein Biomarkers of Escherichia coli Antibiotic Multiresistance Identified by MALDI Mass Spectrometry. Biology, 9(3), 56. https://doi.org/10.3390/biology9030056








